Summary
The proepicardial organ is an important transient structure that contributes cells to various cardiac lineages. However, its contribution to the coronary endothelium has been disputed, with conflicting data arising in chick and mouse. Here we resolve this conflict by identifying two proepicardial markers, Scleraxis (Scx) and Semaphorin3D (Sema3D), that genetically delineate heretofore uncharacterized proepicardial subcompartments. In contrast to previously fate mapped Tbx18/WT-1-expressing cells that give rise to vascular smooth muscle, Scx and Sema3D-expressing proepicardial cells give rise to coronary vascular endothelium both in vivo and in vitro. Furthermore, Sema3D+ and Scx+ proepicardial cells contribute to the early sinus venosus and cardiac endocardium, respectively, two tissues linked to vascular endothelial formation at later stages. Taken together, our studies demonstrate that the proepicardial organ is a molecularly compartmentalized structure, reconciling prior chick and mouse data and providing a more complete understanding of the progenitor populations that establish the coronary vascular endothelium.
Introduction
The proepicardial organ (PEO) is a transient extra-cardiac cluster of cells that arises as an outgrowth of the coelomic mesothelium at the ventro-caudal base of the developing heart. Shortly after it forms, cells from the PEO migrate away from the body wall and onto the surface of the heart (Ishii et al., 2010; Manner, 1992; Nahirney et al., 2003) where most of these cells will give rise to the epicardium while a subset of them invade the underlying heart tube and contribute to various lineages within the developing heart itself (Gourdie et al., 2000). Recently, the epicardium has also been implicated as a resident progenitor cell population for cardiomyocyte repair in adult tissues (Smart et al., 2011).
Lineage tracing studies have led to contradictory findings for the fates of PEO cells. While avian studies using diI labeling, retroviral tracing, and quail-chick chimeras have established the PEO as a source of both vascular smooth muscle and endothelial cells (Guadix et al., 2006; Manner, 1999; Mikawa and Gourdie, 1996; Gourdie et al., 2000; Perez-Pomares et al., 2002), fate mapping studies in mice have not identified a significant proepicardial contribution to the endothelium (Cai et al., 2008; Zhou et al., 2008). These previous fate maps in mouse, using the well characterized proepicardial markers Tbox18 (Tbx18) and Wilms Tumor-1 (WT-1), suggested that the PEO gives rise to smooth muscle, myocardium, and fibroblasts but few or no endothelial cells. In contrast, a recent study demonstrated that some murine coronary endothelial cells arise from both the endothelial lining of the sinus venosus (the main vein that returns blood to the embryonic heart) and from the cardiac endocardium, with no contribution to these structures from Tbx18+ proepicardial cells (Red-Horse et al., 2010).
In this report, we reconcile the apparently divergent findings previously reported in mouse and chick. We demonstrate that, despite being morphologically homogeneous, the PEO is compartmentalized into genetically distinct subcompartments. In particular, domains marked by expression of the markers Scleraxis (Scx) and Semaphorin3D (Sema3D) are largely non-overlapping with Tbx18 and WT-1 expressing populations. Thus, the previous mouse fate maps utilized genetic tools that mark only a subset of proepicardial cells, thereby excluding important domains of the proepicardium. Using fate mapping studies in both mouse and chick, as well as in vitro analysis, we demonstrate that Scx and Sema3D lineage traced proepicardial cells give rise to endothelial cells, in addition to other cardiac fates. Additionally, at E10.5 Sema3D lineage traced proepicardial cells contribute to the sinus venosus, while Scx lineage traced proepicardial cells contribute to cardiac endocardium, two tissues linked at later stages to the development of the coronary endothelium.
Our study characterizes the PEO as a molecularly heterogeneous structure that contributes to the vascular endothelium in mice. We thereby reconcile chick and mouse data while offering a more complete understanding of the progenitor populations that give rise to the coronary vasculature.
RESULTS
Scx and Sema3D mark proepicardial and epicardial development
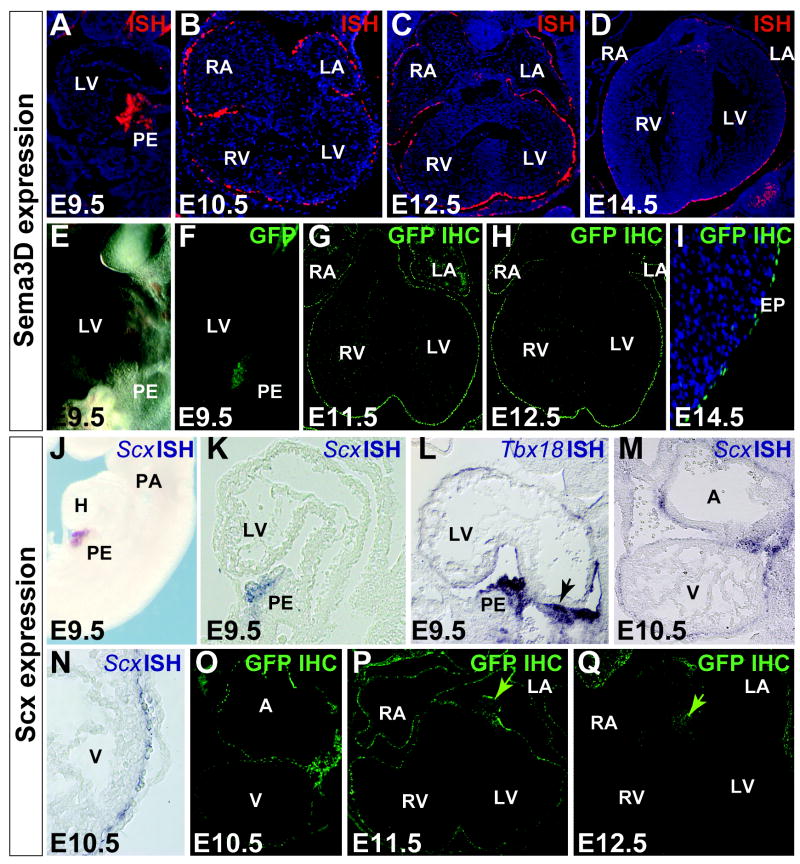
In the course of other studies, we observed strong expression of both Scx and Sema3D in the early and developing PEO (Figure 1). These expression domains in the PEO were faithfully recapitulated, respectively, by a previously characterized ScxGFP transgenic line (Pryce et al., 2007; Levay et al., 2008), and by a GFPCre fusion knock-in into the first coding exon of the endogenous Sema3D locus (Supplemental Figure 1). After confirming that they mark the same cell populations in the heart as detected by in situ hybridization, these lines were used for subsequent visualization of expression domains. Scx expression is first noted in the bilateral anlagen of the developing PEO at E9.0, while Sema3D expression is first detected in the PEO at E9.5. Unlike other proepicardial markers whose early expression extends to either the septum transversum, the endocardial cushions, or both (Figure 1, L), in situ analysis reveals that Scx and Sema3D expression are restricted, within the heart, to only proepicardial cells and migrating epicardial cells prior to E11.5 (Figure 1 and Supplemental Figures 1 and 2).
Figure 1. Sema3D and Scx mark proepicardial and migrating epicardial cells.
A-D, Radioactive RNA section in situ hybridization (ISH) of Sema3D in mouse embryos E9.5-E14.5. Sema3D is restricted to the proepicardium at E9.5 (A) and migrating epicardial cells (B-D) until E12.5 after which expression can also be seen in the valve cushions (D). E-F, bright field (E) and direct GFP fluorescence (F) in Sema3DGFPCre E9.5 mouse embryos. G-I, immunhistochemistry (IHC) with an anti-GFP antibody on Sema3DGFPCre embryos. Sema3DGFPCre recapitulates endogenous Sema3D expression in the early proepicardium and epicardium (F-I). J, whole-mount ISH of Scx in wild type mouse embryos at E9.5. K-L, section ISH to Scx (K) and Tbx18 (L) at E9.5. Scx is restricted to only a subdomain of the PEO as compared to Tbx18 which is expressed throughout the PEO and extends into the septum transversum (notched arrow in L). M-N, RNA ISH to Scx in E10.5 ScxGFP+/- mouse embryos (M) and at higher magnification (N). O-Q, IHC with an anti-GFP antibody in ScxGFP mouse embryo sections, E10.5-E12.5. Scx expression is restricted to the proepicardium and epicardium prior to E11.5 when it comes on only in the developing valves (notched arrow in P and Q). By E12.5, Scx is largely gone from the epicardium (Q). LV/RV – left/right ventricle, LV/RA – left/right atrium, PE-proepicardium, EP-epicardium, A/V- atrium/ventricle. See also Supplemental Figure 1.
