Abstract
Bacterial denitrification results in the loss of fertilizer nitrogen and greenhouse gas emissions as nitrous oxides, but ecological factors in soil influencing denitrifier communities are not well understood, impeding the potential for mitigation by land management. Communities vary in the relative abundance of the alternative dissimilatory nitrite reductase genes nirK and nirS, and the nitrous oxide reductase gene nosZ; however, the significance for nitrous oxide emissions is unclear. We assessed the influence of different long-term fertilization and cultivation treatments in a 160-year-old field experiment, comparing the potential for denitrification by soil samples with the size and diversity of their denitrifier communities. Denitrification potential was much higher in soil from an area left to develop from arable into woodland than from a farmyard manure-fertilized arable treatment, which in turn was significantly higher than inorganic nitrogen-fertilized and unfertilized arable plots. This correlated with abundance of nirK but not nirS, the least abundant of the genes tested in all soils, showing an inverse relationship with nirK. Most genetic variation was seen in nirK, where sequences resolved into separate groups according to soil treatment. We conclude that bacteria containing nirK are most probably responsible for the increased denitrification potential associated with nitrogen and organic carbon in this soil.
Keywords: denitrifier diversity, denitrifier abundance, nitrous oxide flux, long-term field experiment, soil properties, nitrogen fertilizer
1. Introduction
Bacterial denitrification, the process by which nitrate is reduced by respiration to nitrite, nitric oxide, nitrous oxide and nitrogen gas, is a major source of nitrogen losses in fertilized soils. It results in the emission of nitrous oxide (N2O), an important greenhouse gas ca 300 times more radiatively effective than carbon dioxide. The ability to denitrify may be an advantage because nitrate acts as a substitute terminal electron acceptor in place of oxygen and facilitates respiration in anoxic conditions. This is especially relevant where organic carbon substrates (which act as electron donors) are abundant, nitrogen is replete and oxygen is limited. In general, activity increases with soil temperatures and water content, thus waterlogged agricultural soils containing crop and nitrogen (N) fertilizer residues provide conditions highly conducive to denitrification. Factors affecting denitrifying prokaryotes in agricultural soils have been reviewed extensively [1].
Many groups of heterotrophic soil bacteria are known to contain genes required for denitrification, although one-third of sequenced denitrifier genomes lack the gene for nitrous oxide reductase (nosZ) responsible for the final step that results in the release of N2 rather than N2O [2], and the relative abundance of the gene is reported to be a predictor of the ratio of the two gases emitted [3]. Indeed, denitrifiers are more numerous than any other functional groups involved in the N cycle, reported to comprise up to 5 per cent of all soil bacteria [1]. The two alternative genes for nitrite reductase, nirK and nirS, may be present in different but closely-related bacterial species, although they appear incompatible in the same individual [4]. Attempts to identify the environmental factors influencing the prevalence of nirK and nirS in different soil types and ecosystems have shown little consensus in the past, but recent reports indicate that they are linked to soil properties [5]. There are also clear differences between marine and terrestrial communities [6]. Increasing N fertilizer applications have been reported to favour nirK abundance in agricultural soils [1], also nirK diversity increased with soil N in reclaimed wetlands [6,7]. The cytochrome cd1 version of nitrite reductase encoded by nirS is reported to be more common than the Cu-dependent variant encoded by nirK in cultured isolates of environmental bacteria [8], but the preponderance of bacterial species that are as yet uncultured in soil makes the true relative abundance uncertain. Genes for dissimilatory denitrification are found in some eukaryotes and archaea, although their contribution to nitrous oxide emissions in soil is uncertain; furthermore, archaea are outnumbered by bacteria, comprising no more than 1 per cent of prokaryotic cells in soil. The genes are also present in chemoautotrophic nitrifying bacteria and archaea which obtain energy from the oxidation of ammonia to nitrite [2], and may be an essential defence mechanism against accumulation of nitrite [9].
While there have been many studies on the relationship between soil treatments and N2O emissions, and the presence and abundance of genes involved in the denitrification process [1], only a few recent reports combine these aspects [3,5,7]. In this work, we investigated both in an area of arable land at Rothamsted Research, the Broadbalk Wheat Experiment, initiated in 1843 to investigate the long-term consequences of different fertilizer and manure applications, and also the withdrawal of cultivation from part of the site in 1882, leading to the development of a small area of woodland known as the ‘Broadbalk Wilderness’ [10]. We compared the abundance and diversity of nirK, nirS, nosZ and denitrification potentials of the soils, to test the hypotheses that levels of N and organic matter in soil arising from different fertilizer applications will impact the structure of denitrifier communities, increasing overall abundance and activity, and furthermore that the change from arable management to woodland will have a more extreme effect on these communities.
2. Material and methods
(a). Soil sampling
The soil at the Broadbalk sites is a flinty silty clay loam of the Batcombe series (Chromic Luvisol—FAO), slightly alkaline, with clay content of 19–39% and soil organic carbon values of 0.7–3.2%, depending on the different cropping history, mineral and organic fertilizer treatments [11]. The arable plots, which received no N fertilizer (N0), 144 or 288 kg N ha−1 as ammonium nitrate in spring (N144 and N288), or farmyard manure (FYM) at 35 t ha−1 (containing ca 230 kg N ha−1) in early autumn prior to ploughing and planting, were from section 1, which was created in 1926 although these plots have been under continuous wheat cultivation and the same treatments since 1843. The woodland plot taken out of cultivation in 1882 is now maturing towards a mixed deciduous woodland covering ca 0.1 ha. Each arable plot is ca 6 m (N–S) × 30 m (E–W), the woodland is larger: 80 m (N–S) × 15 m (E–W), but because the experiment was set up prior to the introduction of statistics in the design of field experiments, the treatments are not replicated. To account for this, on 21 January 2005, 16 cores (6 cm diameter and 10 cm deep) were taken as eight paired samples at intervals along a W-shaped E–W transect (N–S for the wilderness): these are considered treatment replicates for denitrification potential assays. From the perimeter immediately adjacent to each of these cores, three small cores (1 cm diameter and 10 cm deep) were collected at equidistant points. These three small cores were pooled and sieved to give 16 samples from each treatment. From three of these (representing the E, central and W sections of the plot), a portion was removed to estimate oven-dried weight and the remainder frozen at −80°C for subsequent DNA extraction and quantitative real-time PCR (qPCR) analyses. The remaining samples were pooled to create a composite sample from each treatment for mineral analyses.
