Abstract
Under increasing tensile load, the lifetime of a single catch bond counterintuitively increases up to a maximum and then decreases exponentially like a slip bond. So far, the characteristics of single catch bond dissociation have been extensively studied. However, it remains unclear how a cluster of catch bonds behaves under tensile load. We perform computational analysis on the following models to examine the characteristics of clustered catch bonds: (i) clusters of catch bonds with equal load sharing, (ii) clusters of catch bonds with linear load sharing, and (iii) clusters of catch bonds in micropipette-manipulated cell detachment. We focus on the differences between the slip and catch bond clusters, identifying the critical factors for exhibiting the characteristics of catch bond mechanism for the multiple-bond system. Our computation reveals that for a multiple-bond cluster, the catch bond behaviour could only manifest itself under relatively uniform loading conditions and at certain stages of decohesion, explaining the difficulties in observing the catch bond mechanism under real biological conditions.
Keywords: cell adhesion, catch bond, slid bond, bond lifetime, cell bond dynamics, computational cell mechanics
1. Introduction
Cell adhesion is mediated by a large variety of receptor–ligand bonds. The majority of biological bonds are slip bonds, whose lifetime is shortened by force [1,2]. However, the development of ultrasensitive force probes has led to the finding that the lifetime of certain bonds could also be prolonged by tensile force, a counterintuitive behaviour referred to as catch bond [3–6]. Dembo et al. [7] first introduced the concept of catch bond as a way to rationalize the shear-enhanced adhesion observed for the binding between selectin molecules and leucocyte ligands [8–10].
Despite Dembo's seminal predictions, the first definitive example of catch bonds came more than a decade later. Using custom-made atomic force microscopy, Marshall et al. [3] discovered that, with increasing loading force, the interaction between P-selectin and P-selectin glycoprotein ligand-1 (PSGL-1) experiences an initial increase and a subsequent decrease in its lifetime, indicating a catch to slip transition with increasing force. This transition was subsequently observed for the FimH-mediated attachment of bacteria to host cells [11,12], actin–myosin complexes [13] and integrins [14].
Various studies have been conducted to explain the transition between catch bond and slip bond. The study by Somers et al. [15] on P-selectin bonds suggested a transition of ‘bent’ to ‘extended’ molecular conformation. The work by Thormann et al. [16] on the SP-D-sugar system for the first time revealed its structural transition between dynamic stability and thermodynamic stability. These conformational changes could be indications of the transition between catch bonds and slip bonds. Theoretical studies on catch bonds have also been vigorously conducted, resulting in a variety of models in the literature. Dembo et al. [7] introduced a two-state theory to model intermolecular bonds as transitional springs. But their assumption of an infinite increase in bond strength under increasing tensile force failed to predict the force-induced transition from catch to slip bonds [3]. To explain this transition, the traditional single dissociation pathway model appears insufficient, and two-pathway models based on an envisioned energy landscape of the biological bonds were introduced [17,18]. This interaction energy landscape is thought of as a three-dimensional surface which, when projected onto the direction of force, results in a one-dimensional landscape that captures the essential features of the bond-dissociation mechanisms. The existence of two pathways is supported by both structural studies [15,19] and molecular dynamics simulations [20].
Under the scheme of two-pathway dissociation, several models have been proposed [11,18,21,22]. These models are similar in nature except that they applied different assumptions regarding how the two bound states are initially populated. For example, Evans et al. [21] assumed a rapid equilibration between the two bound states, and their model was validated by a series of experiments on the P-selectin–PSGL-1 bond [21,23,24]. Thomas and colleagues assumed a slow conversion, and their model was successfully employed to explain features of FimH adhesion of the Escherichia coli bond [11,22].
Although the catch-to-slip transition has been intensely studied at the single molecular level, in typical physiological settings, adhesion is often realized by clustering of a number of bonds. Compared with the single-bond system, a cluster of bonds can show more complexities. Firstly, the effect of rebinding becomes significant, because ruptured bonds may re-form while the remaining closed bonds could still hold the cell in place [25–28]. Secondly, the rupture and rebinding of individual bonds will constantly alter the force distribution, resulting in strong cooperative effects [28,29].
Fully unravelling the behaviour of multiple-bond rupture is a highly complicated issue because it is hardly possible to determine the distribution of force among the bonds and the time-course of force applying on each bond. So far, researchers addressed this issue mostly within an ideal scenario where the load is equally shared by a number of parallel bonds. The pioneering theoretical framework was established by Bell [28], who used a deterministic equation to study the stability of adhesion clusters under constant force. Decades later, Erdmann & Schwarz [30,31] developed a stochastic version of Bell's work in order to tackle the same issue for small adhesion clusters. In addition, Evans & Ritchie [32] discussed the dynamics of a bond subjected to linear loading. Seifert [25] extended their discussion to multiple-bond situations, and his analysis has been confirmed by Prechtel et al. [26], who used a biomembrane force probe (BFP) to measure the adhesion strength of living cells. This BFP method was recently adopted by Ligezowska et al. [33] to investigate the influence of divalent ions on the rupture force transition of integrin–ligand bonds. The cooperative behaviour of clusters of slip bonds between two rigid [34] or elastic media [35–37] under different loading conditions have also been extensively investigated. All these previous studies were conducted for the clusters of ordinary slip bonds, while the cooperative behaviour of multiple catch bonds remains to be explored. Here, we present a first attempt to investigate the decohesion behaviour of a catch bond cluster under various loading conditions.
Following Bell's work, we establish and solve the rate equation of the bond number change under a constant force. Our study will focus on the differences between resultant cluster behaviours based on the slip and the catch bond models. Our analysis will start with the simplest case and proceed to cases with increasing sophistications: (i) single slip and catch bond under constant forces; (ii) a cluster of parallel bonds with uniformly distributed loading force; (iii) a cluster of parallel bonds with linearly distributed loading force; and (iv) the micropipette-manipulated detachment of a cell from a substrate surface. Our main objective is to identify the conditions for a cluster of catch bonds to exhibit the characteristics of catch bond behaviour.
2. Models and formulations
2.1. Bond rupture rate
Bell [28] proposed the original model for the force-dependent off-rate of the slip bond,
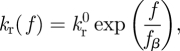 |
2.1 |
where 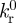 denotes the rupture rate when no force is applied to the bond; the force scale fβ sets the degree of rate increase with force. This model predicts an exponential decay of the slip bond lifetime with the increase in loading force.
denotes the rupture rate when no force is applied to the bond; the force scale fβ sets the degree of rate increase with force. This model predicts an exponential decay of the slip bond lifetime with the increase in loading force.
