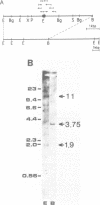
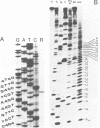
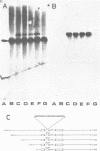
Abstract
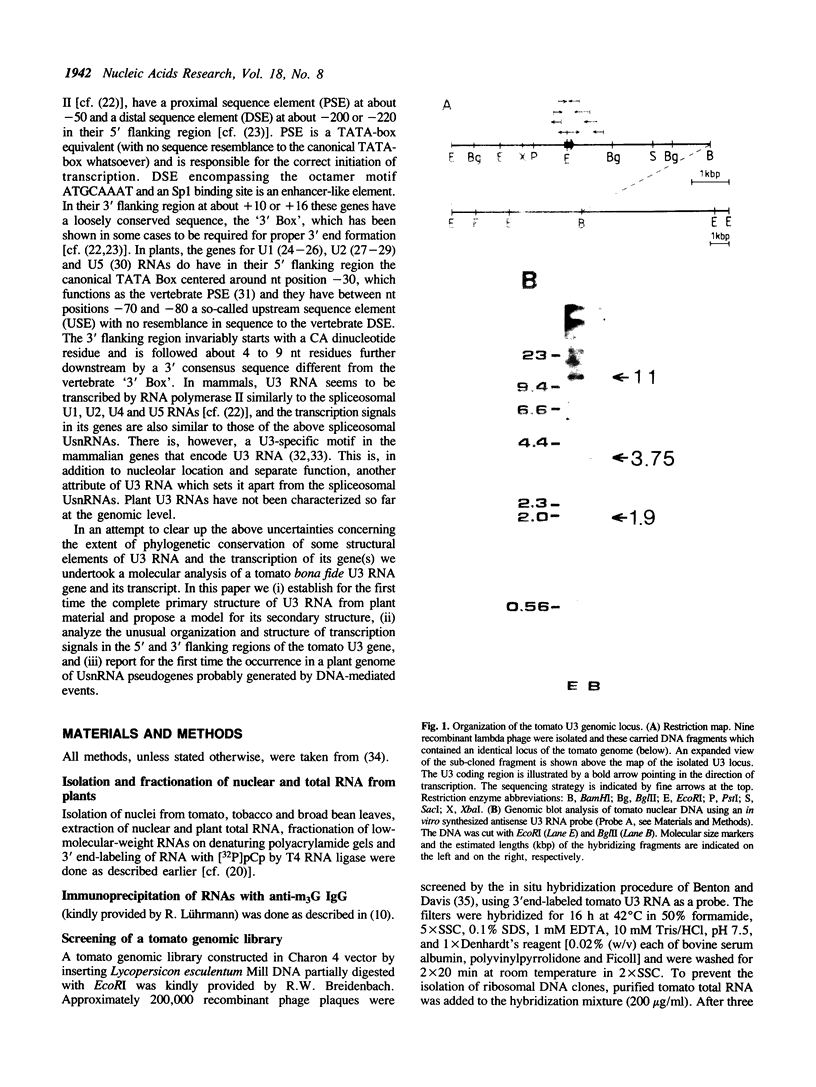
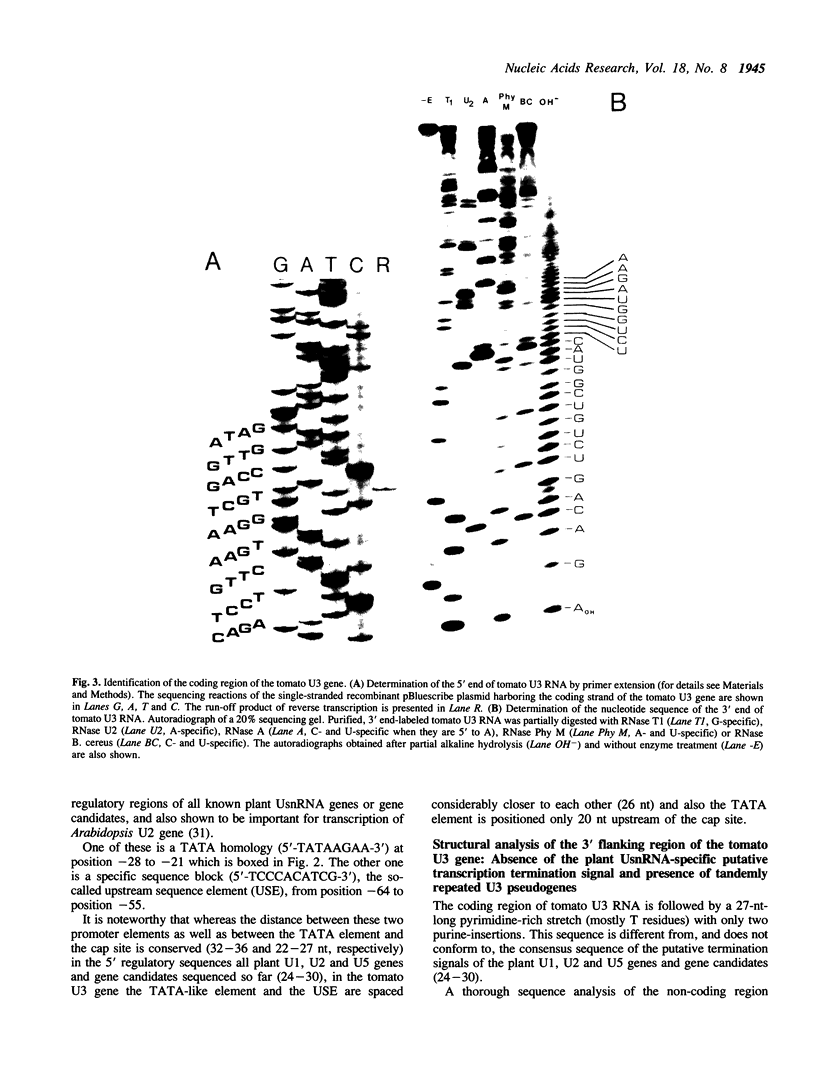
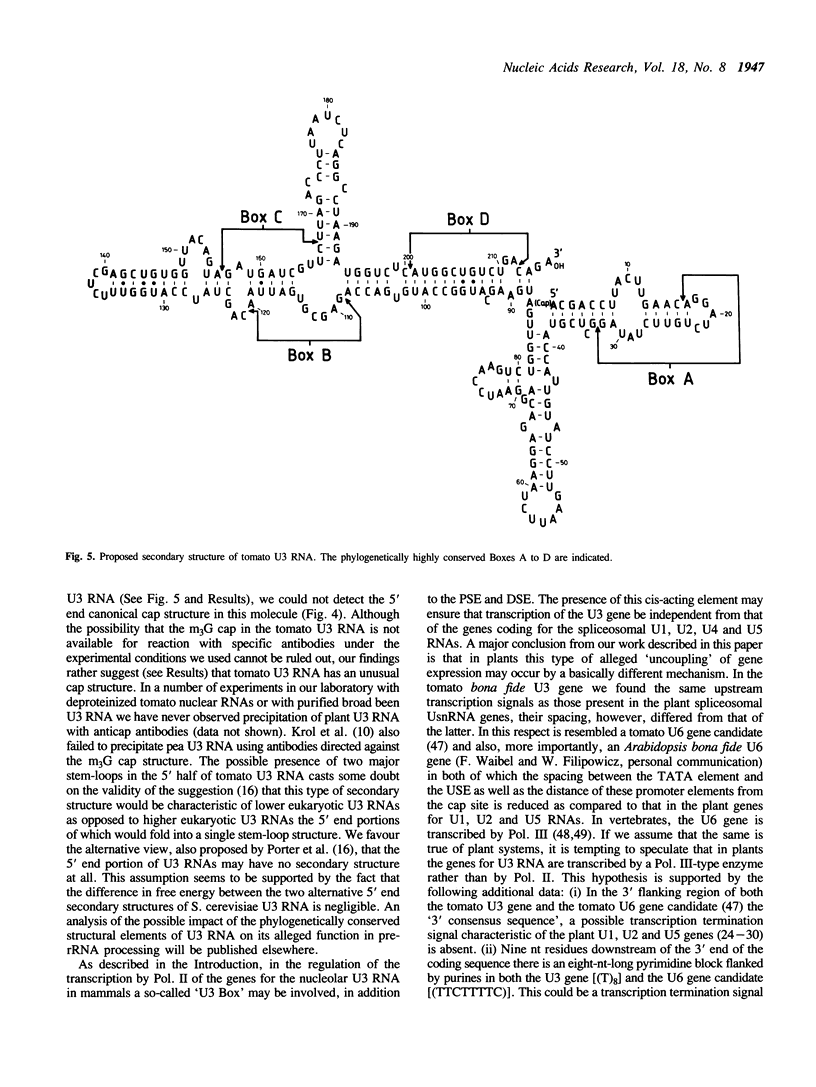
By screening a tomato genomic library with a tomato U3 RNA probe, we detected a U3 genomic locus whose coding region was determined by primer extension (5' end) and direct RNA sequencing of purified U3 RNA from tomato (3' end). Tomato U3 RNA is 216 nucleotides long, contains all the four evolutionarily highly conserved sequence blocks (Boxes A to D), has at its 5' end a cap not precipitable with anti-m3G antibodies and can be folded into a peculiar secondary structure with two stem-loops at its 5' end. A tagged derivative of the U3 gene was faithfully expressed in transgenic tobacco plants. In the 5' flanking region both plant-specific UsnRNA transcription signals [the TATA-like sequence and the upstream sequence element (USE)] were present, but were positioned closer to each other and also to the cap site in the U3 gene than in the genes for the plant spliceosomal UsnRNAs studied so far. The 3' flanking region of the tomato U3 gene lacked the consensus sequence of the putative termination signal established for the plant spliceosomal UsnRNA genes and contained a pyrimidine-rich tract (R1) followed by four tandemly repeated U3 pseudogenes (U3.1 ps to U3.4 ps) flanked by slightly altered forms (R2 to R5) of R1 and