Abstract
Nitrous oxide reductase (N2OR) catalyses the final step of the denitrification pathway—the reduction of nitrous oxide to nitrogen. The catalytic centre (CuZ) is a unique tetranuclear copper centre bridged by inorganic sulphur in a tetrahedron arrangement that can have different oxidation states. Previously, Marinobacter hydrocarbonoclasticus N2OR was isolated with the CuZ centre as CuZ*, in the [1Cu2+ : 3Cu+] redox state, which is redox inert and requires prolonged incubation under reductive conditions to be activated. In this work, we report, for the first time, the isolation of N2OR from M. hydrocarbonoclasticus in the ‘purple’ form, in which the CuZ centre is in the oxidized [2Cu2+ : 2Cu+] redox state and is redox active. This form of the enzyme was isolated in the presence of oxygen from a microaerobic culture in the presence of nitrate and also from a strictly anaerobic culture. The purple form of the enzyme was biochemically characterized and was shown to be a redox active species, although it is still catalytically non-competent, as its specific activity is lower than that of the activated fully reduced enzyme and comparable with that of the enzyme with the CuZ centre in either the [1Cu2+ : 3Cu+] redox state or in the redox inactive CuZ* state.
Keywords: nitrous oxide reductase, Marinobacter, denitrification, isolation in presence of oxygen, fully oxidized CuZ centre, purple form
1. Introduction
Nitrous oxide (N2O) is a potent long-lived greenhouse gas, the atmospheric concentration of which has increased over past years. In addition, the potential of N2O as a global warming gas relative to that of CO2 is 298 for a 100 year period, and this gas is also responsible for stratospheric ozone depletion [1].
Different sources have been responsible for this increase, such as industry and wastewater treatment, but bigger contributors are the soil and seawater because of microbial (fungal and bacterial) processes. N2O is produced by bacteria through denitrification and nitrification, which are part of the biogeochemical nitrogen cycle.
While nitrification is a biochemical pathway that takes place under aerobic conditions, denitrification occurs mainly under strictly anaerobic or microaerobic conditions. In the first case, N2O is produced as a by-product of ammonium oxidation, whereas in denitrification, bacteria reduce inorganic forms of nitrogen and N2O is formed as an intermediate by-product, and in fact this biological process can also consume N2O. Although denitrification can decrease the level of nitrous oxide by reducing it to nitrogen gas, different environmental conditions, such as pH, oxygen and nitrogen levels, and temperature, may lead to its accumulation and release into the atmosphere [2,3]. Therefore, there is a particular interest in understanding how the reduction of nitrous oxide to inert N2 is carried out in nature [4]. The enzyme involved in the denitrification pathway and responsible for catalysing this reaction is nitrous oxide reductase (N2OR), which can be isolated from α-, β- and γ-proteobacteria [5].
Interestingly, another enzyme of the nitrogen biogeochemical cycle, nitrogenase, is able to carry out the in vitro reduction of N2O to N2 [6,7] and Escherichia coli has also been shown to be able to reduce N2O, even if this reaction might not have any physiological implications and the enzyme responsible for the catalysis has not been identified [8]. Recently, another copper enzyme, a multi-copper oxidase, from Pyrobaculum aerophilum has been shown to be able to catalytically reduce N2O, although its in vivo function is still unclear [9].
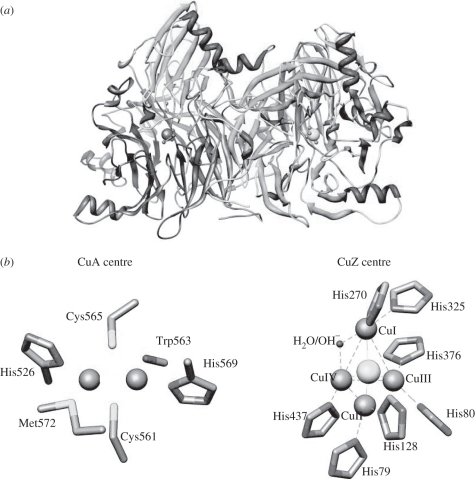
N2OR, a periplasmic enzyme in Gram-negative bacteria, was isolated for the first time many years ago [10], but its structure has only been solved more recently, revealing that it is a functional dimer, containing two copper centres per monomer (figure 1a) [11]. The CuA centre located in a cupredoxin-like domain can be assumed to be the electron-transferring centre, as it is proposed to receive electrons from small electron carrier proteins and transfer them to the catalytic centre—CuZ centre [11]. The CuA centre is a binuclear copper centre, similar to that found in cytochrome c oxidase [12] (figure 1b).
Figure 1.
(a) Representation of Marinobacter hydrocarbonoclasticus nitrous oxide reductase functional dimer. The dimer is coloured according to subunit, with one monomer coloured grey and the other dark grey, and CuA and CuZ centres coloured grey. (b) CuA and CuZ centres are coloured grey and in CuZ centre the copper ions are numbered I, II, III and IV. The sulphur ion in CuZ centre is shown as a light grey sphere. The figure was created with Chimera using 1QNI.
The CuZ centre has a unique structure as it comprises a tetranuclear copper cluster bridged by sulphur in a tetrahedral arrangement [11] (figure 1b). In the past, many studies have focused on the complex chemistry of the catalytic CuZ centre, to identify and spectroscopically characterize its different redox forms, and how these redox states are involved in the catalytic cycle.
Previously, Marinobacter hydrocarbonoclasticus (formerly known as Pseudomonas nautica [13]) N2OR was purified in the presence of oxygen from a strictly anaerobic culture and two forms of the enzyme were characterized, named purple and blue. The difference between these enzyme forms was mainly the oxidation state of the CuA centre, which was in the oxidized state ([Cu1.5+ : Cu1.5+]) and reduced state ([Cu+ : Cu+]), respectively (table 1). In these isolated forms, the CuZ centre was in ‘the resting state’, i.e. the [1Cu2+ : 3Cu+] redox state, denoted by some authors as CuZ* [15,16]. This form presents an absorption spectrum characterized by a band at 640 nm characteristic of the resting CuZ centre together with contributions at 480, 550 and 800 nm of the CuA centre in the [Cu1.5+ : Cu1.5+] state [17]. The proportion of the contribution of CuA centre bands depended on the amount of this centre that was oxidized.
Table 1.