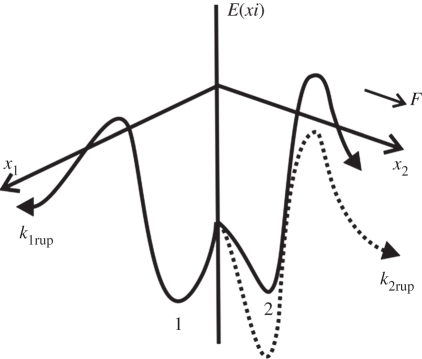
For a catch bond, we adopted Evans' two bound states, two pathway model [21]. As illustrated in figure 1, Evans et al. hypothesized the existence of two bound states from which two dissociation pathways originate. They also assumed a rapid equilibrium between the two bound states so that the bound partition is modified with the applied force. Five parameters were introduced in this model. The dissociation via the two pathways occurs with rates k1rup and k2rup. It was assumed that the fast pathway off-rate k1rup was constant, while the slow pathway, on the other hand, followed the Bell model with k2rup increasing exponentially with force. The dominant dissociation pathway is determined by the occupancy ratio of the two bound states, which is in equilibrium at all times with a small difference in energy between state 2 and state 1, ΔE21. According to the Boltzmann distribution, this energy difference sets the equilibrium occupancy ratio, Φ0 = exp (ΔE21/kBT) of state 1 to state 2 at zero force. However, the force applied to the bond causes a shift in the energy, resulting in a change in the energy difference between the two states. The occupancy ratio of the two states, then, changes exponentially with the applied force with a scale of f12. So, although pathway 1 may dominate at low forces where equilibrium favours occupancy of state 1, at higher forces, pathway 2 dominates as equilibrium shifts to favour occupancy of state 2. For bonds exhibiting this type of catch behaviour, the rupture rate is given by Evans et al. [21],
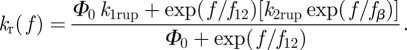 |
2.2 |
Figure 1.
Schematic of conceptual energy landscape of Evans' model [21]. The dissociation pathways x1 and x2 are indicated by the two solid lines. These two pathways originate from two bound states, 1 and 2, respectively. At low forces, state 1 is more favourable; as force on the bond increases, state 2 also becomes more favourable. The dissociation via pathway 1 is fast and has a constant rate k1rup. The dissociation rate along pathway 2 is lower at small forces, but increases significantly with increasing forces by lowering its energy barrier (as denoted by the dashed line), leading to an exponential increase of the dissociation rate k2rup.
With this expression, the rupture rate experiences a decrease at first as force rises and then an exponential increase after a specific critical value of force.
2.2. Formulations of four case studies
In the present paper, the rupture behaviour of both single bond and multiple bonds is discussed and compared. In the following, case 1 represents a single-bond scenario, while cases 2–4 represent three scenarios of multiple-bond systems.
Case 1: in single-bond scenarios, the ruptured bond has little chance to rebind because of the elastic recoil of the force transducer [27,28]. Therefore, its lifetime under a constant force is simply the reciprocal of the rupture rate (equations (2.1) and (2.2)).
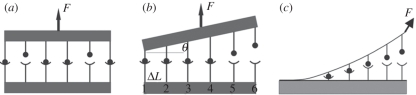
Case 2: as shown in figure 2a, an adhesion cluster consists of N0 receptor–ligand pairs; the ligands are connected to a flat and rigid substrate while receptors are confined to another rigid plate. Both plates are assigned a unit area. When the substrate is fixed in position while the other rigid plate is pulled by a constant loading force F, these bonds can be either ruptured or rebound. Since the two plates are kept parallel to each other, F is shared equally between the closed bonds. Therefore, we can establish the rate equation of the time-dependent change of the bond number [38].
| 2.3 |
Figure 2.
Schematic of bond clusters under constant force F. (a) F is equally shared by all closed bonds. (b) An inclined angle θ is kept between two rigid plates, so that the force is linearly distributed on each row of bonds. (c) The detachment of a BFP-loaded cell from the substrate surface; the force at the adhesion front is nonlinearly distributed on each bond.
where N0 is the initial number of closed bonds; N is the number of closed bonds at time t; f is the force on each closed bond, hence f = F/N. The rebinding of ruptured bonds needs to be considered in the multiple-bond system [28,30,31], with rebinding rate, Sun et al. [38],
| 2.4 |
where constant 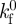 is the rate coefficient; δ is the separation distance between a receptor and ligand pair; δb is the characteristic length for bond formation. The rebinding rate kf was found to be dependent on the receptor–ligand separation [34], a characteristic length, δb, was introduced to describe the effect of the separation on bond-formation rate [38]. In the current computation, the bond is treated as a linear spring, with δ proportional to the bond force f as
is the rate coefficient; δ is the separation distance between a receptor and ligand pair; δb is the characteristic length for bond formation. The rebinding rate kf was found to be dependent on the receptor–ligand separation [34], a characteristic length, δb, was introduced to describe the effect of the separation on bond-formation rate [38]. In the current computation, the bond is treated as a linear spring, with δ proportional to the bond force f as
| 2.5 |
where k is the spring constant.
Case 3: the equally loaded bond cluster is a particular case, while in most biological systems, the force is not uniformly shared by each bond. This raises the question as to whether the spatial distribution of f would influence the decohesion behaviour of bond clusters. We studied a simplified scenario as illustrated in figure 2b where the ligands and receptors are still confined to two flat rigid plates. The substrate is a unit area square plate which is fixed in space, while the upper plate is pulled up by a constant force F. Different from the previous case, now the upper plate is inclined, and the angle (denoted by θ) between the two plates is kept unchanged during the pulling. As a result, the force on the individual bond is no longer uniform and its distribution depends linearly on the spatial arrangement of the bonds. For the present work, we only consider a special arrangement that for a total number of N0 the bonds are aligned in a number of rows which are equally spaced, and each row contains the same number of bonds. As the bond is treated as a linear spring, the force on the individual bonds is linearly related with its lateral position along the substrate:
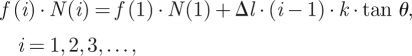 |
2.6 |
where i denotes the row number; Δl is the distance between two rows; k is the bond stiffness; f(i) is the force on single bond which is located at the ith row; N(i) is the number of closed bonds at the ith row. From the force equilibrium of the whole system, we have another equation,
| 2.7 |
The overall bond number,
| 2.8 |
Case 4: finally, we establish a model of more physiological relevance: the BFP-based cell detachment model [39–44]. As illustrated in figure 2c, a red cell is partially aspirated by a micropipette, serving as a force transducer as well as the model cell for adhesion; the adhesion strength can be measured from the aspiration pressure and the membrane extension. Opposite to the pipette entrance, the cell membrane is doped with controlled receptors, and is brought to and then retracted from a rigid flat surface, which is decorated with uniformly distributed ligands. Later the adhesion and detachment would occur in sequence.