most probably generated by DNA-mediated events. Our results are in line with the conjecture that the enzyme transcribing the tomato U3 gene has different structural requirements for transcriptional activity than the enzyme transcribing plant U1, U2 and U5 genes.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abel S., Kiss T., Solymosy F. Molecular analysis of eight U1 RNA gene candidates from tomato that could potentially be transcribed into U1 RNA sequence variants differing from each other in similar regions of secondary structure. Nucleic Acids Res. 1989 Aug 11;17(15):6319–6337. doi: 10.1093/nar/17.15.6319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An G. Development of plant promoter expression vectors and their use for analysis of differential activity of nopaline synthase promoter in transformed tobacco cells. Plant Physiol. 1986 May;81(1):86–91. doi: 10.1104/pp.81.1.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton W. D., Davis R. W. Screening lambdagt recombinant clones by hybridization to single plaques in situ. Science. 1977 Apr 8;196(4286):180–182. doi: 10.1126/science.322279. [DOI] [PubMed] [Google Scholar]
- Brown J. W., Waugh R. Maize U2 snRNAs: gene sequence and expression. Nucleic Acids Res. 1989 Nov 25;17(22):8991–9001. doi: 10.1093/nar/17.22.8991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvet J. P., Pederson T. Base-pairing interactions between small nuclear RNAs and nuclear RNA precursors as revealed by psoralen cross-linking in vivo. Cell. 1981 Nov;26(3 Pt 1):363–370. doi: 10.1016/0092-8674(81)90205-1. [DOI] [PubMed] [Google Scholar]