The different redox forms of nitrous oxide reductase (N2OR).
| CuA centre | CuZ centre | CuZ* centre | |
|---|---|---|---|
| fully oxidized/as-isolated (purple form) | [Cu1.5+–Cu1.5+]3+ | [2Cu2+–2Cu+S]4+ | |
| ascorbate-reduced | [Cu1+–Cu1+]2+ | [2Cu2+–2Cu+S]4+ | |
| dithionite-reduced | [Cu1+–Cu1+]2+ | [1Cu2+–3Cu+S]3+ | |
| fully oxidized/as-isolated (purple/pink forma) | [Cu1.5+–Cu1.5+]3+ | [1Cu2+–3Cu+S]3+ | |
| dithionite-reduced/ascorbate-reduced (blue formb) | [Cu1+–Cu1+]2+ | [1Cu2+–3Cu+S]3+ | |
| fully reduced/activated formc | [Cu1+–Cu1+]2+ | [4Cu+S]2+ | |
aAlso named as a purple form by some authors.
bIn the ‘blue form’ of N2OR, the CuZ centre is found mainly in the CuZ* state, which has also been named ‘resting CuZ’ [14]. The two dithionite-reduced forms of N2OR are different, although both copper centres have the same redox state.
cThis form has only been shown to be formed from CuZ*.
Although the spectroscopic, electronic and structural properties of the CuZ centre in the resting state have been extensively characterized [14], this state is redox inactive in the sense that it cannot be oxidized or easily reduced, and it is also catalytically inactive, as its specific activity is very low when compared with that of the enzyme with the CuZ centre in the fully reduced state [18,19].
However, this resting form of the enzyme can be activated through prolonged incubation with a strong reducing agent, such as reduced methyl viologen [18], during which the CuZ centre is completely reduced to [4Cu+]. It has been hypothesized that this long activation process corresponds to a rearrangement in the coordination sphere of the CuZ centre, which is required for full activity [14].
Recently, an intermediate in the catalytic cycle of the enzyme has been identified, CuZ°, which is proposed to have the CuZ centre in the same redox state as the resting state [1Cu2+ : 3Cu+], but differs by exhibiting full activity. This species has a short half-life, with a rate of decay to the resting form of 0.3 min−1 [20].
However, from the first purifications of Pseudomonas stutzeri N2OR, it became clear that this enzyme can be isolated in different forms with the CuZ centre in different redox states depending on the purification conditions [21]. N2OR purified in the absence of oxygen from Pseudomonas stutzeri, Achromobacter xylosoxidans, Paracoccus denitrificans and Paracoccus pantotrophus [15,21–23] exhibits an intense absorption at 550 nm. This form, named ‘purple’ (because of its colour), presents an oxidized CuA centre because the electron paramagnetic resonance (EPR) spectra exhibit the typical seven-line hyperfine pattern in the g‖ region, and the CuZ centre in the oxidized [2Cu2+ : 2Cu+] state (table 2). In contrast, N2OR purified in the presence of oxygen from these bacterial sources is in a different form, named ‘pink’ [24], which is similar to M. hydrocarbonoclasticus N2OR also isolated in the presence of oxygen [17].
Table 2.
Properties of the purple form of nitrous oxide reductase (N2OR) isolated from different organisms.
| Paracoccus denitrificans | Paracoccus pantotrophus | Pseudomonas stutzeri | Achromobacter xylosoxidans | |
|---|---|---|---|---|
| anaerobic purification | yes | yes | yes | yes |
| visible spectrum | ||||
| ε550 nma (M−1 cm−1) (dimer) | 10 600 | 8000 | 14 000 | 10 500 |
| A550 nm/A640 nm | 1.7 | 1.9 | 2.1 | 1.8 |
| EPR | ||||
| g‖ | 2.224 | 2.181 | 2.180 | 2.180 |
| g⊥ | 2.042 | 2.038 | 2.030 | 2.019 |
| A (gauss) | 35 | 38 | 38 | 40 |
| specific activity (U mg−1)b | 122 | 3 | 4 | 6 |
| reference | [22] | [15] | [21] | [23] |
aValue determined based on the total protein content for the fully oxidized purple N2OR.
bThe specific activity was determined using benzyl viologen (Pseudomonas stutzeri and Paracoccus denitrificans) or methyl viologen (Paracoccus pantotrophus and Achromobacter xylosoxidans) as electron donor.
The reduction of N2OR in the purple form by sodium dithionite proceeds in two kinetic steps: a fast phase in which the absorbance at 540 nm disappears almost within seconds, owing to the reduction of the CuA centre (this form can also be obtained by reduction of the purple form with sodium ascorbate), and a slower phase that takes place over the course of minutes or hours, in which the CuZ centre, as [2Cu2+–2Cu+], is reduced to [1Cu2+–3Cu+] [5,25].
The EPR spectrum of the purple form (CuA centre oxidized and CuZ centre as [2Cu2+–2Cu+]) exhibits a well-defined seven-line hyperfine split axial signal, whereas that of the enzyme with CuA reduced and the CuZ centre as CuZ* (redox inactive [1Cu2+–3Cu+] state) has a broad and poorly resolved four-line hyperfine split axial signal [26]. The purple form of N2OR after reduction of the CuA centre (with the CuZ centre in the [2Cu2+–2Cu+] state) shows an extremely weak EPR signal and must be considered EPR-silent, although it still exhibits an absorption band at 540 nm and a small contribution at 640 nm. In fact, magnetic circular dichroism (MCD) data confirmed that the CuZ centre in that form is not a ferromagnetically coupled (S = 1) centre but must be anti-ferromagnetically coupled (S = 0) [16].
It is also important to mention that Paracoccus pantotrophus N2OR purified in the presence of oxygen exhibits a CuZ*/CuZtotal ratio of 0.66, whereas this ratio is 0.29 when the enzyme is isolated in the absence of oxygen [15]. Therefore, it seems that CuZ centre has always been present as CuZ* in all the reported preparations of N2OR, regardless of the presence or absence of oxygen during purification.
It has been argued that the oxidized form of the CuZ centre (in the [2Cu2+–2Cu+] state) is catalytically important, and its redox potential was estimated to be E′° = +60 mV, for the reduction of [2Cu2+ : 2Cu+] to [1Cu2+ : 3Cu+]. However, this is still a subject of discussion and contradictions, as its catalytic activity seems to be very low, and similar to that of N2OR with the CuZ centre either in the dithionite-reduced state (redox active [1Cu2+–3Cu+]) or as CuZ* (redox inactive [1Cu2+–3Cu+]) [15].