2.3. Numerical methods
For case 1, as discussed in the previous section, a bond's lifetime under a constant force is simply the reciprocal of its rupture rate (equations (2.1) and (2.2)). For case 2, the deterministic computation of the rate equation (equation (2.3)) could yield the numerical solutions of closed bond number versus time t. For case 3, the time-dependent bond number N(t) is computed in an incremental procedure. In each time step Δt, we calculate N(i) and f(i) from equations (2.6) and (2.7); then based on the values of N(i) and f(i), and the rate equation (2.3), the change of bond number at each row ΔN(i) can be derived, and hence the N(i) is updated; the updated values of N(i) are then used to calculate f(i) for the next time step. The iteration proceeds until N < 1, which implies the complete detachment of the cluster. For case 4, an axisymmetric model has been developed following the assembly of previous BFP experiments [39–45]: a pre-swollen, spherical-shaped red blood cell was used as the force transducer; an elastic orthotropic membrane model was chosen to model the composite lipid bilayer of the membrane; hydrostatic fluid elements were introduced to analyse the mechanical response of the fluid-filled cavity inside the cell. The simulations were performed in three phases: firstly, a portion of the cell was aspirated into the micropipette by a suction pressure of magnitude ΔP. Secondly, the pipette-holding cell is brought into contact with the substrate. Cell adhesion was simulated during the membrane–surface touch, and the cell would spread on the substrate in an incremental manner until the whole system reached force equilibrium. Finally, the pipette was pulled back with a specified loading. The cell was therefore forced to move upward and eventually detach from the substrate. The deformation of the cell was computed by finite-element method based on the ABAQUS platform [46]. For details of the model and calculation, please refer to the previous work [43,44].
2.4. System parameters
The values of related parameters in cases 1–3 were chosen based on existing studies [21,45–48]: for the Bell model, the zero-force rupture rate 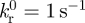 ; for the Evans' model, k1rup = 12 s–1, k2rup = 1 s–1; f12 = 6 pN, ΔE12 = 5 kBT; in both the Bell and the Evans models, we employed the same values of parameters as follows: the force scale fβ = 18 pN; the bond forward reaction rate coefficient
; for the Evans' model, k1rup = 12 s–1, k2rup = 1 s–1; f12 = 6 pN, ΔE12 = 5 kBT; in both the Bell and the Evans models, we employed the same values of parameters as follows: the force scale fβ = 18 pN; the bond forward reaction rate coefficient 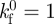 µm2 s−1; the characteristic length for bond-formation rate δb = 0.01 µm; the bond spring constant k = 10−3 N m–1. In case 4, the cell dimensions were acquired from previous typical BFP experiments [39–43]: the thickness of cell wall h = 20 nm; the cell radius R0 = 3.53 µm; the micropipette inner radius Rp = 0.95 µm; the initial ligand density ρ1 = 5000 µm−2 and initial receptor density ρr0 = 30 µm−2; the time step Δt = 10 ms. The aspirated cell serves as a linear spring, whose stiffness can be calculated from the aspiration pressure. In the present case, this pressure is Δp = 500 Pa; thus the stiffness is 640 pN µm–1. Therefore, the loading force F can be systematically adjusted by changing the retraction distance.
µm2 s−1; the characteristic length for bond-formation rate δb = 0.01 µm; the bond spring constant k = 10−3 N m–1. In case 4, the cell dimensions were acquired from previous typical BFP experiments [39–43]: the thickness of cell wall h = 20 nm; the cell radius R0 = 3.53 µm; the micropipette inner radius Rp = 0.95 µm; the initial ligand density ρ1 = 5000 µm−2 and initial receptor density ρr0 = 30 µm−2; the time step Δt = 10 ms. The aspirated cell serves as a linear spring, whose stiffness can be calculated from the aspiration pressure. In the present case, this pressure is Δp = 500 Pa; thus the stiffness is 640 pN µm–1. Therefore, the loading force F can be systematically adjusted by changing the retraction distance.
3. Results
3.1. Lifetime of single bond
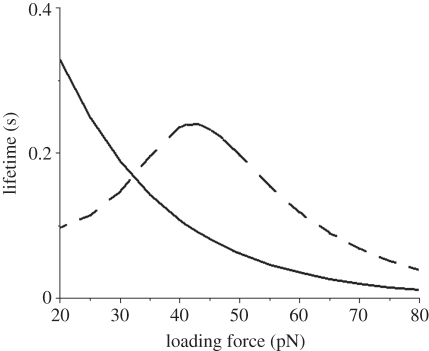
We calculated the single-bond lifetime versus loading force, which is plotted in figure 3. It is seen that for the slip bond model, its lifetime decreases exponentially with increasing force, while the lifetime of catch bond experiences a biphasic change and a peak value at an intermediate force. Owing to a higher rupture rate at low forces, the lifetime of catch bond is initially shorter than that of the slip bond. As force increases, however, its lifetime lengthens and eventually exceeds that of a slip bond. Upon reaching the peak value, it starts to decrease in a similar exponential pattern to the slip bond, indicating a transition from catch-to-slip bond at the high force regime.
Figure 3.
Single-bond lifetimes versus the loading force for both slip and catch bond models. Dashed line represents the Evans model and solid line represents the Bell model.
3.2. Lifetime of parallel multiple bonds with uniformly distributed force
3.2.1. Critical force fc for cluster decohesion
In a multiple-bond cluster, the rebinding rate plays an important role in stabilizing the adhesion contact. It was shown by Bell that the cluster remains stable up to a critical force fc [28]. Therefore, it is necessary to perform a stability analysis before studying the decohesion [30,31]. Although the explicit expressions are not available, with the assigned values of system parameters given above, we could obtain the numerical solutions. Taking the initial bond number N0 = 30, we obtained the critical force for the slip bond cluster, Fc ≈ 216 pN by using Bell's expression of kr (equation (2.1)); and the critical force for the catch bond cluster, Fc ≈ 113 pN by using Evans' rate expression (equation (2.2)). These two critical values define the upper limit above which a rapid transition occurs from adhesion to decohesion. When the force is below the critical value, only a fraction of the bonds break and a new equilibrium state is then established, while application of force which exceeds the critical value will initiate a complete rupture of all bonds.