Though both of these genes mark the PEO and early epicardium they begin to differ in their temporal expression by E12.5. At E12.5 Scx expression is greatly decreased in the epicardium (Figure 1Q), and by E13.5 it is undetectable by in situ hybridization (data not shown). In contrast, strong epicardial expression of Sema3D persists beyond E14.5 (Figure 1I).
To assess the spatial overlap of these genes, previously characterized ScxAP transgenic mice (Pryce et al., 2007) were crossed to Sema3DGFPCre mice. Immunohistochemistry (IHC) for AP and GFP revealed that while Scx and Sema3D represent partially overlapping populations they are not entirely congruent (Supplemental Figure 2A-C).
The proepicardium is a heterogeneous structure with genetically distinct subcompartments
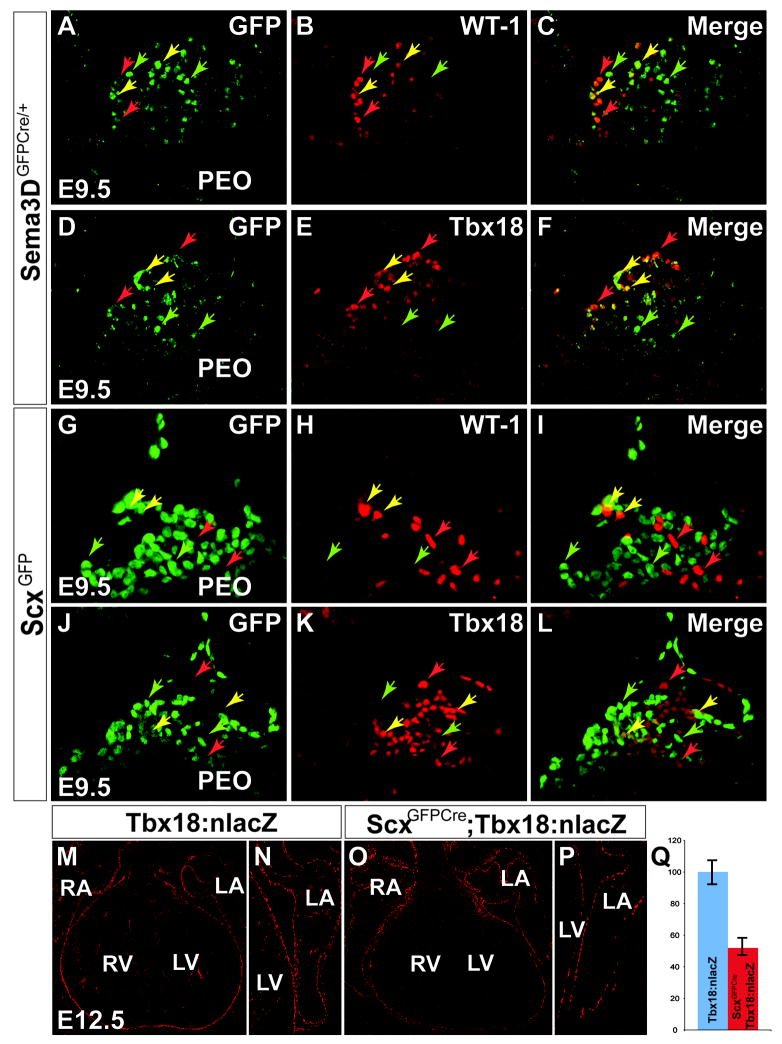
Two additional markers, Tbx18 and WT-1, have been previously reported to label proepicardial cells (Kraus et al., 2001; Zhou et al., 2008). We examined the relative expression domains of Scx, Sema3D, Tbx18 and WT-1 in the PEO at E9.5 (Figure 2). Intriguingly, we found that the expression domains of both Scx and Sema3D are largely distinct from those of Tbx18 and WT-1. In the case of Sema3D, antibodies to GFP, Tbx18, and WT-1 were used to stain histological sections from E9.5 Sema3DGFPCre mice revealing that many Sema3D+ proepicardial cells do not express either Tbx18 or WT-1. Manual quantification revealed that only 29% (140/486) of Sema3D expressing proepicardial cells co-express either Tbx18 or WT-1, while 71% (346/486) singularly express Sema3D (n=3 averaged) (Figure 2A-F). Likewise, E9.5 ScxGFP embryos were assayed by co-IHC to GFP and WT-1, revealing that many Scx expressing cells are distinct from WT-1 expression (Figure 2G-I). To assess the relationship between Scx and Tbx18 expression, ScxGFP mice were crossed to Tbx18nLacZ knock-in mice (previously denoted Tbx18-floxed nLacZ/nGFP (Cai et al., 2008)) and embryos were assayed for expression of GFP and LacZ at E9.5 (Figure 2J-L). Like Sema3D cells, Scx expressing cells are largely distinct from those expressing Tbx18. Of an average 556 Scx expressing cells per PEO, only 33.1% (184/556) coexpressed either Tbx18 or WT-1, while 66.9% (372/556) singularly expressed Scx (n=3, averaged).
Figure 2. Sema3D and Scx expressing proepicardial subdomains are largely distinct from Tbx18 and WT-1 expression.
A-F, magnified view of the PEO of Sema3DGFPCre E9.5 mouse embryos with IHC to GFP (A,D), WT-1 (B) and Tbx18 (E). The merged view of GFP and WT-1 (C) or Tbx18 (F) is shown. A portion of Sema3D expressing proepicardial cells are distinct from the WT-1 and Tbx18 expressing populations, as depicted by the green arrows. Red arrows represent cells expressing only WT-1(C) or Tbx18 (F). Yellow arrows represent cells that are double positive. G-L, proepicardia of ScxGFP E9.5 mouse embryos with IHC to GFP (G,J), WT-1 (H) and Tbx18 (K). Merged images (I,L) show that Scx expression is restricted to a subdomain largely distinct from WT-1 or Tbx18. Arrows represent single positive or double positive cells as described above. M-N, IHC to ß-galactosidase on Tbx18floxednLacZ mice at E12.5 (N, higher magnification of M). O-P, IHC to ß-galactosidase on Tbx18floxednLacZ;ScxCre mice at E12.5 (P, higher magnification of O). Q, by E12.5, 49% of Tbx18 expressing cells have never coexpressed Scx. Error bars represent the mean ± S.D. p≤0.05. RA/LA – right/left atrium, RV/LV – right/left ventricle. See also Supplemental Figure 2.
To further confirm the genetic distinction between Scx and Tbx18, previously characterized ScxGFPCre BAC transgenic mice (Blitz et al., 2009) (Supplemental Figure 2D-O) were crossed to mice harboring a floxed nLacZ allele driven from the Tbx18 promoter (denoted Tbx18floxednLacZ) (Cai et al., 2008). In Tbx18floxednLacZ embryos, all Tbx18 expressing cells should express LacZ unless a Cre mediated recombination event occurs. Therefore, in the presence of ScxCre, only Tbx18 expressing cells that have never expressed ScxCre retain their LacZ expression. To quantify the relative overlap of Tbx18 expression with that of ScxCre, serial sections were manually quantified from Tbx18floxednLacZ embryos (n=4, averaged) and compared to ScxCre;Tbx18floxednlacZ embryos in which cells that express both ScxCre and Tbx18floxednlacZ should have lost expression of the lacZ reporter cassette (n=4, averaged). Even as late as E12.5, 49% of Tbx18 expressing epicardial cells in ScxCre;Tbx18floxednlacZ embryos retain their lacZ expression (Figure 2M-Q), indicating that these Tbx18 expressing cells have never expressed ScxCre. Taken together, our results indicate that Scx and Sema3D identify distinct populations of proepicardial cells including many that would have been overlooked during previous attempts to determine epicardial-derived lineages using Tbx18 or WT-1 as genetic markers (Cai et al., 2008; Zhou et al., 2008).