The redox active purple form, in which the CuZ can be oxidized and reduced between the [2Cu2+ : 2Cu+] and [1Cu2+ : 3Cu+] states, had not been observed in M. hydrocarbonoclasticus. Here, we report, for the first time, the isolation of the purple form of N2OR from this organism. Moreover, N2OR was isolated not only from a microaerobic culture in the presence of nitrate but also from a strictly anaerobic culture purified in the presence of oxygen. This is the first report of a purple N2OR isolated in the presence of oxygen. The purple form of M. hydrocarbonoclasticus N2OR was characterized using visible and EPR spectroscopies and the catalytic activity of the enzyme with the CuZ centre in different redox states was determined.
2. Material and methods
(a). Preparation of the different nitrous oxide reductase forms
Marinobacter hydrocarbonoclasticus 617 (previously known as Pseudomonas nautica 617 [13]) was grown in a 10 l fermenter at 30°C in artificial seawater, with yeast extract (0.1%) and lactate (6%) as carbon and energy sources [27]. The medium was supplemented with a Starkey oligoelement solution [28] (0.2 ml l−1 of culture) and 0.2 mM sodium nitrate (I. Fernandes, I. Moura, P. Paes de Sousa & S. R. Pauleta 2011, unpublished data). The bacterium was grown for 48 h with pH being continuously monitored and maintained at 7.5. A low aeration rate (0.2 v/v min) and an agitation speed of 150 rpm (central axis with flat-blade turbines) were used.
The periplasmic extract was obtained by diluting five times the resuspended cells into Tris–HCl (10 mM; pH 7.6), EDTA (0.5 mM) for 30 min. The soluble extract was obtained by centrifugation for 45 min at 125 000g (Beckman L-70 ultracentrifuge), and loaded onto a DE52 Fast Flow chromatographic column (GE Healthcare), equilibrated with Tris–HCl (10 mM; pH 7.6). The proteins were eluted with a gradient between 0 and 500 mM NaCl in Tris–HCl (10 mM; pH 7.6). The fractions containing N2OR were combined and concentrated in a diaflo apparatus (Millipore) over a 30 kDa cut-off membrane. This fraction was then loaded onto a gel filtration column (Superdex 200, GE Healthcare) equilibrated with Tris–HCl (300 mM; pH 7.6). The fractions containing the pure N2OR were combined, concentrated and stored at −80°C until further use.
The purity of the fractions was checked throughout the purification by SDS–PAGE, and N2OR concentration was determined using an extinction coefficient at 640 nm, for the dithionite-reduced form, of 7.1 mM−1 cm−1, for the sample named ‘anaerobic2’ [17]. Total protein was quantified using the bicinchoninic acid (BCA) assay (Sigma) and bovine serum albumin as protein standard [29]; the total copper content was determined using the 2,2′-biquinoline assay [30].
N2OR from different anaerobic cultures was prepared and purified in the presence of oxygen as previously reported [17].
(b). Spectroscopic methods
N2OR in different redox states was prepared by reduction with sodium ascorbate or sodium dithionite or by oxidation with potassium ferricyanide in Tris–HCl (100 mM; pH 7.6).
UV–visible absorption spectra were recorded in a Shimadzu UV-1800 spectrophotometer using 1 cm quartz cells. X-band EPR spectra were recorded on a Bruker EMX spectrometer equipped with a rectangular cavity (model ER 4102ST) and an Oxford Instruments continuous-flow cryostat at 30 K temperature. EPR spectra were simulated using the program WIN-EPR SimFonia (v. 1.2) from Bruker Instruments.
Samples for EPR spectroscopy were prepared with 70 μM enzyme in Tris–HCl (100 mM; pH 7.6). Spin quantification was performed at the same temperature under non-saturating conditions, using a sample of dithionite-reduced N2OR (1 spin). Experimental conditions are described in the figure legends.
(c). Activity assay
The activity assay was performed using the procedure previously described [31], by adding N2OR (100 nM) in different redox forms as the last reactant into a cuvette already containing reduced methyl viologen and N2O in Tris–HCl (100 mM, pH 7.6). The specific activity of the enzyme is expressed in units per milligram, which corresponds to micromoles of N2O reduced per minute per milligram of N2OR dimer.
3. Results and discussion
(a). Microaerobic culture of Marinobacter hydrocarbonoclasticus
The periplasmic fraction of M. hydrocarbonoclasticus grown under microaerobic conditions in the presence of nitrate, contained a large amount of nitrite reductase cytochrome cd1 and N2OR, thus indicating that this bacterium can be classified as an aerobic denitrifier, similar to Paracoccus denitrificans and Pseudomonas stutzeri [32,33].
(b). Characterization of the purple form of nitrous oxide reductase from Marinobacter hydrocarbonoclasticus
(i). Spectroscopic characterization
The ‘purple’ form of M. hydrocarbonoclasticus N2OR was purified in the presence of oxygen from two different cultures. In one case, M. hydrocarbonoclasticus was grown under microaerobic conditions in the presence of nitrate (‘aerobic sample’), and in the other this bacterium was grown under strict anaerobic conditions (‘anaerobic1 sample’). The purified enzyme from these two cultures has similar features (see below).
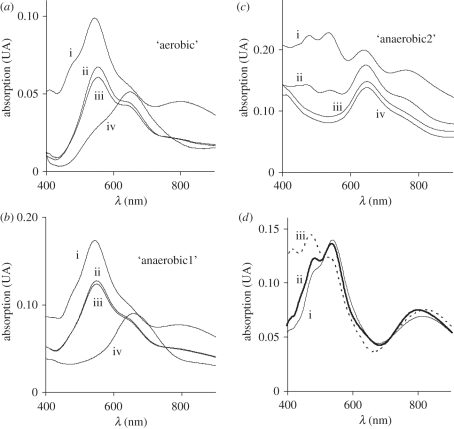
These N2OR preparations present a UV–visible spectrum characterized by an absorption maximum at 550 nm with a shoulder at 635 nm (figure 2a(ii), b(ii)) and a very small absorption band around 800 nm. After oxidation with potassium ferricyanide, the absorption maximum at 550 nm shifts to 540 nm and the absorption band at 800 nm becomes more intense (figure 2a(i),b(i)). This is evidence for the CuA centre being almost completely reduced in as-isolated N2OR (approx. 10% and 3% of CuA centre is oxidized in as-isolated ‘aerobic’ and ‘anaerobic1’ N2OR, respectively), and becoming oxidized upon addition of potassium ferricyanide.