3.2.2. Cluster lifetime
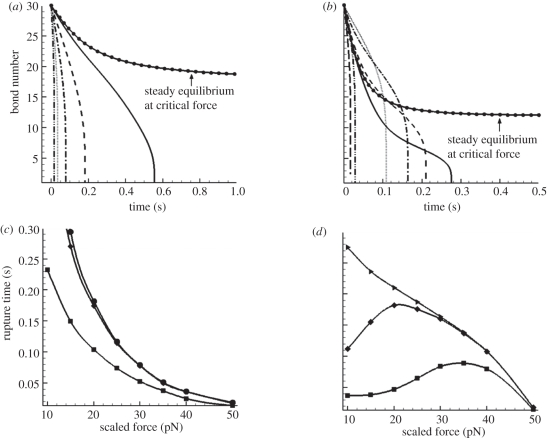
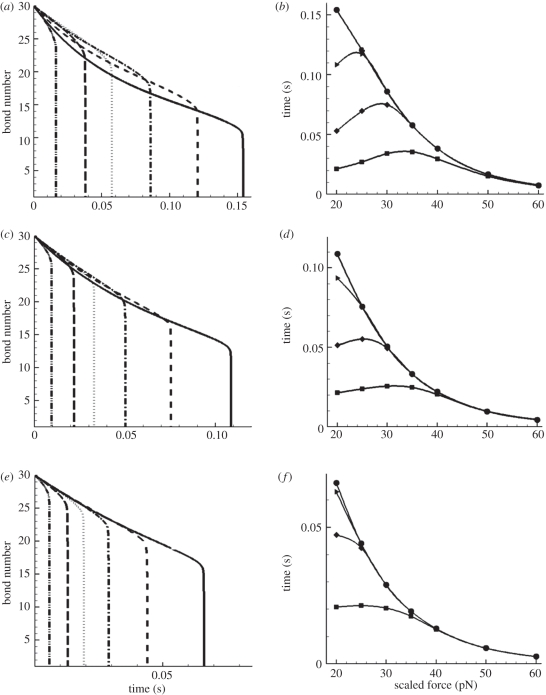
From equation (2.3), we could obtain the numerical solutions of closed bond number versus time t. Figure 4a,b exhibit these solutions at different loading forces for both slip bond and catch bond, respectively. For the convenience of comparing, we use the scaled quantity 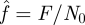 instead of the overall load F.
instead of the overall load F.
Figure 4.
Lifetime analyses for uniformly loaded bond clusters. (a,b) Change of bond number versus time t at different loading forces: (a) slip bond model (solid line with dots, 7.1 pN; solid line, 10 pN; dashed line, 20 pN; dashed dotted line, 30 pN; dotted line, 40 pN; dash double-dotted line, 50 pN); (b) catch bond model (solid line with dots, 3.7 pN; solid line, 10 pN; short dashed line, 20 pN; dashed dotted line, 30 pN; dotted line, 40 pN; long dashed line, 50 pN; dash double-dotted line, 60 pN). The curves with the circle symbols denote the equilibrium status. (c,d) Rupture time versus the loading force at different decohesion stages: (c) slip bond model (lines with squares denote time for one-third bonds broken, lines with diamonds denote time for two-third bonds broken and lines with circles denote lifetime); (d) catch bond model (lines with squares denote time for one-third bonds broken, lines with diamonds denote time for two-third bonds broken and lines with triangles denote lifetime).
The differences between the two types of bonds are obvious. First, they have different critical forces 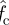 for cluster decohesion:
for cluster decohesion: 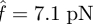 for the slip bond and
for the slip bond and 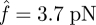 for the catch bond. Hence, the loading forces are 213 pN for the slip bond cluster and 111 pN for the catch bond cluster, respectively. These values are in good agreement with our calculations in the previous section. Second, and more importantly, their bond number decays in distinct manners. The decreasing rate (the slope of the curve) of slip bond cluster changes monotonically with increasing applied force from
for the catch bond. Hence, the loading forces are 213 pN for the slip bond cluster and 111 pN for the catch bond cluster, respectively. These values are in good agreement with our calculations in the previous section. Second, and more importantly, their bond number decays in distinct manners. The decreasing rate (the slope of the curve) of slip bond cluster changes monotonically with increasing applied force from 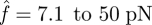 (figure 4a). In contrast, the catch bond cluster displays a crossover of the curves, indicating a biphasic change of the force-dependent decohesion rate (figure 4b). In the early stage of bond breaking (fewer than seven bonds broken), it is seen that the case with
(figure 4a). In contrast, the catch bond cluster displays a crossover of the curves, indicating a biphasic change of the force-dependent decohesion rate (figure 4b). In the early stage of bond breaking (fewer than seven bonds broken), it is seen that the case with 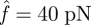 possesses the longest rupture time, which is consistent with the single-bond results shown in figure 3. However, as the broken bond number increases, the actual force f on each bond increases, thus the longest rupture time happens at lower values of
possesses the longest rupture time, which is consistent with the single-bond results shown in figure 3. However, as the broken bond number increases, the actual force f on each bond increases, thus the longest rupture time happens at lower values of 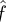 , for example at
, for example at 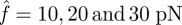 . As the number of broken bonds further increases, the actual force f on each bond is so large that these bonds are actually in slip bond regime, for example at
. As the number of broken bonds further increases, the actual force f on each bond is so large that these bonds are actually in slip bond regime, for example at 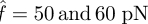 ; thus, this crossover of the curves eventually disappears.
; thus, this crossover of the curves eventually disappears.
For a bond cluster, its lifetime is usually identified by the time when only the last bond remains [28,30,31]. The lifetimes for the slip and catch bond clusters at different forces are shown in figure 4c (the curve with circles) and 4d (the curve with triangles), respectively. It is seen that there is no sign of catch bond characteristics as the lifetime monotonically decreases for both slip and catch bond clusters; the latter decreases in a much more gradual manner than the former. However, when we plot the time courses during which the bond number decreases by one-third and two-thirds of its initial value, these two partial decohesions exhibit the typical biphasic pattern and a maximum lifetime at intermediate loading forces.
The above finding is an interesting contrast to the conventional single-bond experiments [3,21]. Differing from the single catch bond, the multiple-bond cluster ruptures in a more complicated process, which involves two stages: initially, the decohesion is modulated by catch bond mechanism; then as the loading forces on the remaining bonds increase, the slip-bond mechanism begins to take lead. As a result, the decohesion of a catch bond cluster demonstrates two features: a biphasic pattern of rupture time for partial decohesion, and a monotonic decrease of lifetime for complete decohesion.