Scx and Sema3D lineage traced cells differ in their relative contributions to downstream fates
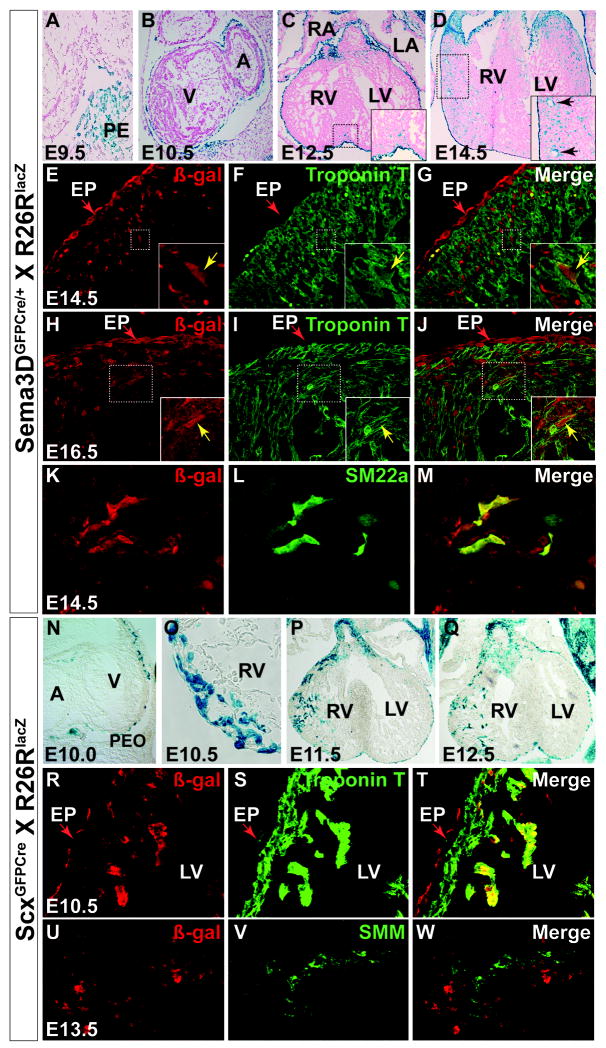
The fact that Scx and Sema3D expression mark only partially overlapping subpopulations raised the possibility that these subcompartments could give rise to descendents with distinct cell fates. To explore this possibility, we crossed the ScxGFPCre and Sema3DGFPCre lines separately to the lineage reporter R26RLacZ (Soriano, 1999). Sections were analyzed for ß-galactosidase activity (identifying Scx or Sema3D descendents) by X-gal staining and by IHC to identify different cell types from E9.5-E14.5 (Figure 3).
Figure 3. Sema3D and Scx lineage tracing in the embryonic heart.
A-D, section X-gal staining of Sema3DGFPCre;R26RLacZ embryonic hearts, E9.5-E14.5. Migrating epicardial cells are seen on the surface of the heart at E10.5 (B) with cells first noted within the heart in the interventricular septum at E12.5 (C, inset) and spread throughout all four chambers (D, inset) and developing vasculature (black arrows in inset) by E14.5. E-J Co-immunostaining with ß-gal as a marker of Sema3D lineage traced cells (E, H inset) and Cardiac Troponin T (F, I inset) at E14.5 and E16.5. The merged images (G, J inset) show that Sema3D lineage traced cells give rise to cardiomyocytes. Shown here are double positive cells in the right ventricle. Note the developed sarcomere morphology in (J). Bright yellow cells in (G, lower magnification) are autofluorescent red blood cells. K-M, Co-immunostaining with ß-gal (K) and smooth muscle actin (L) at E14.5 shows that large numbers of Sema3D lineage traced cells give rise to vascular smooth muscle cells (merged image in M). N-Q, Section X-gal staining of ScxCre;R26RLacZ mouse embryos E10-E12.5. Scx lineage traced cells are present on the surface of the heart at E10 (N) and by E10.5 are observed within the heart. R-T, Co -immunostaining with ß-gal (R) and Cardiac Troponin T (S) shows that Scx lineage traced cells contribute to the cardiomyocte lineage (merged image shown in T). U-W, Co-immunostaining with ß-gal (U) and Smooth Muscle Myosin (V) reveals that unlike Tbx18, WT-1, and Sema3D, Scx lineage traced cells do not contribute in large quantities to the smooth muscle fate by E13.5 (merged image in W) though contribution is observed at later stages. PE-proepicardium, A/V – atrium/ventricle, RA/LA – right/left atrium, RV/LV – right/left ventricle. See also Supplemental Figure 3.
As noted previously, and consistent with previous studies (Levay et al., 2008), Scx and Sema3D expression were not observed in any cells within the heart prior to E11.5, at which point Scx expression is restricted to the endocardial cushions (Figure 1P) (Levay et al., 2008), followed by expression of Sema3D only in the cushions at E12.5 (Figure 1C-D). Prior to E11.5 their expression within the heart is restricted to only the PEO and migrating epicardial cells (Figure1 and Supplemental Figures 1 and 2). Hence LacZ expression within the heart during these stages most likely reflects the migration of cells that previously expressed Scx or Sema3D, respectively, outside of the heart rather than de novo expression within it.
In keeping with their endogenous expression, lineage traced cells of both populations are identified in the PEO at E9.5 and are present in a mosaic pattern throughout the epicardium by E10.5 (Figure 3A-B, N). However, these two populations differ in the timing of their entrance into the heart as well as their relative contributions to downstream lineages.
Sema3D-lineage derived cells are first identified within the heart at E12.5 adjacent to the epicardial surface in the region of the left ventricle and interventricular septum (Figure 3C and inset) despite the absence of any active expression in this tissue. By E14.5, many Sema3D-derived cells are found in the heart, with an epicardial-to-endocardial gradient (Figure 3D). In contrast to Tbx18 and WT-1, Sema3D lineage derived cells were observed to only rarely give rise to cardiomyocytes by E16.5, with manual quantification identifying fewer than 0.36%, or 18/5000 Sema3D lineage traced cells co-expressing Cardiac Troponin T by E16.5 (Figure 3E-J and Supplemental table 1). In contrast, 410/4400, or 9.3% of Sema3D derivatives that give rise to smooth muscle as indicated by co-expression of the smooth muscle marker SM22 (Figure 3K-M). Sema3D derived cells were also noted to give rise to fibroblasts (Supplemental Figure 3A-C).
Like Sema3D, Scx lineage traced cells are visible in a mosaic pattern in the epicardium beginning at E10, yet in contrast to Sema3D, Scx lineage-traced cells are present in the heart walls as early as E10.25-10.5 in cells adjacent to the epicardium (Figure 3O). These cells initially enter the heart via the right ventricle and are present in increasing numbers through E12.5 (Figure 3N-Q) with cells visible in both ventricles by E13.5 (data not shown).
When we examined the cell types that arise from this lineage of former Scx-expressing proepicardial cells, we observed that, like the previously described Tbx18 and WT-1 derived cells, Scx lineage-traced cells give rise to cardiomyocytes at E10.5 (Figure 3R-T). By E14.5, small numbers of Scx lineage traced myocytes are normally distributed throughout the heart as indicated by immunostaining with Cardiac Troponin T (cTnT) and Connexin 43 (Cx43) (Supplemental Figure 3D-E).