Figure 2.
UV–visible spectra of nitrous oxide reductase purified in the presence of oxygen from (a) an aerobic culture; (b) an ‘anaerobic1’ culture and (c) an ‘anaerobic2’ culture. In each panel the spectra of the (i) fully oxidized, (ii) as-isolated, (iii) ascorbate-reduced and (iv) dithionite-reduced forms of the enzyme are presented. (d) Difference spectra between fully oxidized and the ascorbate-reduced forms for the (i) ‘aerobic’ (solid line) (ii) ‘anaerobic1’ (thick line) and (iii) ‘anaerobic2’ (dotted line) N2OR samples.
The absorption spectrum of the CuA centre of N2OR can be obtained by subtracting the ascorbate-reduced N2OR spectrum from the ferricyanide-oxidized N2OR spectrum (figure 2d). Analysis of these spectra shows that the CuA centre has three main absorption bands at 480, 540 and 800 nm, and that the absorption bands observed at 550 and 635 nm in the as-isolated spectrum of N2OR correspond to the CuZ centre in the [2Cu2+ : 2Cu+] oxidation state (see below). Hence, this is the first time that this redox form of N2OR (the purple form) has been isolated in the presence of oxygen and also from a microaerobic culture in the presence of nitrate. In fact, even though Alcaligenes xylosoxidans and Paracoccus denitrificans N2OR have also been isolated with the CuA centre reduced and the CuZ centre in the [2Cu2+ : 2Cu+] redox state, in these cases the enzymes were isolated under exclusion of oxygen from bacteria grown under anaerobic denitrifying conditions and purified under exclusion of oxygen [22,23].
The spectrum of the as-isolated purple N2OR after 1 min reduction with sodium ascorbate exhibits absorption bands at 550 nm with a shoulder at 635 nm (figure 2a(iii),b(iii)), which is similar to that described for the equivalent N2OR forms from Pseudomonas stutzeri or Paracoccus pantotrophus N2OR [15,21].
The reduction of the purple M. hydrocarbonoclasticus N2OR with sodium dithionite reduces the CuA centre to [Cu+ : Cu+] and the CuZ centre to [1Cu2+ : 3Cu+] (figure 2a(iv),b(iv)). The visible spectrum of this form, with an absorption band with maximum at 655 nm (‘aerobic’ sample) or at 660 nm (‘anaerobic1’ sample), is different from that of the enzyme with CuZ* in the [1Cu2+ : 3Cu+] redox state (figure 2c(iii); maximum at 640 nm). Moreover, the absorption band is also broader in the spectrum of the enzyme with a [1Cu2+ : 3Cu+] CuZ centre than that of [1Cu2+ : 3Cu+] CuZ*, with an increased contribution at higher wavelengths.
These spectral characteristics are different from those presented by dithionite-reduced N2OR isolated in the presence of oxygen from different bacterial sources [15,21] and also different from those previously reported for M. hydrocarbonoclasticus N2OR (which had the CuZ centre in the [1Cu2+ : 3Cu+] CuZ* state and differed in the amount of oxidized CuA centre) [17]. In fact, as previously shown by Prudencio et al. [17], N2OR isolated in the presence of oxygen and obtained from an anaerobic culture of M. hydrocarbonoclasticus shows absorption bands at 480, 540, 640 and 800 nm when oxidized with ferricyanide. Thus, the CuA centre is partially oxidized (480, 540 and 800 nm absorption bands, see above) and the CuZ centre is in the [1Cu2+ : 3Cu+] CuZ* state (640 nm) (figure 2c(ii)). This form of N2OR has been named ‘pink’ owing to its colour, and in the present work corresponds to the ‘anaerobic2’ N2OR sample.
Regarding the redox behaviour of the ‘anaerobic2’ N2OR, sodium ferricyanide is able to fully oxidize the CuA centre (figure 2c(i)) which is 40 per cent oxidized in the as-isolated form, whereas both sodium ascorbate and sodium dithionite fully reduce the CuA centre, with the CuZ centre remaining unchanged and in the CuZ* [1Cu2+ : 3Cu+] state (figure 2c(iii)(iv)). It is also important to mention that the degree of oxidation of the CuA centre varies among different preparations, whereas the CuZ centre has always been found mainly in the resting state.
Nevertheless, a N2OR preparation from another anaerobic culture/purification in the presence of oxygen (‘anaerobic1’ N2OR sample) shows the typical spectroscopic features of the ‘purple’ form, as described above (figure 2b). In this preparation, the fully oxidized form (figure 2b(i)) has a 550/640 nm absorbance ratio of 1.7, a value identical to that obtained for the ‘aerobic’ preparation and higher than the absorbance ratio of the ‘anaerobic2’ sample of 1.1 (figure 2c). A similar absorbance ratio was reported for Achromobacter xylosoxidans purple N2OR (1.8) [23] and for Paracoccus pantotrophus purple N2OR (1.9) [15] (table 2).
Moreover, the dithionite-reduced purple forms of M. hydrocarbonoclasticus N2OR not only present a distinct visible spectrum (as mentioned) but also have different redox behaviour. Although complete reduction with sodium dithionite is very slow, as observed for purple N2ORs isolated from other organisms [5,15], this reduction is reversible (data not shown), as addition of potassium ferricyanide allows fast regeneration of the fully oxidized purple form characterized by the spectrum in figure 2a(i). This indicates that the CuZ centre of purple N2OR can be reduced and oxidized between [2Cu2+ : 2Cu+] and [1Cu2+ : 3Cu+] redox states, in contrast to the CuZ centre in the CuZ* [1Cu2+ : 3Cu+] state, which cannot be oxidized.
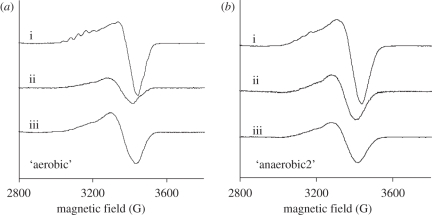
The EPR spectra of the different redox states of the two distinct N2OR preparations (‘aerobic’ and ‘anaerobic2’ N2ORs) were analysed (figure 3). The total spin was quantified for each redox state (table 3), assuming that dithionite-reduced N2OR presents a total spin of 1. This assumption is based on the fact that in this state the CuZ centre contains one unpaired electron and is in the [1Cu2+ : 3Cu+] state, whereas the CuA centre is reduced [Cu1+ : Cu1+] and does not contribute to the EPR spectrum [34].