3.3. Lifetime of multiple bonds with linearly distributed force
In the case of multiple bonds with linearly distributed force, the initial bond number N0 is still assigned to be 30. The bonds are aligned in six equally spaced rows with five bonds in each row. The substrate is a square surface of 1 µm2, so the distance between two rows is l = 0.2 µm. The incremental time step is Δt = 10−4 s. Varying the incline angle θ, we calculated the time-dependent bond number N(t) for different loadings. Figure 5a,c,e shows the bond number changing with time at tan θ = 0.1, tan θ = 0.2 and tan θ = 0.3, respectively. It is clearly shown that with increasing θ, the crossover among different curves is reduced, indicating the deceasing trend in catch bond behaviour. This trend is further demonstrated in figure 5b,d,f, which shows the times for different decohesion stages versus the loading levels at three cases of tan θ = 0.1, tan θ = 0.2 and tan θ = 0.3. A detailed exploration of force distribution on each row of bonds would help in explaining this diminishing effect. Take the case of 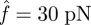 for example, for tan θ = 0.1, the initial distribution of f(i) from the first to the sixth row is 20, 24, 28, 32, 36 and 40 pN; for tan θ = 0.3, f(i) = 0, 12, 24, 36, 48 and 60 pN. Evidently, the latter case gives rise to a wider distribution of f. Because the catch bond mechanism spans only a narrow range within the lower force regime (figure 3), a wider distribution of force results in a smaller portion of catch bonds. For instance, for tan θ = 0.1, at the beginning of decohesion, all bonds are in the catch bond regime, while for tan θ = 0.3, only two-thirds of the bonds are in the catch bond regime (f(i) = 0, 12, 24, 36 pN), and this portion will further decrease during the detachment. Hence, when the two force-bearing plates are not parallel but have an inclined angle, the catch and slip mechanisms are averaged between the different rows. The larger the angle, the more dominant the slip-bond mechanism, and the less effect of the catch bond mechanism.
for example, for tan θ = 0.1, the initial distribution of f(i) from the first to the sixth row is 20, 24, 28, 32, 36 and 40 pN; for tan θ = 0.3, f(i) = 0, 12, 24, 36, 48 and 60 pN. Evidently, the latter case gives rise to a wider distribution of f. Because the catch bond mechanism spans only a narrow range within the lower force regime (figure 3), a wider distribution of force results in a smaller portion of catch bonds. For instance, for tan θ = 0.1, at the beginning of decohesion, all bonds are in the catch bond regime, while for tan θ = 0.3, only two-thirds of the bonds are in the catch bond regime (f(i) = 0, 12, 24, 36 pN), and this portion will further decrease during the detachment. Hence, when the two force-bearing plates are not parallel but have an inclined angle, the catch and slip mechanisms are averaged between the different rows. The larger the angle, the more dominant the slip-bond mechanism, and the less effect of the catch bond mechanism.
Figure 5.
Lifetime analysis for linearly loaded bond clusters. (a,c,e) Bond number versus time t at different loading levels—(a) tan θ = 0.1; (c) tan θ = 0.2 and (e) tan θ = 0.3 (solid lines, 20 pN; short dashed lines, 25 pN; dashed dotted lines, 30 pN; dotted lines, 35 pN; long dashed lines, 40 pN; dash double-dotted lines, 50 pN). (b,d,f) Rupture time versus loading force at different stages of decohesion—(b) tan θ = 0.1; (d) tan θ = 0.2; and (f) tan θ = 0.3 (lines with squares denote time for one-sixth bonds broken, lines with diamonds denote time for one-third bonds broken and lines with triangles denote time for one-half bonds broken; lines with circles denote lifetime).
3.4. Micropipette-manipulated detachment of a cell from a substrate surface
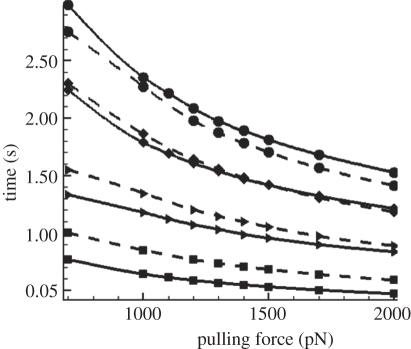
We varied the loading forces by changing the pipette aspiration pressure. In figure 6, we plot the time courses at four different decohesion stages, with 10, 30, 60 and 100 per cent (the complete detachment) of bonds broken versus the loading force. In the initial stage, the rupture time of catch models (denoted by solid lines) is lower than that of slip models (denoted by dashed lines), implying a higher rupture rate of the catch bond at the low force regime. As decohesion proceeds, the force f on each bond increases, and the rupture rate of catch bond decreases; hence its rupture time gradually exceeds that of the slip bond model.
Figure 6.
Rupture time of BFP-manipulated cell detachment versus loading force at four different decohesion stages: catch bond model is denoted by solid lines, while slip bond model is denoted by dashed lines. Lines with squares denote time for 10% bonds broken, lines with triangles denote time for 30% bonds broken and lines with diamonds denote time for 60% bonds broken; lines with circles denote lifetime.
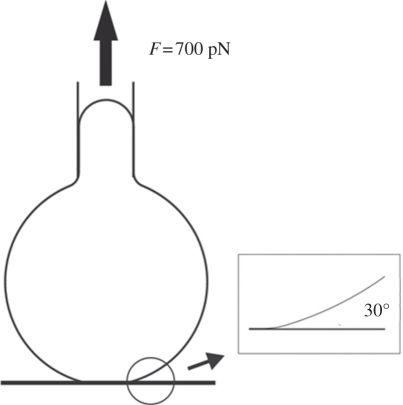
It can be seen that the times at all four stages decrease monotonically with pulling forces, indicating that overall there are no catch bond characteristics during the cell detachment. The difference between the two types of bonds is surprisingly small, compared with the parallel multiple-bond system (figure 4). This can be explained by the highly non-uniform distribution of force at the detachment front arising from the compliance of the cell membrane. When the cell is pulled by an upward force F, the load is primarily concentrated at the peripheral region of the adhesion patch; this region can be sketched as a ring of finite width. Because of the softness of the membrane, this ring is largely bended and the bonds within it are highly non-uniformly loaded. Our calculation results (see figure 7 and its inset) show that even at the early stage of detachment, the angle between the membrane and the substrate surface is around 30°. Since tan(30°) = 0.577, which is much larger than these studied in §3.2 where the upper plate is rigid, the force distribution in the current situation is even wider. Therefore, the catch bond effect is significantly weakened, and both models are basically controlled by the slip-bond mechanism even right from the beginning of the detachment.
Figure 7.
The calculated configuration of the BFP-manipulated cell detachment from a substrate surface, and the close-up view of the cell–substrate contact front. This configuration was obtained at the beginning of the detachment with a loading force F = 700 pN.
4. Discussion
Most previous studies on catch bond are focused on the single-bond system, while multiple-bond situations, which are more physiologically relevant, have been rarely considered. Our present work presents a detailed study on the effects of catch bond mechanism within multiple-bond systems. To describe the dual response of rupture rate to tensile forces, we employed the Evans' two-state, two-pathway model. We performed numerical studies based on several scenarios, including a single-bond system, a multiple-bond system with uniform loading, a multiple-bond system with linearly distributed loading and the micropipette-controlled detachment of a cell from a substrate.
Our results revealed that the multiple-bond system manifests its catch mechanism in a distinct manner compared with the single-bond system. Because of the temporal change of force on each bond, the biphasic time–force relationship is obvious only during partial decohesions, while towards the end of the lifetime, the monotonic slip-bond mechanism takes control. Another interesting observation is drawn from the cases where loading force is also spatially distributed. It is demonstrated that the cooperative act of all the loaded bonds plays an essential role for the cluster to realize its catch manifestations. A wider spatial distribution of the force leads to less cooperativity of the bonds, consequently causing a less pronounced catch behaviour of the system.