However, in contrast to Tbx18 (Cai et al., 2008) and WT-1- derived cells (Zhou et al., 2008) and the Sema3D-derived cells described above, Scx lineage-traced cells only rarely express smooth muscle markers prior to E13.5 (Figure 3U-W). Colocalization of these signals is noted more frequently beginning at E14.5 when active expression of Scx is noted in the smooth muscle of the aorta and pulmonary artery (Supplemental Figure 3F-J). Additionally, some fibroblasts were noted to arise from Scx lineage traced cells (Supplemental Figure 3K-M). To quantify the relative contribution of Scx lineage traced cells to these fates postnatally, ScxCre;R26Rtdtomato lineage traced hearts were isolated from mouse pups on postnatal day 4 and treated with antibodies to Troponin T and Smooth Muscle Myosin. Analysis by FACS indicated that 6.6% of Scx lineage traced cells (1755/26291 cells) give rise to cardiomyocytes while 7.8% (2138/27315) give rise to smooth muscle cells postnatally (Supplemental Figure 4A-B). The relative contributions of Scx and Sema3D lineage traced cells to various cardiac fates is summarized in Supplemental table 1.
Scx and Sema3D lineage traced cells give rise to coronary endothelium
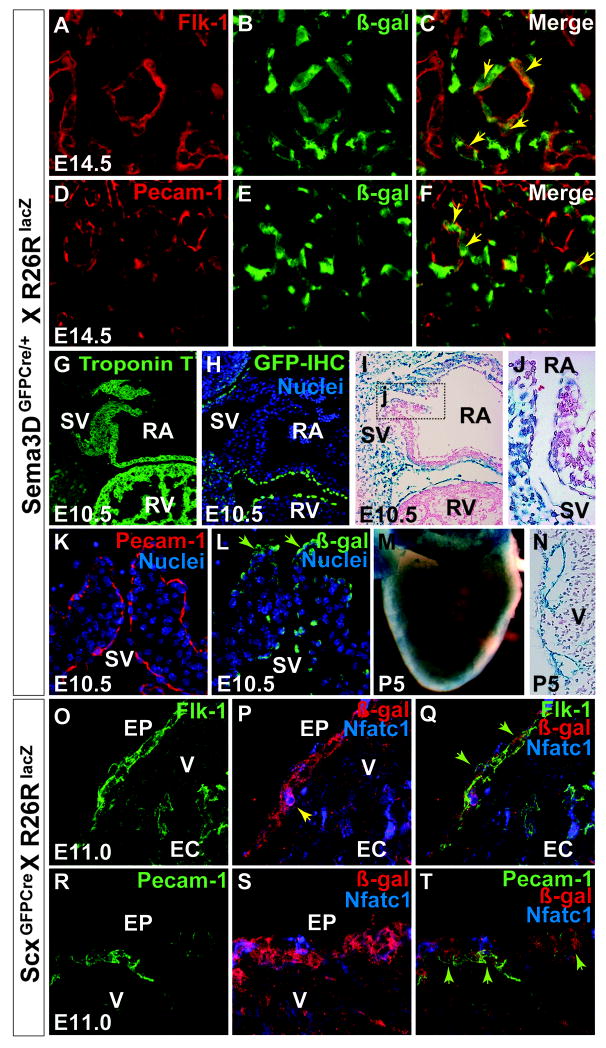
In addition to the cell fates described above, our fate mapping studies of both Sema3D and Scx proepicardial populations identified robust labeling of coronary endothelium (Figure 4). Although active expression of Scx or Sema3D was not observed to colocalize with endothelial markers within the proepicardium (Supplemental Figure 4D-E) each of these proepicardial progenitor populations were noted to give rise to endothelial descendants within the heart. In the case of Sema3D, co-immunostaining of the endothelial markers Flk-1 (Figure 4A-C) and Pecam (Figure 4D-F and Supplemental Figure 4I-L) together with ß-gal showed that Sema3D derivatives contribute to endothelial cells by E14.5. By E16.5, 6.9% (386/5623) of Sema3D lineage traced cells give rise to endothelial cells as assessed by manual quantification (Supplemental table 1).
Figure 4. Endothelial cells derive from Sema3D and Scx lineage traced proepicardial cells.
A-F, Co-immunostain of Flk-1 (A) and Pecam-1 (D) with ß-gal (B,E indicating Sema3D lineage traced cells) in Sema3DGFPCre;R26RLacZ E14.5 vessels shows that Sema3D lineage traced cells give rise to vascular endothelial cells (C is the merged image of A-B, F is the merged image of D-E). Notched arrows in (C,F) indicate double positive cells. These images are from the free wall of the right ventricle. G-H, the sinus venosus (SV) is morphologically distinct by E10.5 (G), but immunostaining for GFP on Sema3DGFPCre embryos shows no expression of Sema3D in this structure (H). I-J, Sema3DGFPCre;R26RLacZ embryos with X-gal stain of the SV (J is a magnified view of the boxed region in I) and immunostaining with Pecam-1 (K) and ß-gal (L) on adjacent sections of the SV valve leaflets shows that Sema3D lineage traced cells contribute to the endothelial lining of the SV (Notched arrows in L). M, X-gal stain of P5 hearts shows Sema3D lineage traced cells in the vascular tree. N, Transverse section of (M). O-T, Section immunostaining on ScxCre;R26RLacZ hearts at E11.0. Immunostaining of Flk-1 (O) and Pecam-1 (R), ß-gal (P,S – red cells) and Nfatc1 (P,S – blue cells). Nfatc1 marks endocardium, and is used for exclusionary purposes. (Q) and (T) are merged composites of (O-P) and (R-S), respectively. Notched arrows in (Q) and (T) show that Scx lineage traced epicardial cells give rise to endothelial cells on the surface of the heart as indicated by ß-gal/Flk1 or ß-gal/Pecam-1 double positive stain and a lack of Nfatc1 staining. Additionally, notched arrow in (P) shows that a few Scx lineage traced cells colocalize with Nfatc1, indicating that Scx lineage traced cells can also give rise to endocardial cells. SV-sinus venosus, RA - right atrium, RV- right ventricle, V-ventricle EP- epicardium, EC-endocardium. See also Supplemental Figure 4.
Interestingly, Sema3D derivatives also populate the endothelial lining of the sinus venosus (SV) by E10.5 (Figure 4G-L), a region previously implicated to contribute to coronary endothelial cells 24 hours later, beginning at E11.5 (Red-Horse et al., 2010). Importantly, Sema3D itself is not expressed in the sinus venosus endothelium at E11.5 or prior (Figure 4H) with no overlap of Sema3D and VE-Cadherin (expressed in the endothelial lining of the SV) detectable at these stages (Supplemental Figure 4F-H), suggesting that these fate-mapped cells are derived from the PEO.
In the case of Scx, co-immunostaining with β-gal, Flk-1 and Pecam revealed that Scx lineage-traced cells give rise to a few coronary endothelial cells as early as E11 with more abundant detection as development continues (Figure 4O-T and Supplemental Figure 4M-U). This is earlier than the time when coronary endothelium derived from the sinus venosus was identified in a prior report (Red-Horse et al., 2010). Importantly, consistent with previous reports (Cossette and Misra, 2011; Kalman et al., 1995; Red Horse et al., 2010) the endothelial cells that we observed are first detected on the epicardial surface of the heart and by E12.5, are present within the heart in an epicardial to endocardial gradient. However, As Flk-1 and Pecam may also mark endocardial cells (a population distinct from endothelial cells) we sought to label endocardial cells for exclusionary purposes. Because Nfatc1 is endocardial specific in the context of cardiac development, an anti-Nfatc1 antibody was used to confirm that β-gal+/Pecam+/Nfatc1- cells were in fact endothelial (Figure 4P,S).
In addition to giving rise to endothelial cells, colocalization with Nfatc1 showed that some Scx lineage traced cells give rise to small numbers of endocardial cells by E11 (Figure 4 P,S), another cell type that, like the sinus venosus, has been shown to contribute to coronary endothelial cells at E12.5 (Red-Horse et al., 2010; Nemer and Nemer, 2002). However, immunohistochemistry suggests that this contribution comprises only a small population of endocardial cells within the heart. This is not surprising in that endocardial tubes are present within the heart prior to the existence of the PEO, thus the majority of endocardial cells likely arise from an alternative source. However, this small contribution may represent an important transient link between the PEO and the endothelial cell fate.