Figure 3.
EPR spectra of N2OR purified in the presence of oxygen from (a) an ‘aerobic’ and (b) an ‘anaerobic2’ culture. In each panel the spectra of the (i) fully oxidized, (ii) as-isolated minus the CuA centre contribution and (iii) dithionite-reduced forms of the enzyme are presented. Instrumental parameters: modulation amplitude (5 G); microwave frequency (9.66 GHz); temperature (30 K).
Table 3.
Spin quantification of the Marinobacter nitrous oxide reductase (N2OR) EPR spectra.
| ‘aerobic’ N2OR | ‘anaerobic2’ N2OR | |
|---|---|---|
| fully oxidized | 1.4 | 2.2 |
| as-isolated CuA contribution | 0.4 | 1.0 |
| dithionite reduced | 1.0 | 1.0 |
In all preparations, the fully oxidized form of N2OR has an EPR spectrum dominated by the features of the CuA centre (figure 3a,b(i)), as observed in the visible spectrum of these samples. This EPR spectrum is characterized by the typical seven-line hyperfine splitting pattern, with g‖ = 2.18, g⊥ = 2.04 and A‖ ≈ 40 G, regardless of the preparation. In the case of the ‘anaerobic2’ N2OR sample, the seven-line hyperfine splitting is less resolved as a result of the larger contribution of the resting CuZ centre characterized by a four-line hyperfine split axial EPR signal [5] (see below).
The dithionite-reduced N2OR has a broad axial EPR signal (figure 3a,b(iii)). The EPR spectrum of the N2OR with mainly CuZ* (‘anaerobic2’) is characterized by g‖ = 2.16, g⊥ = 2.04 and A‖ ≈ 60 G [34]. At this stage, without further deconvolution of the spectrum of the dithionite-reduced N2OR sample with 60 per cent CuZ and 40 per cent CuZ*, it is difficult to determine the characteristic spectrum of CuZ in the [1Cu2+ : 3Cu+] state, although it should be similar to that reported by Oganesyan et al. [35].
The EPR spectrum of the CuA centre can be obtained by subtracting the EPR spectrum of the fully oxidized N2OR from that of the as-isolated enzyme. The contribution of the CuA centre was then subtracted from the EPR spectrum of the as-isolated purple N2OR. For the ‘aerobic’ N2OR sample, this difference spectrum is shown in figure 3a(ii), and has a spin quantification of 0.4 (table 3). Taking into account that this N2OR sample is proposed to have the CuZ centre mainly in the [2Cu2+ : 2Cu+] state, and that this state is diamagnetic, the amount of spin that is quantified must correspond to the amount of CuZ centre in the [1Cu2+ : 3Cu+] state. Therefore, the purple form of N2OR isolated in the presence of oxygen (‘aerobic’ N2OR) has 40 per cent of the CuZ centre as the CuZ* [1Cu2+ : 3Cu+] state, and 60 per cent in the CuZ [2Cu2+ : 2Cu+] state. As observed in other anaerobic preparations of purple N2OR from other organisms, the CuZ centre is not in a single redox state and the amount of CuZ centre in the paramagnetic [1Cu2+ : 3Cu+] state is comparable with that reported in those studies [15].
In contrast, the spin contribution of the CuA centre to the fully oxidized EPR spectrum of ‘anaerobic2’ N2OR is 1, and thus in this sample the CuZ centre is present almost completely in the paramagnetic CuZ* [1Cu2+ : 3Cu+] state, as previously observed [17].
(ii). Specific activity of the different redox forms of N2OR
In addition to the spectroscopic characterization, it is also important to evaluate the specific activity that is associated with the CuZ centre in the [2Cu2+ : 2Cu+] redox state. This state is redox active, in contrast to the redox inactive CuZ centre in the CuZ* [1Cu2+ : 3Cu+] state.
The modified activity assay described by Dell' Acqua et al. [31] enables the separation of the slow activation (complete reduction of CuZ centre to [4Cu+]) accomplished with reduced methyl viologen from the catalytic activity. Therefore, with this assay, it will be possible to determine the specific activity of N2OR in the different redox states: fully oxidized (CuA as [Cu1.5+ : Cu1.5+] and CuZ as [2Cu2+ : 2Cu+]); ascorbate-reduced (CuA as [Cu+ : Cu+] and CuZ as [2Cu2+ : 2Cu+]); dithionite-reduced (CuA as [Cu+ : Cu+] and CuZ as [1Cu2+ : 3Cu+]) and fully reduced (CuA as [Cu+ : Cu+] and CuZ as [4Cu+]). The specific activity of N2OR in these different redox forms is shown in table 4.
Table 4.
Specific activity of Marinobacter nitrous oxide reductase (N2OR) isolated from the different growth conditions. The table also shows the main redox state of the CuZ centre in each redox state. asc, ascorbate; dth, dithionite.
| redox state | ‘aerobic’ |
‘anaerobic1’ |
‘anaerobic2’ |
|||
|---|---|---|---|---|---|---|
| activity (U mg−1) | redox state CuZ centrea | activity (U mg−1) | redox state CuZ centrea | activity (U mg−1) | redox state CuZ centrea | |
| fully oxidized | 0.4 | 60% [2Cu2+ : 2Cu+] | 0.4 | 60% [2Cu2+ : 2Cu+] | 1.1 | [1Cu2+ : 3Cu+] |
| asc-reduced | 0.3 | 60% [2Cu2+ : 2Cu+] | 0.3 | 60% [2Cu2+ : 2Cu+] | 0.9 | [1Cu2+ : 3Cu+] |
| dth-reduced | 0.2 | [1Cu2+ : 3Cu+] | 0.3 | [1Cu2+ : 3Cu+] | 0.2 | [1Cu2+ : 3Cu+] |
| fully reduced | 88.0 | [4Cu+] | 96.1 | [4Cu+] | 92.4 | [4Cu+] |
aThe CuZ centre content for ‘aerobic’ and ‘anaerobic2’ N2OR samples was determined based on the EPR spectra, whereas the one indicated for ‘anaerobic1’ was estimated by comparing its visible spectral features with those of the ‘aerobic’ sample.