The current simulation results allow us to make an interesting comment on the variations between P- and L-selectin-mediated cell-rolling dynamics. P-selectin and L-selectin share similar structures which are characterized as catch bonds [9], but with two notable differences: (i) the transition from catch to slip behaviour of L-selectin bond occurs at a higher force than P-selectin bond, and the width of the transition of L-selectin is also much greater [9,49]; (ii) the binding interface of L-selectin is much stiffer than the P-selectin interface [49]. In view of these two differences and observations from the present study, we may reason that the multiple L-selectin bonds may exhibit a more obvious catch behaviour than the P-selectin bonds. Intriguingly, most studies [9,49–52] did reveal that the shear threshold of L-selectin-mediated leucocyte rolling is much more pronounced than the P-selectin-mediated rolling. Therefore, our study may provide an explanation for this discrepancy from the perspective of the cooperative effect of catch bonds.
Several theoretical refinements can be made on the basis of the present models. Firstly, in our current continuum framework, the fluctuations of single-bond lifetime and nonlinear effects are not included. These effects are negligible in the current cases, but could be significant for small adhesion clusters where several bonds are involved. For the latter case, stochastic models are a better option [30,31]. Also, it is noted that most of the existing researches on catch bonds have been primarily focused on model development at the single-bond level. On the other hand, the cooperative behaviour of multiple slip bonds under various loading and environmental conditions have been investigated [34–37,53]. The present work highlights the importance of the cooperative behaviour of multiple catch bonds, providing a new perspective to understand the collective behaviour of catch bonds. Finally, in modelling the BFP-controlled cell detachment, the effects of membrane modulus is worth an in-depth analysis. In the current model, the membrane within the adhesion patch is of the same elastic modulus as the non-adhered region. However, in real physiological situations, a mature adhesion region always consists of a number of small adhesion clusters, which are called focal adhesions [54,55]. Focal adhesion, which consists of a large number of proteins, is much stiffer than normal membrane. Hence, focal adhesions could be a way for cell to realize its catch bond mechanism, because with a higher elastic modulus within the adhesion region, the bonds could act in a more cooperative way [37]. However, the actual modulus change within focal adhesions is a very complicated issue, and needs to be studied in great detail in future works.
5. Conclusions
In this paper, we examine the effect of catch mechanism on decohesion of multiple-bond systems. Computational analyses reveal that, in a multiple-bond system, catch bond behaviour could only manifest itself in limited loading conditions (uniform loading among bonds) and certain stages (partial decohesion at early stages). These observations may explain the fact that the deterministic experimental evidence of catch bonds [3] came much later than its predictions [7]. These discoveries also provide a new framework for studying catch bonds: in multiple-bond systems, only testing the final lifetime of decohesion is inadequate to obtain the ‘catch’ behaviour; both loading conditions and fraction of ruptured bonds are of great importance in manifesting the characteristics of catch bond clusters.
References
- 1.Evans E. 2001. Probing the relation between force—lifetime—and chemistry in single molecular bonds. Annu. Rev. Biophys. Biomol. Struct. 30, 105–128 10.1146/annurev.biophys.30.1.105 (doi:10.1146/annurev.biophys.30.1.105) [DOI] [PubMed] [Google Scholar]
- 2.Hammer D. A., Tirrel M. 1996. Biological adhesion at interfaces. Annu. Rev. Mater. Sci. 26, 651–691 10.1146/annurev.ms.26.080196.003251 (doi:10.1146/annurev.ms.26.080196.003251) [DOI] [Google Scholar]
- 3.Marshall B. T., Long M., Piper J. W., Yago T., McEver R. P., Zhu C. 2003. Direct observation of catch bonds involving cell-adhesion molecules. Nature 423, 190–193 10.1038/nature01605 (doi:10.1038/nature01605) [DOI] [PubMed] [Google Scholar]
- 4.Barsegov V., Thirumalai D. 2005. Dynamics of unbinding of cell adhesion molecules: transition from catch to slip bonds. Proc. Natl Acad. Sci. USA 102, 1835–1839 10.1073/pnas.0406938102 (doi:10.1073/pnas.0406938102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prezhdo O. V., Perverzev Y. V. 2009. Theoretical aspects of the biological catch-bond. Acc. Chem. Res. 42, 693–703 10.1021/ar800202z (doi:10.1021/ar800202z) [DOI] [PubMed] [Google Scholar]
- 6.Thomas W. E., Trintchina E., Forero M., Vogel V., Sokurenko E. V. 2002. Bacterial adhesion to target cells enhanced by shear force. Cell 109, 913–923 10.1016/S0092-8674(02)00796-1 (doi:10.1016/S0092-8674(02)00796-1) [DOI] [PubMed] [Google Scholar]
- 7.Dembo M., Torney D. C., Saxaman K., Hammer D. A. 1988. The reaction-limited kinetics of membrane-to-surface adhesion and detachment. Proc. R. Soc. Lond. B 234, 55–83 10.1098/rspb.1988.0038 (doi:10.1098/rspb.1988.0038) [DOI] [PubMed] [Google Scholar]
- 8.Finger E. B., Purl K. D., Alon R., Lawrence M. B., Andrian U. H., Springer T. A. 1996. Adhesion through L-selectin requires a threshold hydrodynamic shear. Nature 379, 266–269 10.1038/379266a0 (doi:10.1038/379266a0) [DOI] [PubMed] [Google Scholar]
- 9.Sarangapani K. K., Yago T., Klopocki A. G., Lawrence M. B., Fieger C. B., Rosen S. D., McEver R. P., Zhu C. 2004. Low force decelerates L-selectin dissociation from P-selectin glycoprotein ligand-1 and endoglycan. J. Biol. Chem. 279, 2291–2298 10.1074/jbc.M310396200 (doi:10.1074/jbc.M310396200) [DOI] [PubMed] [Google Scholar]