(b). Nitrous oxide flux measurements
Each large core was weighed then pre-incubated for 3 days at 15°C in adapted 1 l Kilner glass jars. At this point, 15 ml 0.153 M KNO3 solution was added to eight cores (N+) from each subplot to give the equivalent rate of 100 kg N ha−1; 15 ml sterile distilled water (SDW) was added to the remaining eight cores (N−). Jars were sealed and 20 ml gas samples removed after 13 h and then at 24 h intervals. When N2O flux from the large cores was predicted to be at a maximum based on past experience [12], 4 days after adding KNO3, three large cores (corresponding to the small cores described above) were removed from each treatment (i.e. N+ or N−), broken up and sieved for chemical analyses, with a portion frozen for DNA extraction, PCR, denaturing gradient gel electrophoresis (DGGE) and sequence analyses (the remaining cores for each treatment provided further N2O flux measurements that confirmed that the peak had been reached). The N2O content of the gas samples was measured using an Ai93 gas chromatograph with an electron capture detector; the mineral N content of the soil samples was measured by extracting 62.5 g soil in 200 ml 2 M KCl for 2 h and analysing the extract for ammonium- and nitrate-N using a Skalar SANPLUS System.
(c). DNA amplification for DGGE
DNA was extracted from 250 mg soil samples from each of the three paired replicate soil cores per treatment (with or without added N) at estimated peak N2O emission, using the PowerSoil DNA isolation kit (MO BIO Laboratories, Inc.) including bead beating at 5.5 m s−1 for 30 s (FastPrep Instrument). From each of these DNA extracts, PCR was performed in duplicate (giving six DGGE profiles per treatment) to provide analytical replication, using specific primers (table 1 in the electronic supplementary material): sections of the genes for 16S rRNA, nirK, nirS and nosZ were amplified and resolved using DGGE (details in the electronic supplementary material) and their relative intensities estimated [14]. Gel profiles were analysed using Phoretix software (nonlinear dynamics); the statistical package MVSP v. 3.13d (Kovach Computing Services) provided principal component analysis (PCA) and Shannon diversity indices (H′) of these data.
(d). DNA sequence analysis
Sequencing effort concentrated on nirK PCR products because DGGE indicated many variants of the gene that differed between treatments, in contrast to nirS and nosZ. Bands that appeared to represent either a distinct sequence variant of nirK present in only one treatment, or a common type present in all treatments, based on their relative migration on the DGGE gels, were excised, re-amplified, cloned using StrataClone PCR cloning kit (Agilent Technologies UK Ltd) and sequenced by ABI Prism BigDye Terminator Cycle Sequencing (Applied Biosystems). To validate the methods, a small number of nirS and nosZ PCR products were also sequenced.
DNA sequence information from the nirK products (submitted to GenBank as accessions HQ873874– HQ873925), together with nirK sequences from culture-independent environmental studies and cultured bacteria in the National Centre for Biotechnology Information (NCBI) database (http://www.ncbi.nlm.nih.gov/), chosen for closest sequence similarity using the NCBI BLAST tool, were used to construct a phylogenetic tree. The corresponding sequence from a Neisseria gonorrhoeae surface protein gene with nitrite reductase activity was used as an out-group. Sequences were aligned using MUSCLE [15], and phylogenetic analysis was performed using the maximum-likelihood program DNAML (http://evolution.gs.washington.edu/phylip.html [16]) with 1000 bootstraps and a translation/transversion parameter of 1.0 as determined using PUZZLE [17] and the resulting tree was viewed in TreeView.
(e). Quantitative real-time PCR
DNA samples were extracted from each of the three replicate soil cores pre-incubation, and each PCR reaction was performed in triplicate, as described (above and table 1 in the electronic supplementary material). To generate the standard curves for estimating gene copy number in qPCR, serial dilutions were made of an aliquot of amplified product generated from DNA extracted from the soil with the relevant primer set, the gel was purified and quantified using PicoGreen dsDNA Quantification kit (Molecular Probes). This was to minimize the bias inherent in the usual approach, where one or only a few genes are used to standardize complex communities [18]. Problems arise because the degenerate primers have only partial homology to the gene variants used to generate the standard curve, and there are differences in the amplification efficiencies of the range of sequences internal to the primers in the environmentally extracted DNA samples. The qPCR assays were run on an Applied Biosystems 7900HT Fast Real-Time PCR System in a 384 well format with 10 µl final volume per well. The reaction consisted of 10 ng template DNA (quantified by PicoGreen fluorescence), 1X QuantiTect SYBR Green Master Mix (Qiagen) and primer concentrations (see table 1 in the electronic supplementary material). All DNA preparations were checked for the absence of inhibitors prior to PCR and all results were analysed using LinRegPCR program v. 11.1 [19,20] to confirm the efficiency of amplification and the absence of inhibition.
(f). Statistical analysis
The significance of differences in abundance for each gene was assessed in Excel using Anova, and comparisons between individual treatments made using the t-test, as appropriate.
To assess the significance of correlations in the data, a non-parametric analysis in Genstat v. 13 (VSN International Ltd., Hemel Hempstead, UK) was undertaken, generating a Spearman's rank correlation matrix.