The results obtained clearly show that none of these redox forms has an activity comparable with the specific activity that is reached with the fully reduced form of the enzyme, and that there are no significant differences between the redox forms of the different preparations of N2OR.
A similar result was obtained for N2OR from the organisms listed in table 2—as the specific activity of these enzymes is very low compared with the high specific activity of the fully reduced enzyme—with the exception of Paracoccus denitrificans purple N2OR in which the as-isolated enzyme has a high specific activity [22].
These results show that the [2Cu2+ : 2Cu+] state of the CuZ centre, that is the main state of the CuZ centre in the two preparations of M. hydrocarbonoclasticus purple N2OR, is not catalytically competent, even though the CuZ centre is in state that is redox active and thus can be reversibly reduced to [1Cu2+ : 3Cu+]. A similar conclusion has also been reached by Thomson and co-workers [15], although in that case the purple N2OR form was isolated in the absence of oxygen.
It is biologically relevant to correlate these different redox forms with the in vivo cellular capacity of N2OR to reduce N2O. In the recent work of Zumft et al. [36], it is reported that in Paracoccus denitrificans, the in vitro N2O reducing capacity is higher in cells from which N2OR is isolated with the CuZ centre mainly as CuZ [2Cu2+ : 2Cu+] then when it is mainly in the CuZ* [1Cu2+ : 3Cu+] state. However, after isolation, both enzyme forms have similarly low specific activities (3.6 U mg−1). Therefore, at this moment, it is still not possible to conclusively say which of the enzyme forms is the active one in vivo, because the fully reduced form still has higher specific activity than any of the as-isolated forms (when the activity of the purified enzyme is compared using the same assay).
In conclusion, N2OR purified either in the absence or in the presence of oxygen and with CuZ centre in either the [2Cu2+ : 2Cu+] or [1Cu2+ : 3Cu+] redox states, is not fully active. An activation process involving the complete reduction of the CuZ centre is required to turn the enzyme into an active state. It is possible to hypothesize that N2OR might be inactivated by a conformational rearrangement during the purification process, which makes it imperative to obtain structural information on the active fully reduced form of N2OR to be able to identify these conformational changes, different coordination spheres or rearrangements.
4. Conclusion
Here, we present the first characterization and purification of a purple form of N2OR isolated from M. hydrocarbonoclasticus. For the first time, N2OR from this bacterium was isolated from a culture grown under non-strictly anaerobic conditions, which indicates that the denitrification pathway can occur in the presence of oxygen and thus that this bacterium can be classified as an aerobic denitrifier.
Moreover, the spectroscopic properties of the purple form of M. hydrocarbonoclasticus N2OR were compared with those from other bacterial sources, showing that the catalytic centre, CuZ, is present as a mixture of two species: 60 per cent as CuZ in the [2Cu2+ : 2Cu+] state and 40 per cent as CuZ* in the [1Cu2+ : 3Cu+] state.
The activity assay allowed us to conclude that also the oxidized [2Cu2+ : 2Cu+] state of the CuZ centre is not catalytically competent, because the activity of the enzyme with CuZ centre mainly in that state (either fully oxidized, as-isolated or ascorbate-reduced) is much smaller than that of the enzyme with the CuZ centre in the fully reduced state and of the same order of magnitude of specific activity as that of the enzyme with the CuZ centre in the CuZ* [1Cu2+ : 3Cu+] state.
Up to now, only the fully reduced state [4Cu+] and the CuZ° states have been identified as being catalytically competent and not requiring activation, CuZ° being an intermediate in the catalytic cycle of N2OR [20]. However, it is important to point out that in the proposed catalytic cycle of N2OR, there is indeed a [2Cu2+ : 2Cu+] state of the CuZ centre, as the first intermediate after N2O release [37], which has neither yet been observed nor characterized.
This study also shows that in order to obtain N2OR in the purple form, it does not have to be purified in the absence of oxygen and that the enzyme is not damaged by its presence, as upon activation the specific activities of the enzyme which was isolated with the CuZ centre mainly as CuZ (‘aerobic’) or as CuZ* (‘anaerobic2’) are similar.
However, we believe that the storage conditions of the cellular extract have an influence on the enzyme redox state (even they are if stored at −80 °C), as N2OR purified from cellular extracts stored for long periods at −80°C have the CuZ centre mainly in the [1Cu2+ : 3Cu+] state (compare ‘anaerobic1’ and ‘anaerobic2’). In addition, the number of purification steps and duration of these steps also influence the redox state of the enzyme, in particular the CuZ centre.
These differences seem to indicate a conformational change of the enzyme, especially around the CuZ centre, to a resting non-active state upon storage. This conformational change could be the result of a rearrangement in the coordination sphere of CuA or CuZ centres or in the pathway between these two, which might be identified by comparing the structures of N2OR in the different redox states, because the presently available three-dimensional structures have the CuZ centre in the CuZ* [1Cu2+ : 3Cu+] state.
During revision of this manuscript, the structure of a purple form of N2OR from Pseudomonas stutzeri was published [38]. This structure shows an additional sulphur bridging CuI and CuIV atoms, forming a [4Cu : 2S] cluster. According to these new data, we can now assume that the differences of the properties between CuZ and CuZ* might be partly due to a different cluster composition.
Acknowledgements
We thank Fundação para a Ciência e Tecnologia for financial support to I.M. (PTDC/QUI/64638/2006 and PTDC/QUI-BIQ/116481/2010) and to S.D. (SFRH/BD/30414/2006). S.D. also thanks C.I.R.C.M.S.B. for financial support.