Given that endothelial cells have not previously been shown to arise from the PEO in mouse, we wanted to confirm that the lineage traced cells that we detect do in fact arise from the epicardial expressing populations rather than from cardiac endothelial cells themselves expressing Cre at sub-detectable levels. We therefore turned to quantitative PCR as a more sensitive detection method. Ventricles from E12.5 wild-type embryonic hearts were dissociated and labeled with a biotinylated anti-Pecam antibody to identify endothelial cells. These labeled cells were then isolated by means of streptavidin conjugated beads. Quantitative RT-PCR was performed to assess the relative expression levels of Sema3D and Scx in the cardiac endothelial cell population. For each of these markers, non-cardiac expression in wild type embryos was used as a positive control, and absence of expression in E12.5 Scx-/- and Sema3D-/- embryos was used as a negative control. We did not detect Scx or Sema3D expression above negative control levels within endothelial cells (Supplemental Figures 1O and 2P).
Postnatally, Scx lineage traced endothelial cells are present primarily in the ventricles with only small numbers detected in the atria. These cells contribute to both arteries and veins as marked by neuropilin-1 and Ephrin B2 staining (Supplemental Figure 4P-U) with a preferred contribution to arterioles and venules.
To quantify the relative contribution of Scx lineage traced cells to the endothelium postnatally, ScxCre;R26Rtdtomato lineage traced hearts were isolated from mouse pups on postnatal day 4 and treated with antibodies to Pecam and Nfatc1 (to label endocardial cells for purposes of exclusion). Quantitative FACS analysis revealed a distinguishable population of Scx+/Pecam+/Nfatc1- endothelial cells accounting for a minority of all endothelial cells (Supplemental Figure 4C and Supplemental table 1).
Cross species transplantation verifies the ability of murine PEO cells to give rise to coronary endothelium in vivo
In situ and immunohistochemical analysis of Scx and Sema3D expression did not detect active expression of these markers within the heart other than the cardiac valves prior to E12.5, nor did we detect any leaky Cre expression (Supplemental Figures 1 and 2). This is critical to the interpretation that the LacZ expressing cells within the endothelium and other heart tissues prior to these stages reflects cells exposed to ScxCre and Sema3DGFPCre activity earlier, within the PEO. Nevertheless, to directly demonstrate that PEO cells have the potential to differentiate into endothelium, we isolated murine proepicardial cells and grafted them into the developing chick heart to test their developmental potential.
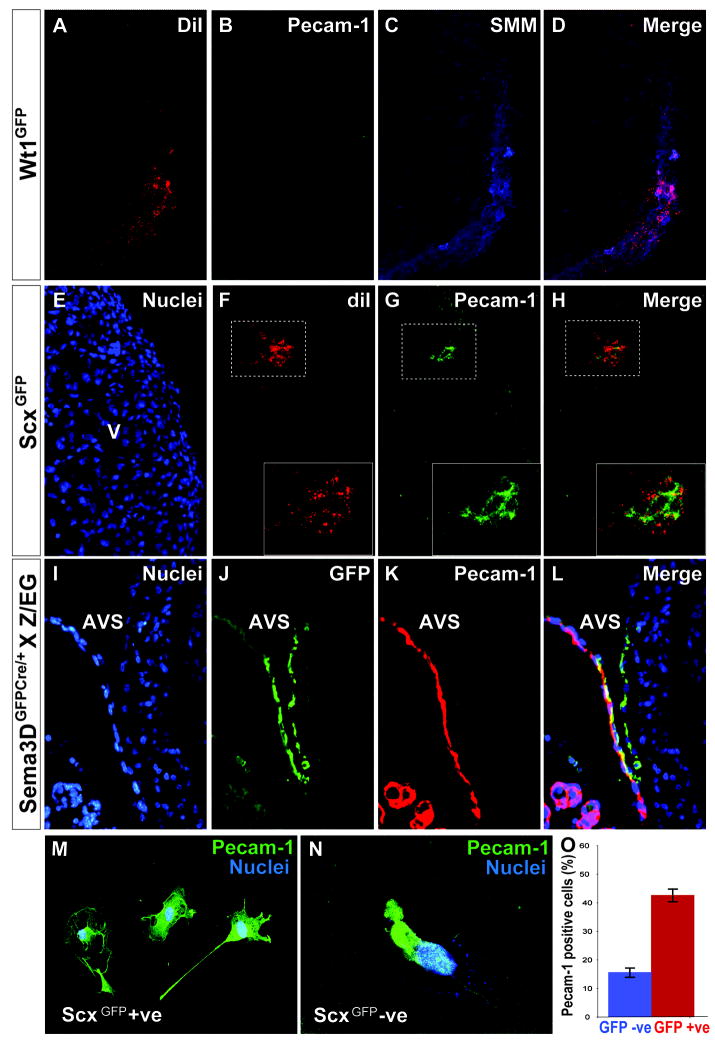
To satisfy ourselves that grafted cells would faithfully recapitulate endogenous cell fate decisions, we examined the WT-1 proepicardial subpopulation that has been previously fate-mapped. WT-1 lineage traced cells do not commonly contribute to the endothelium in mouse; however, they do contribute significantly to smooth muscle (Zhou et al., 2008). We dissected proepicardia from WT-1GFPCre mice at E9.5 (a time when the only cells expressing GFPcre are within the proepicardial organ) and isolated GFP expressing cells via FACS. We then colabeled these cells with diI. GFP+/diI labeled cells (~1000/specimen) were grafted into Hamburger and Hamilton (HH) stage 16 chick proepicardia in ovo (developmentally equivalent to E9.5 mouse proepicardial development). Following a 72 hour reincubation period, chick hearts were harvested at HH stages 26-28 and examined for the presence of diI labeled cells, denoting cells of mouse proepicardial origin. We detected no contribution of WT-1 proepicardial cells to the endothelial cell fate (n=0/13 transplants, though these cells often gave rise to smooth muscle cells (n=8/13) (Figure 5A-D). These results are consistent with previous in vivo WT-1 fate mapping studies done in mouse (Zhou et al., 2008), suggesting that the murine proepicardial cells faithfully differentiate in the chick heart microenvironment.
Figure 5. Mouse PEO cells can give rise to endothelial cells both in vivo and in vitro.
A-L, sections though a chick heart that has been transplanted with E9.5 mouse PEO cells. Prior to transplantation, mouse cells were co-labeled with diI (A, F). WT-1 transplanted cells do not express Pecam-1 (B) but do express Smooth Muscle Myosin (C). Merged image shown in (D). E-H, transplants with E9.5 proepicardia from ScxGFP mice. DAPI staining of a chick ventricle is shown in (E). Transplanted Scx cells give rise to endothelium as marked by Pecam-1 (G). Merge of diI and Pecam shown in (H). Insets in panels (F-H) show higher magnifications of the boxed regions. I-L, Transplants from Sema3DGFPCre+;Z/EG mice. DAPI staining of the chick atrioventricular sulcus (I). Sema3D lineage traced cells express GFP following Cre recombination at E9.5 (J). These cells are detected within the heart and are seen to co-express Pecam-1 (K). Merged image of (J) and (K) shown in (L). M-N, E9.5 Scx expressing PEO cells can give rise to endothelial cells in culture as indicated by Pecam-1 expression. The PEO was sorted into a Scx+ fraction (M) and a Scx- fraction (N). Comparative quantification of each fraction is shown in (O). Error bars represent the mean ± S.D. p≤ 0.05. V-ventricle, AVS – atrioventricular sulcus.
We next turned to Scx and Sema3D labeled murine proepicardial cells at E9.5. Each of these two populations was independently isolated and separately transplanted into developing chick hearts in ovo. In the case of Scx, the protocol followed was identical to that used for the WT-1 expressing cells. By HH stages 26-28, small numbers of DiI labeled cells were noted both in the epicardium and within the heart (Figure 5E-F). The presence of diI+/Pecam+ cells in 7/11 viable transplants confirmed the ability of Scx PEO derived cells to assume an endothelial cell fate in vivo (Figure 5G-H). The murine proepicardial origin of these cells is further validated by the fact that anti-Pecam antibodies do not cross react with chick endothelial cells.