3. Results
(a). Soil properties and nitrogen fluxes
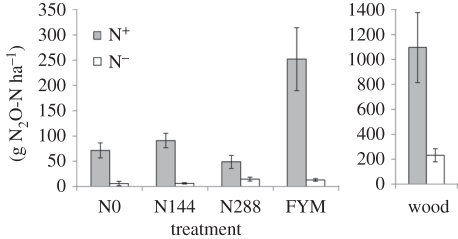
The soil pH values were similar across all sites and slightly alkaline, as expected for a site overlying chalk: the N0 treatment had the highest pH and contained significantly less mineral N (p < 0.005) than the fertilized treatments, which is unsurprising as it has received no N fertilizer for more than 160 years (table 1). However it and the other Broadbalk arable treatments receive 35–45 kg N ha−1 yr−1, and the woodland possibly twice as much, via atmospheric deposition [12]. The mineral N in the N288 and FYM treatments was similar, significantly more than in the N144 treatment, but much less than in the woodland soil (p < 0.005). Soil organic C increased with soil N in the N0, N144 and N288 treatments, but there was proportionally more in the FYM and woodland treatments, so that the C : N ratio was higher (ca 12 : 1) in the woodland soil and slightly higher in the FYM treatment (ca 11 : 1) than in the other three (ca 10 : 1); table 1. Soil moisture at sampling showed a similar trend to organic C and was inversely related to bulk density. Nitrous oxide emissions from cores amended with KNO3 (N+) were significantly elevated in the FYM and woodland soils (5- and 20-fold, respectively; p < 0.005) compared with those in the N0, N144 or N288 treatments, which were not significantly different (p > 0.1). In the unamended soils (N−), the control and N144 had similar very small emissions, significantly less (p < 0.005) than the N288 and FYM soils, which exceeded them fourfold, and the woodland soil which exceeded them 10-fold (figure 1).
Table 1.
Broadbalk soil properties: N144 and N288 plots receive N as ammonium nitrate, FYM receives farmyard manure; arable plots are under continuous wheat cultivation (section 1).
| plot | fertilizer |
moisture: stones removed (% w/w ± s.e.)a | od soil bulk density (g cm−3)b | total N: NO3 + NH4 0–23 cm (kg N ha−1 ± s.e.)a | soil pH in H2Ob | N (%)b | organic C (%)b | C : N | |
|---|---|---|---|---|---|---|---|---|---|
| N | other | ||||||||
| N0 (plot 05) | nil | PKMg | 25 (0.3) | 1.2 | 3.7 (0.2) | 8.10 | 0.094 | 0.90 | 9.57 |
| N144 (plot 08) | 144 kg ha−1 (April) | PKMg | 27 (1.3) | 1.2 | 6.2 (0.5) | 7.30 | 0.112 | 1.13 | 10.09 |
| N288 (plot 16) | 288 kg ha−1 (April) | PKMg | 25 (0.2) | 1.2 | 8.3 (0.5) | 7.80 | 0.122 | 1.20 | 9.84 |
| FYM (plot 2.2) | 35 t ha−1 (Aug–Sept) | nil | 32 (0.6) | 1.1 | 8.1 (0.3) | 7.82 | 0.266 | 2.83 | 10.64 |
| woodland (wilderness) | nil | nil | 41 (0.6) | 0.9 | 25.0 (0.1) | 7.70 | 0.29 | 3.45 | 11.73 |
aod, oven dried. Measured at January sampling.
bData from [13] and P. Poulton (2010, personal communication).
Figure 1.
Cumulative N2O flux from soil cores for the first 4 days. Error bars indicate s.e. of mean; note different scale on the vertical axis for woodland soil (wood); N+ denotes KNO3 added to cores compared with basal rate from N− control cores without KNO3 addition.
(b). Gene abundance
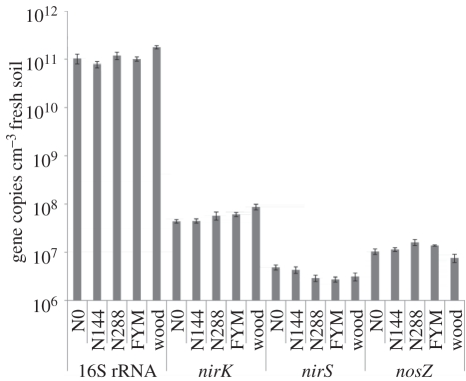
Microbial abundance is often reported as cells or genes per gram dried soil, but because we wanted to link microbial parameters to the N2O flux measured in undisturbed cores of field soil with data expressed as g ha−1, our abundance data have been calculated as gene copies cm−3 soil rather than g−1 soil (figure 2) to facilitate comparison of the five soil treatments with different bulk densities (table 1). Nevertheless, results calculated as cm−3 and g−1 soil were not significantly different (p < 0.001). The number of 16S rRNA genes was similar in all the arable soils (8–12 × 1010 copies cm−3), although there were significantly more in the woodland soil (18 × 1010 copies cm−3) compared with the others (p < 0.005). There was more variation within and between different denitrification genes with nirK more abundant and nirS least abundant in all plots. For nirS, the plots with least N (N0 and N144) had the most gene copies (4–5 × 106 cm−3), compared with 2–3 × 106 cm−3 copies for the other plots, the differences being significant (p < 0.005). Differences were also significant (p < 0.05) for nirK genes: the mean copy number increased with increasing N so that the N0 plot had fewest copies (4 × 107 cm−3) and the woodland plot the most (9 × 107 cm−3). Differences in the abundance of the N2O reductase gene nosZ were not statistically significant overall, although woodland soil with fewest gene copies (7 × 106 cm−3) was significantly different (p < 0.01) from the N288 treatment with most (16 × 106 cm−3; figure 2). The ratio of nosZ to the nitrite reductase genes (nirK and nirS) indicated a similar numerical relationship in all treatments (1 : 4) except woodland where it was threefold lower (1 : 12).
Figure 2.
Abundance of denitrification and 16S rRNA genes in different plots estimated by qPCR; error bars indicate s.e.
Further analysis (table 2) indicated that the total abundance of the 16S rRNA genes increased with soil organic C and N (p < 0.1) and confirmed that N2O emissions were significantly correlated with these parameters whether or not KNO3 had been added (p < 0.05; <0.001, respectively). The correlation between nirK gene abundance, soil N and C and N2O emissions was also highly significant (p < 0.001), but nosZ was not apparently correlated with these environmental variables. The abundance of nirK and 16S rRNA genes was significantly correlated (p < 0.05); nirS and nirK showed a significant negative correlation (p < 0.05), as did nirS and nosZ (p < 0.1). This may indicate different ecological drivers for the abundance of organisms containing these genes.
Table 2.