References
- 1.Solomon S., Qin D., Manning M., Chen Z., Marquis M., Averyt K. B., Tignor M., Miller H. L. 2007. Climate change 2007: The physical science basis. Contribution of working group I to the fourth assessment report of the intergovernmental panel on climate change. Cambridge, UK: Cambridge University Press [Google Scholar]
- 2.Sorai M., Yoshida N., Ishikawa M. 2007. Biogeochemical simulation of nitrous oxide cycle based on the major nitrogen processes. J. Geophys. Res. 112, G01006. 10.1029/2005jg000109 (doi:10.1029/2005jg000109) [DOI] [Google Scholar]
- 3.Capone D. G. 1991. Aspects of the marine nitrogen cycle with relevance to the dynamics of nitrous and nitric oxide. In Microbial production and consumption of greenhouse gases (eds Roders J. E., Whitman W. E.), pp. 255–275 Washington, DC: American Society of Microbiology [Google Scholar]
- 4.Richardson D., Felgate H., Watmough N., Thomson A., Baggs E. 2009. Mitigating release of the potent greenhouse gas N2O from the nitrogen cycle—could enzymic regulation hold the key? Trends Biotechnol. 27, 388–397 10.1016/j.tibtech.2009.03.009 (doi:10.1016/j.tibtech.2009.03.009) [DOI] [PubMed] [Google Scholar]
- 5.Zumft W. G., Kroneck P. M. 2007. Respiratory transformation of nitrous oxide (N2O) to dinitrogen by Bacteria and Archaea. Adv. Microb. Physiol. 52, 107–227 10.1016/S0065-2911(06)52003-X (doi:10.1016/S0065-2911(06)52003-X) [DOI] [PubMed] [Google Scholar]
- 6.Jensen B., Burris R. 1986. N2O as a substrate and as a competitive inhibitor of nitrogenase. Biochemistry 25, 1083–1088 10.1021/bi00353a021 (doi:10.1021/bi00353a021) [DOI] [PubMed] [Google Scholar]
- 7.Christiansen J., Seefeldt L. C., Dean D. R. 2000. Competitive substrate and inhibitor interactions at the physiologically relevant active site of nitrogenase. J. Biol. Chem. 275, 36 104–36 107 10.1074/jbc.M004889200 (doi:10.1074/jbc.M004889200) [DOI] [PubMed] [Google Scholar]
- 8.Kaldorf M., Linne von Berg K., Meier U., Servos U., Bothe H. 1993. The reduction of nitrous oxide to dinitrogen by Escherichia coli. Arch. Microbiol. 160, 432–439 10.1007/BF00245303 (doi:10.1007/BF00245303) [DOI] [PubMed] [Google Scholar]
- 9.Fernandes A. T., Damas J. M., Todorovic S., Huber R., Baratto M. C., Pogni R., Soares C. M., Martins L. O. 2010. The multicopper oxidase from the archaeon Pyrobaculum aerophilum shows nitrous oxide reductase activity. FEBS J. 277, 3176–3189 10.1111/j.1742-4658.2010.07725.x (doi:10.1111/j.1742-4658.2010.07725.x) [DOI] [PubMed] [Google Scholar]
- 10.Zumft W. G., Matsubara T. 1982. A novel kind of multi-copper protein as terminal oxidoreductase of nitrous oxide respiration in Pseudomonas perfectomarinus. FEBS Lett. 148, 107–112 10.1016/0014-5793(82)81253-2 (doi:10.1016/0014-5793(82)81253-2) [DOI] [Google Scholar]
- 11.Brown K., Tegoni M., Prudencio M., Pereira A. S., Besson S., Moura J. J. G., Moura I., Cambillau C. 2000. A novel type of catalytic copper cluster in nitrous oxide reductase. Nat. Struct. Biol. 7, 191–195 10.1038/73288 (doi:10.1038/73288) [DOI] [PubMed] [Google Scholar]
- 12.Kroneck P. M., Antholine W. A., Riester J., Zumft W. G. 1989. The nature of the cupric site in nitrous oxide reductase and of CuA in cytochrome c oxidase. FEBS Lett. 248, 212–213 10.1016/0014-5793(89)80464-8 (doi:10.1016/0014-5793(89)80464-8) [DOI] [PubMed] [Google Scholar]
- 13.Sproer C., Lang E., Hobeck P., Burghardt J., Stackebrandt E., Tindall B. J. 1998. Note: transfer of Pseudomonas nautica to Marinobacter hydrocarbonoclasticus. Int. J. Syst. Bacteriol. 48, 1445–1448 10.1099/00207713-48-4-1445 (doi:10.1099/00207713-48-4-1445) [DOI] [Google Scholar]
- 14.Dell'acqua S., Pauleta S., Moura I., Moura J. J. G. 2011. The tetranuclear copper active site of nitrous oxide reductase: the CuZ center. J. Biol. Inorg. Chem. 16, 183–194 10.1007/s00775-011-0753-3 (doi:10.1007/s00775-011-0753-3) [DOI] [PubMed] [Google Scholar]
- 15.Rasmussen T., Berks B. C., Butt J. N., Thomson A. J. 2002. Multiple forms of the catalytic centre, CuZ, in the enzyme nitrous oxide reductase from Paracoccus pantotrophus. Biochem. J. 364, 807–815 10.1042/BJ20020055 (doi:10.1042/BJ20020055) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Farrar J. A., Zumft W. G., Thomson A. J. 1998. CuA and CuZ are variants of the electron transfer center in nitrous oxide reductase. Proc. Natl Acad. Sci USA 95, 9891–9896 10.1073/pnas.95.17.9891 (doi:10.1073/pnas.95.17.9891) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prudencio M., et al. 2000. Purification, characterization, and preliminary crystallographic study of copper-containing nitrous oxide reductase from Pseudomonas nautica 617. Biochemistry 39, 3899–3907 10.1021/bi9926328 (doi:10.1021/bi9926328) [DOI] [PubMed] [Google Scholar]
- 18.Ghosh S., Gorelsky S. I., Chen P., Cabrito I., Moura J. J. G., Moura I., Solomon E. I. 2003. Activation of N2O reduction by the fully reduced micro4-sulfide bridged tetranuclear CuZ cluster in nitrous oxide reductase. J. Am. Chem. Soc. 125, 15 708–15 709 [DOI] [PubMed] [Google Scholar]
- 19.Chan J. M., Bollinger J. A., Grewell C. L., Dooley D. M. 2004. Reductively activated nitrous oxide reductase reacts directly with substrate. J. Am. Chem. Soc. 126, 3030–3031 10.1021/ja0398868 (doi:10.1021/ja0398868) [DOI] [PubMed] [Google Scholar]