Similar transplants were performed to confirm the ability of Sema3D proepicardial cells to give rise to endothelial cells. Because we are unable to efficiently sort the Sema3D cells due to weak fluorescence of GFP in the targeted mice, we instead harvested the proepicardia from E9.5 Sema3DCre/+; Z/EG mice that permanently express GFP following a Cre recombination event. 2-3 proepicardia were transplanted into each stage 16 (HH) chick embryo and reincubated until stages 26-28 (HH). Pecam positive cells were observed in 4/5 viable transplants. Colocalization of GFP and Pecam confirmed the ability of Sema3D lineage traced proepicardial cells to give rise to an endothelium (Figure 5I-L). Additionally, in both the Sema3D and Scx transplants, some vessels of mixed chick-mouse origin were noted. Together, these results indicate that, unlike the cells of the WT-1-marked compartment, the Scx and Sema3D-expressing cells in the proepicardium have the potential to differentiate into endothelial cells.
Proepicardial cells are competent to differentiate into endothelial cells in vitro
To further verify the developmental capability of murine PEO cells to give rise to endothelial descendants, proepicardia from E9.5 ScxGFP embryos were dissociated and, after FACS, the GFP expressing fraction was collected and cultured in vitro under conditions previously reported to drive cells toward an endothelial cell fate (Moretti et al., 2006). In parallel, the non-GFP expressing fraction of the proepicardium was collected and cultured under matching conditions. After 7 days in culture, 42% of cells in the Scx enriched proepicardial fraction (51/121) gave rise to endothelial cells as identified by anti-Pecam antibody staining. In addition, 16% of the non Scx-expressing cells (16/99) gave rise to endothelial cells under these conditions (Figure 5M-O). This result is not surprising in as much as Scx and Sema3D mark only partially overlapping populations. It is notable, however, that the Pecam positive cells in the Scx enriched fractions were, on average, more highly developed and more closely resembled appropriate endothelial cell morphology than those marked in the non Scx-expressing fraction.
DISCUSSION
Here, we describe expression of Scx and Sema3D, two markers of proepicardial and epicardial development. Our in situ and immunohistochemical characterization shows that each of these markers identifies a population of cells largely distinct from Tbx18 and WT-1. These results indicate that, based on gene expression, there are several molecularly distinguishable cell populations within the developing PEO. We next used recombinase based fate mapping under the control of two independent Cre driver mouse lines to study the cell types derived from the Scx and Sema3D expressing PEO cells. The differences in cell fates between the Tbx18 and WT-1, Scx, and Sema3D cells indicate that these represent distinct, functionally significant, subpopulations. Moreover, the finding that both the Scx and Sema3D populations contribute to the endothelial lineage reconciles previous chick and mouse fate mapping data that has until now been contradictory. In particular, studies in avian species have suggested that proepicardial cells may give rise to endothelial cells (Gittenberger-de Groot et al., 1998; Reese et al., 2002) but the previous fate maps in mice have failed to confirm this data (Cai et al., 2008; Zhou et al., 2008). This seeming disparity arose because prior avian studies, by the techniques utilized, labeled cells throughout the proepicardium while the previous murine studies focused on what we now conclude to be restricted sub-populations that provide little or no endothelial contribution.
The cardiac endocardium and the venous endothelium of the sinus venosus could potentially represent independent sources of coronary endothelial cells to those identified in our studies. However, our fate mapping data indicate that by E10.5 Sema3D lineage traced cells contribute to the sinus venosus, while Scx lineage traced cells contribute to the cardiac endocardium by E11 in the absence of any active expression of either of these markers within these tissues. Previous studies have timed the contribution from the sinus venosus beginning primarily at E11.5 and that of the endocardium at E12.5 and later (Red-Horse et al., 2010), 24-48 hours later than Scx or Sema3D proepicardial cells have begun to contribute to these tissues. Therefore, our data are consistent with the possibility that some Scx/Sema3D precursors traverse through the sinus venosus endothelium en route to the heart and/or transiently contribute to the endocardium before entering the coronary vascular endothelial lineage. It is possible that previous studies of the sinus venosus and endocardium may have unknowingly included cells that originate in the proepicardium while inadvertently overlooking proepicardial derivatives that failed to express Tbx18 or WT-1. At the same time, our observation that Scx lineage traced cells express endothelial markers on the epicardial surface of the heart at E11, prior to any sinus venosus contribution, suggests that some coronary endothelium derived from the proepicardium arises via traditional routes of proepicardial migration (Cosette and Misra, 2011; Hiruma and Hirakow, 1989; Komiyama et al., 1987; Nahirney et al., 2003; Viragh and Challice, 1981).
In any recombinase-based fate mapping strategy, the conclusion reached depends upon the assumption that Cre is expressed only in the tissue being mapped. The possibility always remains that there is leaky expression of Cre below detectable levels in other locations relevant to the mapping, in our case within the heart itself or alternative tissues that contribute to the heart. Despite careful expression analysis by ISH, IHC, and qPCR, we cannot rule out the formal possibility that Sema3D or Scx are expressed at sub-detectable, yet nonetheless significant, levels within relevant tissues. Additionally, at later time points, both lines express in other tissues within the embryo and we cannot technically eliminate the possibility that some other source of Scx or Sema3D expressing cells gives rise to a population in the heart. However, the pattern of expression that we observe in innumerable sections from staged embryos strongly suggests an origin from the epicardial surface with subsequent epicardial-to-endocardial migration. If the cells originated in pharyngeal mesenchyme, for example (as do second heart progenitors or those expressing Isl1Cre or Mef2c-AHF-Cre), then they would be expected to migrate to the ventricles via the anterior or posterior poles of the heart, and we would first identify these indelibly labeled cells in the outflow or inflow tracts. But this is not the pattern that we see. If the labeled cells originated via the circulation from a distant site, then we would expect to see them first on the endocardial surface, or adjacent to coronary vessels, but we do not. The earliest endothelial cells that we mark are first noted directly on the epicardial surface of the heart at E11 and derive from lineage traced epicardial cells. For these reasons, we believe our data strongly supports the conclusion that the labeled cells arise from the PEO.
Because an endothelial lineage had not previously been reported to arise from the PEO in mice, we sought to verify the competence of murine PEO cells to contribute to coronary endothelium in two ways: mouse to chick proepicardial transplants as well as an in vitro cell culture assay. In both experiments we utilized PEO cells that actively express Scx or Sema3D at E9.5, hence in each case all endothelial cells that arose must have originated from E9.5 mouse PEO cells. These experiments confirm the competence of PEO lineage traced cells to give rise to endothelial cells both in vivo and in vitro. Furthermore, mouse to chick proepicardial transplants utilizing WT-1 expressing cells failed to give rise to endothelial cells but were able to give rise to smooth muscle cells as marked by smooth muscle markers. This important control supports the notion that the PEO is comprised of genetically distinct subcompartments that differ in their downstream fates.
In addition to observing endothelial cells in our in vitro cultures arising from the ScxGFP expressing fractions, we also saw smaller numbers of endothelial cells in cultures of GFP- cells. A likely explanation is that, given that the Scx and Sema3D expressing PEO populations only partially overlap, the endothelial cells in the GFP- fraction arise from the Sema3D cells that were excluded during FACS. However, our results do not rule out the possibility that there exist additional, as yet uncharacterized, subcompartments of the PEO that also have the potential to give rise to endothelial cells. It is also possible that known proepicardial populations (such as Tbx18 or WT-1) are not restricted from the endothelial lineage while in the PEO or in culture though their descendants become restricted from the endothelial lineage in vivo.
These studies demonstrate and begin to define complex inhomogeneous populations of cardiac precursor cells within the PEO. These genetically distinct subcompartments differ in both the routes and the timing of their migration and also give rise to distinct albeit overlapping cell fates, including contributions to the vascular endothelium. Our results establish the complexity of the PEO as a source of multiple progenitor populations while simultaneously offering a more complete understanding of the diversity of tissues that give rise to the coronary vascular endothelium.