Spearman's rank correlation matrix of soil properties. Soil pH, bulk density, total N (NO3 + NH4), % N, % organic C and C : N ratios from table 1; N2O emission rates in soil cores with (N+) and without (N−) added KNO3 shown in figure 1; nirK, nirS, nosZ gene abundance from qPCR as in figure 2.
| 16S rRNA | 1 | 1.00 | |||||||||||
| % N | 2 | 0.50* | 1.00 | ||||||||||
| % organic C | 3 | 0.50* | 1.00*** | 1.00 | |||||||||
| C : N | 4 | 0.20 | 0.90*** | 0.90*** | 1.00 | ||||||||
| N2O (N−) | 5 | 0.70** | 0.90*** | 0.90*** | 0.70** | 1.00 | |||||||
| N2O (N+) | 6 | 0.10 | 0.70** | 0.70** | 0.90*** | 0.40 | 1.00 | ||||||
| total N | 7 | 0.70** | 0.90*** | 0.90*** | 0.70** | 1.00*** | 0.40 | 1.00 | |||||
| bulk density | 8 | −0.45 | −0.89*** | −0.89*** | −0.89*** | −0.67* | −0.89*** | −0.67* | 1.00 | ||||
| nirK | 9 | 0.50* | 1.00*** | 1.00*** | 0.90*** | 0.90*** | 0.70** | 0.90*** | −0.89*** | 1.00 | |||
| nirS | 10 | −0.10 | −0.70** | −0.70** | −0.50* | −0.60* | −0.20 | −0.60* | 0.45 | −0.70** | 1.00 | ||
| nosZ | 11 | −0.30 | −0.10 | −0.10 | −0.30 | 0.00 | −0.60* | 0.00 | 0.45 | −0.10 | −0.60* | 1.00 | |
| pH | 12 | 0.10 | −0.30 | −0.30 | −0.50* | −0.40 | −0.30 | −0.40 | 0.11 | −0.30 | 0.00 | 0.10 | 1.00 |
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 |
*p<0.1; **p<0.05; ***p<0.01.
(c). Gene diversity
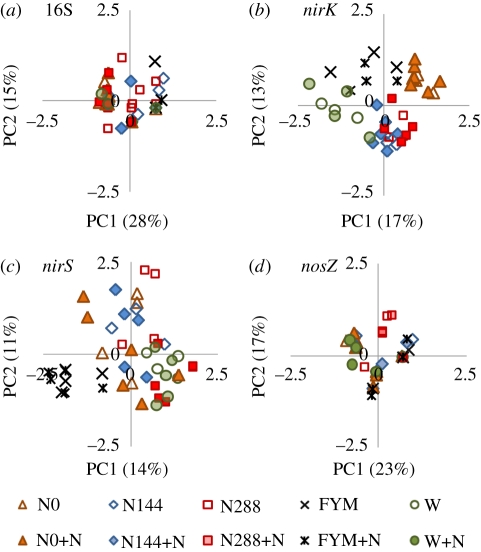
The gene abundance results indicated that the microbial community structure varied with the different treatments. However, only a few obvious differences in DGGE patterns were seen, whether the DNA had been extracted from cores prior to setting up the N2O emission assays or at the peak time of emission. PCA comparing 16S rRNA gene diversity at this time (after 4 days incubation) did not resolve the treatments (figure 3a), confirmed by similar Shannon diversity indices (H′) and numbers of DGGE bands (indicating different phyla): the H′ for different soils ranged from 3.7 to 3.8 and the number of DGGE bands from 62 to 66. The nirK genes grouped according to treatment for the N0, FYM and woodland soils, and separately from the two mineral N fertilized treatments, which grouped together (figure 3b). The nirS genes grouped separately only for the FYM treatment (figure 3c) and there were no clear differences in the distribution of the nosZ gene with different treatments (figure 3d). The differences between treatments accounted for by the first and second principal components were 43, 30, 25 and 40 per cent in figure 3a–d, respectively. The number of denitrification gene groups and the associated H′ were similar for all plots, H′ ranging from 2.9 to 3.1 for nirK and nirS and 1.4 to 1.5 for nosZ; DGGE bands in the replicates for each plot ranged from 18 to 23 for nirK and nirS (mean value 20) with six for nosZ.
Figure 3.
Principal component analysis plots of 16S rRNA and denitrification genes from Broadbalk treatments. W, woodland.
(d). Sequence analysis
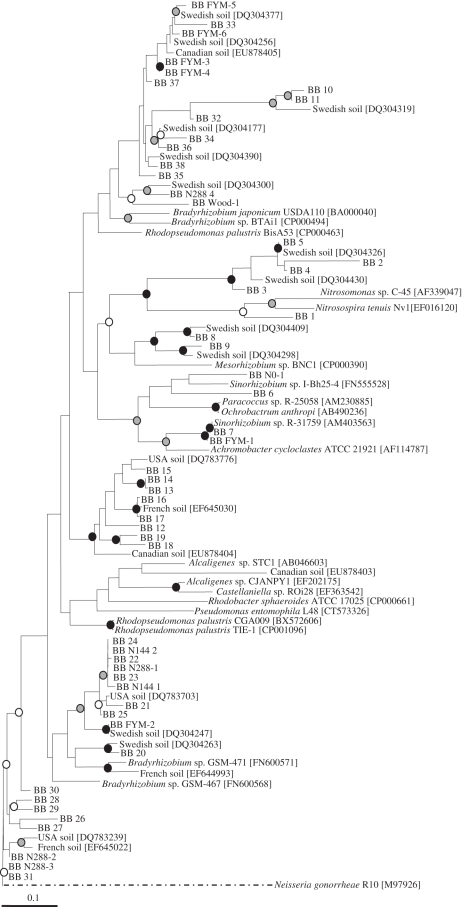
To further investigate the diversity of denitrifying bacteria in the different soils, bands from the nirK DGGE gels (where treatments were differentiated to the greatest extent by PCA) were excised, cloned and sequenced. They were chosen as either unique to one treatment or alternatively common across all treatments. Sequence analysis using BLAST indicated that the closest similarity of the majority was to a variety of ‘unknown’ nirK sequences identified by PCR from agricultural soils. However, phylogenetic analysis demonstrated that the sequences had affinity to a range of well-known denitrifying bacteria, the majority being α or β proteobacteria including Rhodopseudomonas, Bradyrhizobium and Paracoccus, and the nitrifier Nitrosomonas (figure 4). The phylogenetic tree gave no clear indication of sequences grouping according to either their origin or ubiquity, although there was a group of sequences unique to FYM-amended soil that clustered at the top of the tree, and a cluster of sequences unique to nitrate-fertilized soil at the bottom. However, the closest cultured bacteria to both groups were different isolates of Bradyrhizobium spp.