- 20.Dell'acqua S., Pauleta S. R., Paes de Sousa P. M., Monzani E., Casella L., Moura J. J. G., Moura I. 2010. A new CuZ active form in the catalytic reduction of N2O by nitrous oxide reductase from Pseudomonas nautica. J. Biol. Inorg. Chem. 15, 967–976 10.1007/s00775-010-0658-6 (doi:10.1007/s00775-010-0658-6) [DOI] [PubMed] [Google Scholar]
- 21.Coyle C. L., Zumft W. G., Kroneck P. M., Korner H., Jakob W. 1985. Nitrous oxide reductase from denitrifying Pseudomonas perfectomarina. Purification and properties of a novel multicopper enzyme. Eur. J. Biochem. 153, 459–467 10.1111/j.1432-1033.1985.tb09324.x (doi:10.1111/j.1432-1033.1985.tb09324.x) [DOI] [PubMed] [Google Scholar]
- 22.Snyder S. W., Hollocher T. C. 1987. Purification and some characteristics of nitrous oxide reductase from Paracoccus denitrificans. J. Biol. Chem. 262, 6515–6525 [PubMed] [Google Scholar]
- 23.Ferretti S., Grossmann J. G., Hasnain S. S., Eady R. R., Smith B. E. 1999. Biochemical characterization and solution structure of nitrous oxide reductase from Alcaligenes xylosoxidans (NCIMB 11015). Eur. J. Biochem. 259, 651–659 10.1046/j.1432-1327.1999.00082.x (doi:10.1046/j.1432-1327.1999.00082.x) [DOI] [PubMed] [Google Scholar]
- 24.Riester J., Zumft W. G., Kroneck P. M. 1989. Nitrous oxide reductase from Pseudomonas stutzeri. Redox properties and spectroscopic characterization of different forms of the multicopper enzyme. Eur. J. Biochem. 178, 751–762 10.1111/j.1432-1033.1989.tb14506.x (doi:10.1111/j.1432-1033.1989.tb14506.x) [DOI] [PubMed] [Google Scholar]
- 25.Alvarez M. L., Ai J., Zumft W., Sanders-Loehr J., Dooley D. M. 2001. Characterization of the copper-sulfur chromophores in nitrous oxide reductase by resonance Raman spectroscopy: evidence for sulfur coordination in the catalytic cluster. J. Am. Chem. Soc. 123, 576–587 10.1021/ja994322i (doi:10.1021/ja994322i) [DOI] [PubMed] [Google Scholar]
- 26.Farrar J. A., Thomson A. J., Cheesman M. R., Dooley D. M., Zumft W. G. 1991. A model of the copper centres of nitrous oxide reductase (Pseudomonas stutzeri). Evidence from optical, EPR and MCD spectroscopy. FEBS Lett. 294, 11–15 10.1016/0014-5793(91)81331-2 (doi:10.1016/0014-5793(91)81331-2) [DOI] [PubMed] [Google Scholar]
- 27.Alves T., et al. 1999. A cytochrome c peroxidase from Pseudomonas nautica 617 active at high ionic strength: expression, purification and characterization. Biochim. Biophys. Acta 1434, 248–259 10.1016/S0167-4838(99)00188-0 (doi:10.1016/S0167-4838(99)00188-0) [DOI] [PubMed] [Google Scholar]
- 28.Starkey R. L. 1938. A study of spore formation and other morphological characteristics of Vibrio desulphuricans. Arch. Mikrobiol. 8, 268–304 10.1007/BF00407364 (doi:10.1007/BF00407364) [DOI] [Google Scholar]
- 29.Stoscheck C. 1990. Quantitation of protein. Methods Enzymol. 182, 50–68 10.1016/0076-6879(90)82008-P (doi:10.1016/0076-6879(90)82008-P) [DOI] [PubMed] [Google Scholar]
- 30.Felsenfeld G. 1960. The determination of cuprous ion in copper proteins. Arch. Biochem. Biophys. 87, 247–251 10.1016/0003-9861(60)90168-5 (doi:10.1016/0003-9861(60)90168-5) [DOI] [PubMed] [Google Scholar]
- 31.Dell'acqua S., Pauleta S. R., Monzani E., Pereira A. S., Casella L., Moura J. J. G., Moura I. 2008. Electron transfer complex between nitrous oxide reductase and cytochrome c552 from Pseudomonas nautica: kinetic, nuclear magnetic resonance, and docking studies. Biochemistry 47, 10 852–10 862 10.1021/bi801375q (doi:10.1021/bi801375q) [DOI] [PubMed] [Google Scholar]
- 32.Baker S. C., Ferguson S. J., Ludwig B., Page M. D., Richter O.-M. H., van Spanning R. J. M. 1998. Molecular genetics of the genus Paracoccus: metabolically versatile bacteria with bioenergetic flexibility. Microbiol. Mol. Biol. Rev. 62, 1046–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lalucat J., Bennasar A., Bosch R., Garcia-Valdes E., Palleroni N. J. 2006. Biology of Pseudomonas stutzeri. Microbiol. Mol. Biol. Rev. 70, 510–547 10.1128/mmbr.00047-05 (doi:10.1128/mmbr.00047-05) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen P., Gorelsky S. I., Ghosh S., Solomon E. I. 2004. N2O reduction by the mu4-sulfide-bridged tetranuclear CuZ cluster active site. Angew. Chem. Int. Ed. Engl. 43, 4132–4140 10.1002/anie.200301734 (doi:10.1002/anie.200301734) [DOI] [PubMed] [Google Scholar]
- 35.Oganesyan V. S., Rasmussen T., Fairhurst S., Thomson A. J. 2004. Characterisation of [Cu4S], the catalytic site in nitrous oxide reductase, by EPR spectroscopy. Dalton Trans. 7, 996–1002 10.1039/b313913a (doi:10.1039/b313913a) [DOI] [PubMed] [Google Scholar]
- 36.Wunsch P., Körner H., Neese F., van Spanning R. J., Kroneck P. M., Zumft W. G. 2005. NosX function connects to nitrous oxide (N2O) reduction by affecting the Cu(Z) center of NosZ and its activity in vivo. FEBS Lett. 579, 4605–4609 10.1016/j.febslet.2005.07.023 (doi:10.1016/j.febslet.2005.07.023) [DOI] [PubMed] [Google Scholar]
- 37.Gorelsky S. I., Ghosh S., Solomon E. I. 2006. Mechanism of N2O reduction by the mu4-S tetranuclear CuZ cluster of nitrous oxide reductase. J. Am. Chem. Soc. 128, 278–290 10.1021/ja055856o (doi:10.1021/ja055856o) [DOI] [PubMed] [Google Scholar]
- 38.Pomowski A., Zumft W., Kroneck P., Einsle O. 2011. N(2)O binding at a [4Cu:2S] copper–sulphur cluster in nitrous oxide reductase. Nature 477, 234–237 10.1038/nature10332 (doi:10.1038/nature10332) [DOI] [PubMed] [Google Scholar]