Experimental Procedures
Mice
Wild-type analysis was performed on C57BL6 mice (Charles River Laboratories). ScxGFP(Pryce et al., 2007), R26Rtdtomato (Madisen et al.), Rosa26LacZ (Soriano, 1999), and Tbx18:nLacZ/nGFP(Cai et al., 2008) have been described. For details on the generation of Sema3DeGFPcre knock in mice and ScxGFPCre BAC transgenic mice see supplemental information.
Immunostaining
Embryos were dissected in PBS and either transferred immediately to 30% sucrose or fixed in 4% paraformaldehyde for 15 min – 2 hours at 4 degrees. Whole mount hearts were washed 3 X 10 minutes in PBS and incubated with anti-β-galacotosidase antibodies. Embryos were transferred to 30% sucrose for 4 hours-overnight at 4 degrees C. Embryos were embedded on OCT (Tissue Tek) and cut by cryosectioning (7-10 and 80 um). Sections were rehydrated in PBS or TBS, incubated in blocking solution for one hour (5% donkey serum in PBS + 0.1% Triton-X) and incubated with primary antibodies in PBS + 0.1% Triton-X or TBS (without Triton-X) overnight at 4 degrees C. Sections were then washed in PBS-Triton or TBS and incubated with fluorescent conjugated secondary antibodies (1:500) for 2 hours at room temperature. For a complete list of primary antibodies used see supplemental information.
RNA in situ hybridization
Radioactive in situ hybridization was performed for Sema3D on paraformaldehyde-fixed, paraffin- embedded sections according to standard protocols (Singh et al. 2010). A 676 bp long in situ probe was generated containing the 3-untranslated region of the mouse Sema3D gene. The following primers were used to generate this fragment: forward, CAGTACTGTG AGCAGATGTG; reverse, CATTACTGCAGTACACTAGATG. Colorimetric whole mount and section in situ hybridizations for Scx were carried out as previously described (Brent et al., 2003; Murtaugh et al., 1999). Colorimetric sections shown at time points up to E9.5 were stained in whole mount and sectioned following staining. Some in situs at E10.5 and later were performed on ScxGFP+/- embryos in addition to wild type embryos.
Xgal staining
Timed pregnant embryos were harvested in PBS and fixed in either ice cold 4% paraformaldehyde for 15-60 minutes at 4 degrees, or in 25% glutaraldehyde in PBS for 10-20 minutes at room temperature. Staining was performed as previously described (Cai et al., 2008).
Endothelial bead isolation
E12.5 wild type hearts were harvested into PBS and then transferred to PBS + 20% Fetal Bovine Serum + Roche Collagenase A 10mg/ml + Roche collagenase B 10 mg/ml and incubated at 37 degrees for one hour with manual tituration every 15 minutes. Cells were pelleted and resuspended in TBS. Following dissocation, endothelial cells were labeled with biotinylated Rat Anti-Mouse CD31 (BD pharmingen catalog # 553371) antibody and then isolated accordingto manufactures instructions using CELLectionTM biotin binder kit (Invitrogen Catalog #115.33D) or MagPrep Streptavidin beads (Novagen, Catalong # 70716).
Quantitative PCR
Quantitative PCR was performed as previously described (Singh et al., 2010; Singh et al., 2011). Briefly, total RNA was isolated from E12.5 wild-type embryos, Sema3D-knockout embryos, Scx knockout embryos, and endothelial cells isolated from E12.5 wild type hearts, using TRIzol (Invitrogen). RNA was reverse-transcribed using random hexamers and the SuperScript First Strand Synthesis kit (Invitrogen). Gene expression was measured by quantitative RT-PCR (ABI PRISM 7900) using SYBR Green master mix (Applied Biosystems). Signals were normalized to corresponding GAPDH controls. PCR conditions and primer set sequences are available upon request.
Mouse to Chick Proepicardial Transplants
For transplantation of Scx and WT-1 expressing cells, proepicardia were dissected from ScxGFP or WT-1GFPCre embryos at E9.5. Proepicardia were dissociated (PBS + 20% Fetal Bovine Serum + Roche Collagenase A 10mg/ml + Roche collagenase B 10 mg/ml) and incubated at 37 degrees for one hour with manual tituration at 15 minute intervals. Dissociated cells were FACS sorted to isolate GFP+ cells. GFP positive cells were treated with CM-diI (Sigma) for 15 minutes on ice. Cells were then pelleted and washed with PBS 3 x 15 minutes, with cells being spun and pelleted between washes to remove any nonbound diI. Eggs at stage HH16-17 were windowed, and a small opening was made in the external embryonic membranes to allow access to the pericardial cavity. Using a pump ejector, Scx PEO cells were injected into the pericardial cavity surrounding the surface of the heart. Therefore, only the external surface of the heart was exposed to the murine PEO cells. After a 72 hour reincubation period, chick embryos were harvested into cold PBS and then immediately into 30% sucrose and embedded in OCT. Cryosections of 10-20um were treated with an anti-Pecam and anti-Nfatc1 antibody according to method described above (see “immunostaining”). For transplantation of Sema3D expressing cells, whole proepicardia were isolated from E9.5 Sema3DGFPCre+; Z/EG mice. Two propepicardia were placed into the pericardial cavity of each chick embryo adjacent to the chick proepicardium. The staging and timing of the transplants was the same as described above.
Endothelial Cell Culture
ScxGFP+ and GFP- mouse proepicardial cells were isolated as described above (see Mouse to chick Proepicardial Transplants). Immediately after FACS sorting the GFP+ and GFP- fractions were collected and each fraction was plated separately in chamber slides pretreated with 0.1% gelatin. Cells were cultured in the following media: DMEM + 10% FBS + 1% Pen-Strep + 50ng/ml VEGF (R&D Systems, catalog # 494-VE-005) + 150 ug/ml endothelial growth supplement (sigma, catalog # E2759). Cells were cultured for 7 days, with media refreshed on day 4. After 7 days, cells were fixed on the slide with PFA and stained with an anti-Pecam antibody.
FACS Analysis
Individual ScxCre;R26Rtdtomato hearts from P0-P7 pups were dissected and manually titurated in PBS. Tissue was then transferred to dissocation solution (PBS + 20% Fetal Bovine Serum + Roche Collagenase A 10mg/ml + Roche collagenase B 10 mg/ml) and incubated at 37 degrees for one hour with manual tituration at 15 minute intervals. Following dissocation, tissue was treated with red blood cell lysing solution (0.15M NH4Cl, 10 mM KHCO3, 0.1 mM EDTA in ddH20) and resuspended in PF10 (PBS + 10% fetal bovine serum) for antibody staining. For labeling with intracellular markers such as Troponin T and Smooth muscle myosin cells were treated with PBS+0.1% Triton-X for permeabilization. Triton-X was not used for membrane markers such as Pecam. For a list of primary antibodies used in this study see supplemental information. Antibody treated cells were sorted in a FACSAria cell sorter and analyzed using BDFACS Diva software.
Supplementary Material
Highlights.