Figure 4.
Phylogenetic tree of nirK sequences. BB denotes Broadbalk sequences from DGGE bands either common to all plots (BB 1– BB 38) or unique to one treatment (BB FYM-1, etc.). Information on environmental sequences (Swedish soil, etc.) or cultured bacterial genera and species names preceding accession numbers were obtained from NCBI. Bootstrap values: open circles, 70–80%; grey circles, 80–90%; black circles, 90–100%.
Eight different DGGE bands of nirS products from the nitrate-fertilized plots were excised and identified by their closet similarity: four were very similar to the α-proteobacterium Paracoccus denitrificans PD1222 (CP000489); one was most similar to the β-proteobacterium Acidovorax ebreus TPSY (CP001392); one resembled an uncultured isolate from Swedish arable soil (AY583426) and two were similar to an uncultured isolate from Chinese agricultural soil (GQ397301). A single nosZ product identified from DGGE was also most similar to nosZ from P. denitrificans PD1222.
4. Discussion
The observation of increased N2O emissions in soils containing large amounts of organic carbon (FYM and woodland) compared with those unfertilized or fertilized with mineral N has been reported previously [12]. Although overall in this study N2O fluxes were strongly correlated with soil N, with similar N content and basal (N−) emissions for FYM and N288, the flux for the FYM plot with added KNO3 was fivefold greater than that of the N288 plot (table 2). This may reflect the different soil structure (FYM soil was less dense than N288 but retained more moisture), related to its greater organic C content. Similarly, the woodland soil was less dense and moister than the FYM soil, contained more organic C, and its basal (N−) and N-fertilized (N+) N2O fluxes were ca 20- and fivefold higher than N288, respectively. Analysis of all factors showed that bulk density as well as soil C and N was significantly correlated with N2O flux, whether or not KNO3 had been added (table 2).
The 16S rRNA gene is multicopy. Using information on the relative abundance of different bacterial phyla in soils and information of rRNA gene copy number from the rrndb website [21], we estimate the mean copy number in soil communities as four: this is similar to a previous estimate of an upper limit of approximately three for environmental bacteria from all sources [22]. Together with the differential amplification efficiencies arising from different primer sets and nucleotide sequences internal to the primer-binding sites, qPCR can provide only a rough estimate of the relative abundance of different genes, although these concerns do not apply when comparing the abundance of one gene in multiple samples from similar environments. The total bacterial community estimated from 16S rRNA gene abundance in the woodland soil was twofold greater than in the other plots. Woodland soils have been reported to maintain larger microbial communities than those in arable agriculture, and the soil microbial biomass in the Broadbalk woodland soil has been reported to be approximately twice that of the arable plots [23], supporting the qPCR estimate. The Cu-dependent nitrite reductase nirK increased with soil N, with approximately twice the number of copies in the woodland compared with the N0 plot. However, in all plots, c nirK comprised around 0.05 per cent of the estimated number of 16S rRNA genes, equivalent to 0.2 per cent of bacterial cells. Soil Cu has been reported to have an impact on the abundance of bacteria-carrying nirK [24] but this does not explain the Broadbalk results, in which only the FYM treatment, with 37 mg Cu kg−1 soil, had an elevated Cu content compared with 26–28 mg Cu kg−1 soil for all the other treatments including woodland [25,26]. Rather, nirK appears to be present in a relatively constant proportion of all bacteria present in each of the Rothamsted soil treatments. In contrast, the gene for the cytochrome cd1 nitrite reductase nirS was less abundant overall than nirK and showed a different pattern of distribution and an inverse correlation: there were threefold more nirS copies in the N0 treatment compared with the woodland soil indicating that bacteria carrying nirS may be less well adapted to soils with high N than those carrying nirK. Subject to the proviso discussed earlier, the N2O reductase gene nosZ (less abundant than nirK but more than nirS in all soils) was present in 0.04–0.06% of bacteria in the arable soil and less than 0.02 per cent in the woodland soil, thus is likely to be carried by 22–29% of denitrifying bacteria containing nirK in the arable soil but less than 10 per cent in the woodland soil. This is important as nosZ ameliorates one of the most environmentally harmful consequences of denitrification by reducing N2O to N2: the relatively low proportion of denitrifying bacteria in the woodland soil containing nosZ could partly explain the larger N2O flux compared with the arable soils and is consistent with a recent study where soil communities were manipulated to increase the relative abundance of denitrifiers lacking nosZ [27]. The ammonia-oxidizing bacteria are known to have nitrite reductase genes and to generate N2O, either by denitrification or as a consequence of nitrification, but have not been shown to produce N2. However, our data (I.M.C. & P.R.H. 2008, unpublished) indicate that bacterial nitrifier communities in the woodland plot are relatively low compared with the arable plots. We were not able to measure N2 evolution during this experiment and it would be interesting to investigate whether nosZ activity reduced N2O flux in any of the assays, for example by using K15NO3 and measuring 15N2 emissions. The exploitation of isotopomers (compounds differentially labelled with stable isotopes including 18O and 15N) has made it possible to establish which microbial pathways (nitrification or denitrification) have contributed to nitrous oxide emissions from soils [28]. To obtain a full picture of denitrification, it would be necessary to combine isotopomer approaches with the analysis of mRNA (rather than DNA) to establish which genes are active.