- 10.Doggett T. A., Girdhar G., Lawshe A., Schmidtke D. W., Laurenzi I. J., Diamond S. L., Diacovo T. G. 2002. Selectin-like kinetics and biomechanics promote rapid platelet adhesion in flow: the GPIbα-vWF tether bond. Biophys. J. 83, 194–205 10.1016/S0006-3495(02)75161-8 (doi:10.1016/S0006-3495(02)75161-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thomas W., Forero M., Yakovenko O., Nilsson L., Vicini P., Sokurenko E., Vogel V. 2006. Catch-bond model derived from allostery explains force-activated bacterial adhesion. Biophys. J. 90, 753–764 10.1529/biophysj.105.066548 (doi:10.1529/biophysj.105.066548) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tchesnokova V., Aprikian P., Yakovenko O., Larock C., Kidd B., Vogel V., Thomas W., Sokurenko E. 2008. Integrin-like allosteric properties of the catch bond-forming FimH adhesin of Escherichia coli. J. Biol. Chem. 283, 7823–7833 10.1074/jbc.M707804200 (doi:10.1074/jbc.M707804200) [DOI] [PubMed] [Google Scholar]
- 13.Guo B., Guilford W. H. 2006. Mechanics of actomyosin bonds in different nucleotide states are tuned to muscle contraction. Proc. Natl Acad. Sci. USA 103, 9844–9849 10.1073/pnas.0601255103 (doi:10.1073/pnas.0601255103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kong F., García A. J., Mould A. P., Humphries M. J., Zhu C. 2009. Demonstration of catch bonds between an integrin and its ligand. J. Cell Biol. 185, 1275–1284 10.1083/jcb.200810002 (doi:10.1083/jcb.200810002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Somers W. S., Tang J., Shaw G. D., Camphausen R. T. 2000. Insights into the molecular basis of leukocyte tethering and rolling revealed by structures of P- and E-selectin bound to SLe(X) and PSGL-1. Cell 103, 467–479 10.1016/S0092-8674(00)00138-0 (doi:10.1016/S0092-8674(00)00138-0) [DOI] [PubMed] [Google Scholar]
- 16.Thormann E., Dreyer J. K., Simonsen A. C., Hansen P. L., Hansen S., Holmskov U., Mouritsen O. G. 2007. Dynamic strength of the interaction between lung surfactant protein D (SP-D) and saccharide ligands. Biochemistry 46, 12231–12237 10.1021/bi700823k (doi:10.1021/bi700823k) [DOI] [PubMed] [Google Scholar]
- 17.Prezhdo O. V., Perverzev Y. V. 2009. Theoretical aspects of the biological catch bond. Acc. Chem. Res. 42, 693–703 10.1021/ar800202z (doi:10.1021/ar800202z) [DOI] [PubMed] [Google Scholar]
- 18.Pereverzev Y. V., Prezhdo O. V., Forero M., Sokurenko E. V., Thomas W. E. 2005. The two-pathway model for the catch–slip transition in biological adhesion. Biophys. J. 89, 1446–1454 10.1529/biophysj.105.062158 (doi:10.1529/biophysj.105.062158) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yew Z. T., Schlierf M., Rief M., Paci E. 2010. Direct evidence of multidimensionality of free energy landscape. Phys. Rev. E 81, 031923. 10.1103/PhysRevE.81.031923 (doi:10.1103/PhysRevE.81.031923) [DOI] [PubMed] [Google Scholar]
- 20.Martínez L., Sonoda M. T., Webb P., Baxter J. D., Skaf M. S., Polikarpov I. 2005. Molecular dynamics simulations reveal multiple pathways of ligand dissociation from thyroid hormone receptors. Biophys. J. 89, 2011–2023 10.1529/biophysj.105.063818 (doi:10.1529/biophysj.105.063818) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Evans E., Leung A., Heinrich V., Zhu C. 2004. Mechanical switching and coupling between two dissociation pathways in a P-selectin adhesion bond. Proc. Natl Acad. Sci. USA 101, 11 281–11 286 10.1073/pnas.0401870101 (doi:10.1073/pnas.0401870101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thomas W. 2008. Catch bond in adhesion. Annu. Rev. Biomed. Eng. 10, 39–57 10.1146/annurev.bioeng.10.061807.160427 (doi:10.1146/annurev.bioeng.10.061807.160427) [DOI] [PubMed] [Google Scholar]
- 23.Heinrich V., Leung A., Evans E. 2005. Nano-to-microscale mechanical switches and fuses mediate adhesive contacts between leukocytes and the endothelium. J. Chem. Inf. Model. 45, 1482–1490 10.1021/ci0501903 (doi:10.1021/ci0501903) [DOI] [PubMed] [Google Scholar]
- 24.Barbaux S., Poirier O., Pincet F., Hermand P., Tiret L., Deterre P. 2010. The adhesion mediated by the P-selectin P–selectin glycoprotein ligand-1 (PSGL-1) couple is stronger for shorter PSGL-1 variants. J. Leukoc. Biol. 87, 4727–4734 10.1189/jlb.0609408 (doi:10.1189/jlb.0609408) [DOI] [PubMed] [Google Scholar]
- 25.Seifert U. 2000. Rupture of multiple parallel molecular bonds under dynamic loading. Phys. Rev. Lett. 84, 2750–2753 10.1103/PhysRevLett.84.2750 (doi:10.1103/PhysRevLett.84.2750) [DOI] [PubMed] [Google Scholar]
- 26.Prechtel K., Rausch A. R., Marchi-Artzner V., Kantlehner M., Kessler H., Merkel R. 2002. Dynamic force spectroscopy to probe adhesion strength of living cells. Phys. Rev. Lett. 89, 028101. 10.1103/PhysRevLett.89.028101 (doi:10.1103/PhysRevLett.89.028101) [DOI] [PubMed] [Google Scholar]
- 27.Tang C. C., Chu Y. P., Chen H. Y. 2007. Lifetime of ligand–receptor clusters under external force. Phys. Rev. E 76, 061905. 10.1103/PhysRevE.76.061905 (doi:10.1103/PhysRevE.76.061905) [DOI] [PubMed] [Google Scholar]
- 28.Bell G. I. 1978. Models for the specific adhesion of cells to cells. Science 200, 618–627 10.1126/science.347575 (doi:10.1126/science.347575) [DOI] [PubMed] [Google Scholar]
- 29.Boulbitch A. 2003. Enforced unbinding of biomembranes whose mutual adhesion is mediated by a specific interaction. Eur. Biophys. J. 31, 637–642 10.1007/s00249-002-0257-8 (doi:10.1007/s00249-002-0257-8) [DOI] [PubMed] [Google Scholar]
- 30.Erdmann T., Schwarz U. S. 2004. Stability of adhesion clusters under constant force. Phys. Rev. Lett. 92, 108102. 10.1103/PhysRevLett.92.108102 (doi:10.1103/PhysRevLett.92.108102) [DOI] [PubMed] [Google Scholar]
- 31.Erdmann T., Schwarz U. S. 2004. Stochastic dynamics of adhesion clusters under shared constant force and with rebinding. J. Chem. Phys. 121, 8997–9017 10.1063/1.1805496 (doi:10.1063/1.1805496) [DOI] [PubMed] [Google Scholar]