The proepicardium is organized into genetically distinct subcompartments
Scx and Sema3D expressing compartments partially differ in their downstream fates
The Scx and Sema3D expressing compartments both give rise to endothelial cells
Sema3D+ cells contribute to the sinus venosus endothelium among other tissues
Acknowledgments
The authors would like to thank C. Cai for discussion, S. Curtis for technical assistance and reagents for FACS analysis, R. Mathieu and the Boston Children’s Hospital Flow Cytometry Core Facility for help with FACS data interpretation, and S. M. Evans for mouse strains. This work was supported by grants from the National Institutes of Health RO1HD045499 (T.C.K. and C.J.T.) and U01HL100405 (M.K.S., K.D., and J.A.E.). The latter were also supported by the AHA DeHaan Myogenesis Center. K.D. was additionally supported by NIH 5K12HD043245-09.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Blitz E, Viukov S, Sharir A, Shwartz Y, Galloway JL, Pryce BA, Johnson RL, Tabin CJ, Schweitzer R, Zelzer E. Bone ridge patterning during musculoskeletal assembly is mediated through SCX regulation of Bmp4 at the tendon-skeleton junction. Dev Cell. 2009;17:861–873. doi: 10.1016/j.devcel.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brent AE, Schweitzer R, Tabin CJ. A somitic compartment of tendon progenitors. Cell. 2003;113:235–248. doi: 10.1016/s0092-8674(03)00268-x. [DOI] [PubMed] [Google Scholar]
- Cai CL, Martin JC, Sun Y, Cui L, Wang L, Ouyang K, Yang L, Bu L, Liang X, Zhang X, et al. A myocardial lineage derives from Tbx18 epicardial cells. Nature. 2008;454:104–108. doi: 10.1038/nature06969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossette S, Misra R. The identification of different endothelial cell populations within the mouse proepicardium. Dev Dyn. 2011;240:2344–2353. doi: 10.1002/dvdy.22724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gittenberger-de Groot AC, Vrancken Peeters MP, Mentink MM, Gourdie RG, Poelmann RE. Epicardium-derived cells contribute a novel population to the myocardial wall and the atrioventricular cushions. Circ Res. 1998;82:1043–1052. doi: 10.1161/01.res.82.10.1043. [DOI] [PubMed] [Google Scholar]
- Guadix JA, Carmona R, Munoz-Chapuli R, Perez-Pomares JM. In vivo and in vitro analysis of the vasculogenic potential of avian proepicardial and epicardial cells. Dev Dyn. 2006;235:1014–1026. doi: 10.1002/dvdy.20685. [DOI] [PubMed] [Google Scholar]
- Gourdie RG, Cheng G, Thompson RP, Mikawa T. Retroviral cell lineage analysis in the developing chick heart. Methods Mol Biol. 2000;135:297–304. doi: 10.1385/1-59259-685-1:297. [DOI] [PubMed] [Google Scholar]
- Hiruma T, Hirakow R. Epicardial formation in embryonic chick heart: computer-aided reconstruction, scanning, and transmission electron microscopic studies. Am J Anat. 1989;184:129–138. doi: 10.1002/aja.1001840204. [DOI] [PubMed] [Google Scholar]
- Ishii Y, Garriock RJ, Navetta AM, Coughlin LE, Mikawa T. BMP signals promote proepicardial protrusion necessary for recruitment of coronary vessel and epicardial progenitors to the heart. Dev Cell. 19:307–316. doi: 10.1016/j.devcel.2010.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komiyama M, Ito K, Shimada Y. Origin and development of the epicardium in the mouse embryo. Anat Embryol (Berl) 1987;176:183–189. doi: 10.1007/BF00310051. [DOI] [PubMed] [Google Scholar]
- Kraus F, Haenig B, Kispert A. Cloning and expression analysis of the mouse T-box gene Tbx18. Mech Dev. 2001;100:83–86. doi: 10.1016/s0925-4773(00)00494-9. [DOI] [PubMed] [Google Scholar]
- Levay AK, Peacock JD, Lu Y, Koch M, Hinton RB, Jr, Kadler KE, Lincoln J. Scleraxis is required for cell lineage differentiation and extracellular matrix remodeling during murine heart valve formation in vivo. Circ Res. 2008;103:948–956. doi: 10.1161/CIRCRESAHA.108.177238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manner J. The development of pericardial villi in the chick embryo. Anat Embryol (Berl) 1992;186:379–385. doi: 10.1007/BF00185988. [DOI] [PubMed] [Google Scholar]
- Manner J. Does the subepicardial mesenchyme contribute myocardioblasts to the myocardium of the chick embryo heart? A quail-chick chimera study tracing the fate of the epicardial primordium. Anat Rec. 1999;255:212–226. doi: 10.1002/(sici)1097-0185(19990601)255:2<212::aid-ar11>3.3.co;2-o. [DOI] [PubMed] [Google Scholar]
- Mikawa T, Gourdie RG. Pericardial mesoderm generates a population of coronary smooth muscle cells migrating into the heart along with ingrowth of the epicardial organ. Dev Biol. 1996;174:221–232. doi: 10.1006/dbio.1996.0068. [DOI] [PubMed] [Google Scholar]
- Moretti A, Caron L, Nakano A, Lam JT, Bernshausen A, Chen Y, Qyang Y, Bu L, Sasaki M, Martin-Puig S, et al. Multipotent embryonic isl1+ progenitor cells lead to cardiac, smooth muscle, and endothelial cell diversification. Cell. 2006;127:1151–1165. doi: 10.1016/j.cell.2006.10.029. [DOI] [PubMed] [Google Scholar]
- Murtaugh LC, Chyung JH, Lassar AB. Sonic hedgehog promotes somitic chondrogenesis by altering the cellular response to BMP signaling. Genes Dev. 1999;13:225–237. doi: 10.1101/gad.13.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahirney PC, Mikawa T, Fischman DA. Evidence for an extracellular matrix bridge guiding proepicardial cell migration to the myocardium of chick embryos. Dev Dyn. 2003;227:511–523. doi: 10.1002/dvdy.10335. [DOI] [PubMed] [Google Scholar]
- Nemer G, Nemer M. Cooperative interaction between GATA5 and NF-ATc regulates endothelial-endocardial differentiation of cardiogenic cells. Development. 2002;129:4045–4055. doi: 10.1242/dev.129.17.4045. [DOI] [PubMed] [Google Scholar]
- Perez-Pomares JM, Carmona R, Gonzalez-Iriarte M, Atencia G, Wessels A, Munoz-Chapuli R. Origin of coronary endothelial cells from epicardial mesothelium in avian embryos. Int J Dev Biol. 2002;46:1005–1013. [PubMed] [Google Scholar]
- Pryce BA, Brent AE, Murchison ND, Tabin CJ, Schweitzer R. Generation of transgenic tendon reporters, ScxGFP and ScxAP, using regulatory elements of the scleraxis gene. Dev Dyn. 2007;236:1677–1682. doi: 10.1002/dvdy.21179. [DOI] [PubMed] [Google Scholar]
- Red-Horse K, Ueno H, Weissman IL, Krasnow MA. Coronary arteries form by developmental reprogramming of venous cells. Nature. 2010;464:549–553. doi: 10.1038/nature08873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese DE, Mikawa T, Bader DM. Development of the coronary vessel system. Circ Res. 2002;91:761–768. doi: 10.1161/01.res.0000038961.53759.3c. [DOI] [PubMed] [Google Scholar]
- Singh MK, Li Y, Li S, Cobb RM, Zhou D, Lu MM, Epstein JA, Morrisey EE, Gruber PJ. Gata4 and Gata5 cooperatively regulate cardiac myocyte proliferation in mice. J Biol Chem. 2010;285:1765–1772. doi: 10.1074/jbc.M109.038539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh MK, Lu MM, Massera D, Epstein JA. MicroRNA-processing enzyme Dicer is required in epicardium for coronary vasculature development. J Biol chem. 2011;286:41036–41045. doi: 10.1074/jbc.M111.268573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart N, Bollini S, Dubé KN, Vieira JM, Zhou B, Davidson S, Yellon D, Riegler J, Price AN, Lythgoe MF, Pu WT, Riley PR. De novo cardiomyocytes form within the activated adult heart after injury. Nature. 2011;474:640–4. doi: 10.1038/nature10188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Viragh S, Challice CE. The origin of the epicardium and the embryonic myocardial circulation in the mouse. Anat Rec. 1981;201:157–168. doi: 10.1002/ar.1092010117. [DOI] [PubMed] [Google Scholar]
- Zhou B, Ma Q, Rajagopal S, Wu SM, Domian I, Rivera-Feliciano J, Jiang D, von Gise A, Ikeda S, Chien KR, Pu WT. Epicardial progenitors contribute to the cardiomyocyte lineage in the developing heart. Nature. 2008;454:109–113. doi: 10.1038/nature07060. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.