Our results are consistent with other qPCR studies that report the dominance of nirK in forest and rice paddy soils, although different primer sets were used [29,30]. Other studies based on the same primers that we used showed no clear differences in nirS and nirK abundance in different zones of a nutrient-poor glacier foreland [31] and a long-term fertilizer crop trial [5]. This latter experiment involved an acid brown soil (Eutric Cambisol) and the authors concluded that soil pH was one of the major influences, in contrast to our study where soil pH was greater than 7 and similar in all plots. In a farm-scale comparison of integrated and organic management, where soil clay content was around 40 per cent and pH 5.7–7.0, nirK was slightly less abundant than nirS (using the same primers as in our study) and spatial distribution was related to denitrification potential in the field, with soil nitrate and clay implicated as driving factors, although farm management also influenced distribution [24]. This contrasts with our observations of more abundant nirK although we also found a positive correlation between nitrite reductase genes and N2O flux. The explanation for apparently contradictory results may, in some cases, be explained by the use of different primer sets, but a combination of soil properties (soil pH, clay, carbon and nitrogen content) and land use (organic manure or mineral fertilizer applications, tilled or uncultivated soil) appears to exert a major influence on denitrifier communities.
The relative abundance of the various genes, together with their sequence diversity, indicates differences in the structure of the communities of denitrifying bacteria present in each plot. The scale of sampling in this study was insufficient to resolve 16S rRNA gene diversity between treatments, although previously we had demonstrated differences in the community structure of the FYM and N144 plots [14]. Local differences in selective pressure exerted on bacteria in the various soil treatments is likely to influence both the overall genotype (as indicated by the 16S rRNA gene) and individual genes involved in denitrification, since the latter may be subject to horizontal transfer [2].
Comparison of denitrification and 16S rRNA gene sequences indicates that the relationship is complex and that nirK, nirS and nosZ are probably subject to different selective pressures [2,32], which might explain the greater genetic diversity of nirK compared with nirS in the different plots. Despite the apparent grouping of the nirK products by DGGE, sequence analysis showed only slight evidence for any distinct groups. A group of three sequences unique to FYM grouped with other sequences found in all five treatments, other soils and Bradyrhizobium spp. Another group of three sequences unique to mineral N-fertilized soil grouped separately with closest sequence similarity to different Bradyrhizobium spp., along with various soil-derived sequences from this and other studies. Not all sequences resembled Bradyrhizobium—one common sequence was most similar to the nirK from the nitrifying β-proteobacteria Nitrosomonas and Nitrosospira, but the majority were closest to α-proteobacterial groups, which is in agreement with a study of cultured denitrifying bacteria where nirK was most prevalent in α- and nirS in β-proteobacteria [32]. Given the limited sequencing effort, it is not possible to speculate on the distribution of nirS sequences from Broadbalk soils.
Overall, the analysis of the genetic diversity of denitrification genes in the different Broadbalk arable treatments does not provide evidence that distinct dominant populations have developed in response to different long-term treatments. This is in agreement with a previous investigation where there were no great differences in the distribution of 16S rRNA or two genes involved in the N-cycle, amtB (an ammonia transporter) and nifH (nitrogenase) on the N0, FYM and mineral N-fertilized plots [33]. This earlier study found high intra-plot heterogeneity, similar in magnitude to the differences between plots. High heterogeneity within plots masking differences between different treatments may have affected the results presented here. Also, the sampling effort in these studies was relatively small; future metagenomic sequence analysis may provide evidence of differences in the microbial communities resulting from the different treatments.
The woodland plot at Broadbalk is relatively small and subject to the large N deposition enhancement of the edges of woodland; the high N levels may not be typical of pristine natural woodlands [12] but provide an extreme value for comparison with the arable plots. The microbial community is likely to be derived from the same original population as the arable soils and the significant differences in the relative abundance of denitrification genes may provide some insight into the selective pressures at work, with bacteria-carrying nirK genes becoming more abundant in response to increased N. In contrast, bacteria-carrying nirS may be at an advantage in the low N soils.
Fungi are reported to contribute significantly to N2O emissions in some ecosystems, but are also consumers [34,35], and as soil N and C increase, more competition is predicted between fungal assimilation of NO3− and bacterial dissimilation via denitrification, aerobic conditions favouring the former. Our N2O flux measurements were carried out under conditions conducive to denitrification, but were in broad agreement with field measurements [12], the main difference being a less marked increase in fluxes from FYM and woodland soil compared with the other treatments, possibly owing in part to increased fungal assimilation of NO3− under field conditions. The ratio of fungi to bacteria in the Broadbalk woodland soil (2.3 : 1) is higher than that in the arable treatments soils [36], but the correlation of fluxes to the abundance of nirK indicates bacterial denitrification was the main source in these experiments. In the future, this could be addressed by measuring isotopomers, which can potentially discriminate between fungal and bacterial denitrification as the source of N2O, owing to a distinct difference in 15N site preference [37].
In conclusion, soil physiochemical properties (bulk density, organic matter, organic C, N and C : N ratio) have an overriding influence on the potential denitrification activity resulting in increased N2O emissions in soils with high organic matter (FYM and woodland). However, for the first time, we show clear evidence for a relationship between nirK gene abundance, soil C and N and denitrification fluxes, and an inverse correlation between these factors and nirS. We also show evidence for a significant change in the community structure, resulting in a threefold decrease in nosZ compared with the nitrite reductase genes in soil that has developed into woodland (albeit in an unusual situation), with implications for denitrification potential and N2O emissions. The findings provide some guidance to inform soil management: maintaining high rates of soil N and organic C over a long period support the development of microbial communities that produce more N2O when conditions are conducive to denitrification, compared with soils containing less N and organic C. Importantly, there are also structural differences in denitrifier communities in soils with high N and C: they possess proportionally fewer copies of the N2O reductase gene nosZ, so may be less able to close the nitrogen cycle by reducing N2O to N2. This provides further impetus for avoiding excessive inputs of N, whether as fertilizer, manure or atmospheric deposition, and of organic C. Nevertheless, this must be balanced with the need for adequate soil organic matter to maintain soil structure and reduce energy use in soil management (tillage), which of themselves reduce greenhouse gas emissions.