- 32.Evans E., Ritchie K. 1997. Dynamic strength of molecular adhesion bonds. Biophys. J. 72, 1541–1555 10.1016/S0006-3495(97)78802-7 (doi:10.1016/S0006-3495(97)78802-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ligezowska A., Boye K., Eble J. A., Hoffmann B., Klosgen B., Merkel R. 2011. Mechanically enforced bond dissociation reports synergistic influence of Mn2+ and Mg2+ on the interaction between integrin α7β1 and invasion. J. Mol. Recognit. 24, 715–723 10.1002/jmr.1108 (doi:10.1002/jmr.1108) [DOI] [PubMed] [Google Scholar]
- 34.Erdmann T., Schwarz U. S. 2007. Impact of receptor–ligand distance on adhesion cluster stability. Eur. Phys. J. E 22, 123–137 10.1140/epje/e2007-00019-8 (doi:10.1140/epje/e2007-00019-8) [DOI] [PubMed] [Google Scholar]
- 35.Qian J., Wang J., Gao H. 2008. Lifetime and strength of adhesive molecular bond clusters between elastic media. Langmuir 24, 1262–1270 10.1021/la702401b (doi:10.1021/la702401b) [DOI] [PubMed] [Google Scholar]
- 36.Qian J., Wang J., Lin Y., Gao H. 2009. Lifetime and strength of periodic bond clusters between elastic media under inclined loading. Biophys. J. 97, 2438–2445 10.1016/j.bpj.2009.08.027 (doi:10.1016/j.bpj.2009.08.027) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qian J., Gao H. 2010. Soft matrices suppress cooperative behaviors among receptor–ligand bonds in cell adhesion. PLoS ONE 5, e12342. 10.1371/journal.pone.0012342 (doi:10.1371/journal.pone.0012342) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun L., Cheng Q. H., Gao H., Zhang Y. W. 2009. Computational modeling for cell spreading on a substrate mediated by specific interactions, long-range recruiting interactions, and diffusion of binders. Phys. Rev. E 79, 061907. 10.1103/PhysRevE.79.061907 (doi:10.1103/PhysRevE.79.061907) [DOI] [PubMed] [Google Scholar]
- 39.Evans E., Ritchie K., Merkel R. 1995. Sensitive force technique to probe molecular adhesion and structural linkages at biological interfaces. Biophys. J. 68, 2580–2587 10.1016/S0006-3495(95)80441-8 (doi:10.1016/S0006-3495(95)80441-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simson D. A., Ziemann F., Strigl M., Merkel R. 1998. Micropipet-based pico force transducer: in depth analysis and experimental verification. Biophys. J. 74, 2080–2088 10.1016/S0006-3495(98)77915-9 (doi:10.1016/S0006-3495(98)77915-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heinrich V., Ounkomol C. 2007. Force versus axial deflection of pipette-aspirated closed membranes. Biophys. J. 93, 363–372 10.1529/biophysj.107.104091 (doi:10.1529/biophysj.107.104091) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Freund L. B. 2009. The stiffness of a biomembrane force probe vesicle. Math. Mech. Solids. 14, 148–160 10.1177/1081286508092608 (doi:10.1177/1081286508092608) [DOI] [Google Scholar]
- 43.Sun L., Cheng Q. H., Gao H., Zhang Y. W. 2011. A nonlinear characteristic regime of biomembrane force probe. J. Biomech. 44, 662–668 10.1016/j.jbiomech.2010.11.005 (doi:10.1016/j.jbiomech.2010.11.005) [DOI] [PubMed] [Google Scholar]
- 44.Cheng Q. H., Liu P., Gao H. J., Zhang Y. W. 2009. A computational modeling for micropipette-manipulated cell detachment from a substrate mediated by receptor–ligand binding. J. Mech. Phys. Solids 57, 205–220 10.1016/j.jmps.2008.11.003 (doi:10.1016/j.jmps.2008.11.003) [DOI] [Google Scholar]
- 45.Pierrat S., Brochart-Wyart F., Nassoy P. 2004. Enforced detachment of red blood cells adhering to surfaces: statics and dynamics. Biophys. J. 87, 2855–2869 10.1529/biophysj.104.043695 (doi:10.1529/biophysj.104.043695) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.ABAQUS. 2007. User's manual version 6.7. Pawtucket, RI: ABAQUS Inc [Google Scholar]
- 47.Valle-Delgado J. J., Molina-Bolivar J. A., Galisteo-Gonzalez F., Galvez-Ruiz M. J., Feiler A., Rutland M. W. 2006. Adhesion forces between protein layers studied by means of atomic force microscopy. Langmuir 22, 5108–5114 10.1021/la053011k (doi:10.1021/la053011k) [DOI] [PubMed] [Google Scholar]
- 48.Pawar P., Jadhav S., Eggleton C. D., Konstantopoulos K. 2008. Roles of cell and microvillus deformation and receptor–ligand binding kinetics in cell rolling. Am. J. Physiol. Heart Circ. Physiol 295, H1439–H1450 10.1152/ajpheart.91536.2007 (doi:10.1152/ajpheart.91536.2007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Barsegov V., Thirumalai D. 2006. Dynamic competition between catch and slip bonds in selectins bound to ligands. J. Phys. Chem. B 110, 26 403–264 12 10.1021/jp0653306 (doi:10.1021/jp0653306) [DOI] [PubMed] [Google Scholar]
- 50.Zhu C., Yago T., Lou J. Z., Zarnitsyna V. I., McEver R. P. 2008. Mechanisms for flow-enhanced cell adhesion. Ann. Biomed. Eng. 36, 604–621 10.1007/s10439-008-9464-5 (doi:10.1007/s10439-008-9464-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Caputo K. E., Lee D., King M. R., Hammer D. A. 2007. Adhesive dynamics simulations of the shear threshold effect for leukocytes. Biophys. J. 92, 787–797 10.1529/biophysj.106.082321 (doi:10.1529/biophysj.106.082321) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Paschall C. D., Guilford W. H., Lawrence M. B. 2008. Enhancement of L-selectin, but not P-selectin, bond formation frequency by convective flow. Biophys. J. 94, 1034–1045 10.1529/biophysj.106.098707 (doi:10.1529/biophysj.106.098707) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gao H., Qian J., Chen B. 2011. Probing mechanical principles of focal contacts in cell–matrix adhesion with a coupled stochastic–elastic modelling framework. J. R. Soc. Interface 8, 1217–1232 10.1098/rsif.2011.0157 (doi:10.1098/rsif.2011.0157) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Balaban N. Q., Schwarz U. S., Riveline D. 2001. Force and focal adhesion assembly: a close relationship studied using elastic micropatterned substrates. Nat. Cell Biol. 3, 466–472 10.1038/35074532 (doi:10.1038/35074532) [DOI] [PubMed] [Google Scholar]
- 55.Vogel V., Sheetz M. 2006. Local force and geometry sensing regulate cell functions. Nat. Rev. Mol. Cell Biol. 7, 265–275 10.1038/nrm1890 (doi:10.1038/nrm1890) [DOI] [PubMed] [Google Scholar]