Acknowledgements
This research is funded by the Biotechnology and Biological Sciences Research Council (BBSRC). Natalya Buchkina received funding from Rothamsted International. We are indebted to Colin Webster for practical help and advice and to Sue Welham and Rodger White for statistical guidance.
References
- 1.Philippot L., Hallin S., Schloter M. 2007. Ecology of denitrifying prokaryotes in agricultural soil. Adv. Agronomy 96, 249–305 10.1016/S0065-2113(07)96003-4 (doi:10.1016/S0065-2113(07)96003-4) [DOI] [Google Scholar]
- 2.Jones C. M., Stres B., Rosenquist M., Hallin S. 2008. Phylogenetic analysis of nitrite, nitric oxide, and nitrous oxide respiratory enzymes reveal a complex evolutionary history for denitrification. Mol. Biol. Evol. 25, 1955–1966 10.1093/molbev/msn146 (doi:10.1093/molbev/msn146) [DOI] [PubMed] [Google Scholar]
- 3.Philippot L., Cuhel J., Saby N. P. A., Chèneby D., Chronáková A., Bru D., Arrouays D., Martin-Laurent F., Šimek M. 2009. Mapping field-scale spatial patterns of size and activity of the denitrifier community. Environ. Microbiol. 11, 1518–1526 (doi:10.1111/j.1462–2920.2009.01879.x) [DOI] [PubMed] [Google Scholar]
- 4.Zumft W. G. 1997. Cell biology and molecular basis of denitrification. Mol. Biol. Revs. 61, 533–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hallin S., Jones C. J., Schloter M., Philippot L. 2009. Relationship between N-cycling communities and ecosystem functioning in a 50-year-old fertilization experiment. ISME J. 3, 597–605 10.1038/ismej.2008.128 (doi:10.1038/ismej.2008.128) [DOI] [PubMed] [Google Scholar]
- 6.Jones C. M., Hallin S. 2010. Ecological and evolutionary factors underlying global and local assembly of denitrifier communities. ISME J. 4, 633–641 10.1038/ismej.2009.152 (doi:10.1038/ismej.2009.152) [DOI] [PubMed] [Google Scholar]
- 7.Smith J. M., Ogram A. 2008. Genetic and functional variation in denitrifier populations along a short-term restoration chronosequence. Appl. Environ. Microbiol. 74, 5615–5620 (doi:10.1128/AEM.00349–08) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coyne M. S., Arunakumari A., Averill A., Tiedje J. M. 1989. Immunological identification and distribution of dissimilatory heme cd1 and non-heme copper nitrite reductases in denitrifying bacteria. Appl. Environ. Microbiol. 55, 2924–2931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Geets J., Boon N., Verstraete W. 2006. Strategies of aerobic ammonia-oxidizing bacteria for coping with nutrient and oxygen fluctuations. FEMS Microbiol. Ecol. 58, 1–13 10.1111/j.1574-6941.2006.00170.x (doi:10.1111/j.1574-6941.2006.00170.x) [DOI] [PubMed] [Google Scholar]
- 10.Poulton P. R. 2006. Rothamsted research: guide to the classical and other long-term experiments, datasets and sample archive. Harpenden, UK: Rothamsted Research; See http://www.rothamsted.ac.uk/resources/LongTermExperiments.pdf. [Google Scholar]
- 11.Watts C. W., Clark L. J., Poulton P. R., Powlson D. S., Whitmore A. P. 2006. The role of clay, organic carbon and long-term management on mouldboard plough draught measured on the Broadbalk wheat experiment at Rothamsted. Soil Use Manag. 22, 334–341 10.1111/j.1475-2743.2006.00054.x (doi:10.1111/j.1475-2743.2006.00054.x) [DOI] [Google Scholar]
- 12.Goulding K. W. T., Bailey N. J., Bradbury N. J., Hargreaves P., Howe M., Murphy D. V., Poulton P. R., Willison T. W. 1998. Nitrogen deposition and its contribution to nitrogen cycling and associated soil processes. New Phytol. 139, 49–58 10.1046/j.1469-8137.1998.00182.x (doi:10.1046/j.1469-8137.1998.00182.x) [DOI] [Google Scholar]
- 13.Poulton P. R., Pye E., Hargreaves P. R., Jenkinson D. S. 2003. Accumulation of carbon and nitrogen by old arable land reverting to woodland. Global Change Biol. 9, 942–955 10.1046/j.1365-2486.2003.00633.x (doi:10.1046/j.1365-2486.2003.00633.x) [DOI] [Google Scholar]
- 14.Clark I. M., Hirsch P. R. 2008. Survival of bacterial DNA and culturable bacteria in archived soils from the Rothamsted Broadbalk experiment. Soil Biol. Biochem. 40, 1090–1102 10.1016/j.soilbio.2007.11.021 (doi:10.1016/j.soilbio.2007.11.021) [DOI] [Google Scholar]
- 15.Edgar R. C. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1997 10.1093/nar/gkh340 (doi:10.1093/nar/gkh340) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Felsenstein J. 2004. PHYLIP (Phylogeny Inference Package) version 3.6. Distributed by the author. Seattle, WA: Department of Genome Sciences, University of Washington [Google Scholar]
- 17.Strimmer K., von Haeseler A. 1996. Quartet puzzling: a quartet maximum likelihood method for reconstructing tree topologies. Mol. Biol. Evol. 13, 964–969 [Google Scholar]
- 18.Töwe S., Kleineidam K., Schloter M. 2010. Differences in amplification efficiency of standard curves in quantitative real-time PCR assays and consequences for gene quantification in environmental samples. J. Microbiol. Methods 82, 338–341 10.1016/j.mimet.2010.07.005 (doi:10.1016/j.mimet.2010.07.005) [DOI] [PubMed] [Google Scholar]
- 19.Ramakers C., Ruijter J. M., Deprez R. H. L., Moorman A. F. M. 2003. Assumption: free analysis of quantitative real-time polymerase chain reaction (PCT) data. Neurosci. Lett. 339, 62–66 10.1016/S0304-3940(02)01423-4 (doi:10.1016/S0304-3940(02)01423-4) [DOI] [PubMed] [Google Scholar]
- 20.Ruijter J. M., Ramakers C., Hoogaars W. M. H., Karlen Y., Bakker O., van den Hoff M. J. B., Moorman A. F. M. 2009. Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 37, 1–12 10.1093/nar/gkp045 (doi:10.1093/nar/gkp045) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klappenbach J. A., Saxman P. R., Cole J. R., Schmidt T. M. 2001. rrndb: the Ribosomal RNA operon copy number database . Nucleic Acids Res. 29, 181–184 10.1093/nar/29.1.181 (doi:10.1093/nar/29.1.181) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Acinas S. G., Marcelino L. A., Klepac-Ceraj V., Polz M. F. 2004. Divergence and redundancy of 16S rRNA sequences in genomes with multiple rrn operons. J. Bacteriol. 186, 2629–2635 10.1128/JB.186.9.2629-2635.2004 (doi:10.1128/JB.186.9.2629-2635.2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hargreaves P. R., Brookes P. C., Ross G. J. S., Poulton P. R. 2003. Evaluating soil microbial biomass carbon as an indicator of long-term environmental change. Soil Biol. Biochem. 35, 401–407 10.1016/S0038-0717(02)00291-2 (doi:10.1016/S0038-0717(02)00291-2) [DOI] [Google Scholar]
- 24.Enwall K., Throbäck I. N., Stenberg M., Söderström M., Hallin S. 2010. Soil resources influence spatial patterns of denitrifying communities at scales compatible with land management. Appl. Environ. Microbiol. 76, 2243–2250 10.1128/AEM.02197-09 (doi:10.1128/AEM.02197-09) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones K. C., Symon C. J., Johnston A. E. 1987. Retrospective analysis of an archived soil collection. I. Metals. Sci. Total Environ. 61, 131–144 10.1016/0048-9697(87)90363-9 (doi:10.1016/0048-9697(87)90363-9) [DOI] [PubMed] [Google Scholar]
- 26.Blake L., Goulding K. W. T. 2002. Effects of atmospheric deposition, soil pH and acidification on heavy metal contents in soils and vegetation of semi-natural ecosystems at Rothamsted Experimental Station, UK. Plant Soil 240, 235–251 10.1023/A:1015731530498 (doi:10.1023/A:1015731530498) [DOI] [Google Scholar]
- 27.Philippot L., Andert J., Jones C. M., Bru B., Hallin S. 2011. Importance of denitrifiers lacking the genes encoding the nitrous oxide reductase for N2O emissions from soil. Global Change Biol. 17, 1497–1504 10.1111/j.1365-2486.2010.02334.x (doi:10.1111/j.1365-2486.2010.02334.x) [DOI] [Google Scholar]
- 28.Baggs E. M. 2008. A review of stable isotope techniques for N2O source partitioning in soils: recent progress, remaining challenges and future considerations. Rapid Commun. Mass Spectrom. 22, 1664–1672 10.1002/rcm.3456 (doi:10.1002/rcm.3456) [DOI] [PubMed] [Google Scholar]
- 29.Wallenstein M. D., Vilgalys R. J. 2005. Quantitative analyses of nitrogen cycling genes in soils. Pedobiologia 49, 665–672 10.1016/j.pedobi.2005.05.005 (doi:10.1016/j.pedobi.2005.05.005) [DOI] [Google Scholar]
- 30.Yoshida M., Ishii S., Otsuka S., Senoo K. 2009. Temporal shifts in diversity and quantity of nirK and nirS in a rice paddy field soil. Soil Biol. Biochem. 41, 2044–2051 10.1016/j.soilbio.2009.07.012 (doi:10.1016/j.soilbio.2009.07.012) [DOI] [Google Scholar]
- 31.Kandeler E., Deiglmayr K., Tscherko D., Bru D., Philippot L. 2006. Abundance of narG, nirS, nirK, and nosZ genes of denitrifying bacteria during primary successions of a glacier foreland. Appl. Environ. Microbiol. 72, 5957–5962 10.1128/AEM.00439-06 (doi:10.1128/AEM.00439-06) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heylen K., Gevers D., Vanparys B., Wittebolle L., Geets J., Boon N., DeVos P. 2006. The incidence of nirS and nirK and their genetic heterogeneity in cultivated denitrifiers. Environ. Microbiol. 8, 2012–2021 10.1111/j.1462-2920.2006.01081.x (doi:10.1111/j.1462-2920.2006.01081.x) [DOI] [PubMed] [Google Scholar]
- 33.Ogilvie L. A., Hirsch P. R., Johnston A. W. B. 2008. Bacterial diversity of the Broadbalk ‘Classical’ winter wheat experiment in relation to long-term fertilizer inputs. Microb. Ecol. 56, 525–537 10.1007/s00248-008-9372-0 (doi:10.1007/s00248-008-9372-0) [DOI] [PubMed] [Google Scholar]
- 34.de Vries F. T., van Groenigen J. W., Hoffland E., Bloem J. 2011. Nitrogen losses from two grassland soils with different fungal biomass. Soil Biol. Biochem. 43, 997–1005 10.1016/j.soilbio.2011.01.016 (doi:10.1016/j.soilbio.2011.01.016) [DOI] [Google Scholar]
- 35.Gorfer M., et al. 2011. Community profiling and gene expression of fungal assimilatory nitrate reductases in agricultural soil. ISME J. 5, 1771–1783 10.1038/ismej.2011.53 (doi:10.1038/ismej.2011.53) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jenkinson D. S., Powlson D. S., Wedderburn R. W. M. 1976. The effects of biocidal treatments on metabolism in soil—III. The relationship between soil biovolume, measured by optical microscopy, and the flush of decomposition caused by fumigation. Soil Biol. Biochem. 8, 189–202 10.1016/0038-0717(76)90003-1 (doi:10.1016/0038-0717(76)90003-1) [DOI] [Google Scholar]
- 37.Sutka R. L., Adams G. C., Ostrom N. E., Ostrom P. H. 2008. Isotopologue fractionation during N2O production by fungal denitrification. Rapid. Commun. Mass. Spectrom. 22, 3989–3996 10.1002/rcm.3820 (doi:10.1002/rcm.3820) [DOI] [PubMed] [Google Scholar]