- Denison R. A., Weiner A. M. Human U1 RNA pseudogenes may be generated by both DNA- and RNA-mediated mechanisms. Mol Cell Biol. 1982 Jul;2(7):815–828. doi: 10.1128/mcb.2.7.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein P., Reddy R., Busch H. Multiple states of U3 RNA in Novikoff hepatoma nucleoli. Biochemistry. 1984 Nov 6;23(23):5421–5425. doi: 10.1021/bi00318a007. [DOI] [PubMed] [Google Scholar]
- Freier S. M., Kierzek R., Jaeger J. A., Sugimoto N., Caruthers M. H., Neilson T., Turner D. H. Improved free-energy parameters for predictions of RNA duplex stability. Proc Natl Acad Sci U S A. 1986 Dec;83(24):9373–9377. doi: 10.1073/pnas.83.24.9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie C., Patterson B. Spliceosomal snRNAs. Annu Rev Genet. 1988;22:387–419. doi: 10.1146/annurev.ge.22.120188.002131. [DOI] [PubMed] [Google Scholar]
- Hanley B. A., Schuler M. A. Nucleotide sequence of a pea U2 snRNA gene. Nucleic Acids Res. 1989 Dec 11;17(23):10106–10106. doi: 10.1093/nar/17.23.10106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helms C., Graham M. Y., Dutchik J. E., Olson M. V. A new method for purifying lambda DNA from phage lysates. DNA. 1985 Feb;4(1):39–49. doi: 10.1089/dna.1985.4.39. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Hughes J. M., Konings D. A., Cesareni G. The yeast homologue of U3 snRNA. EMBO J. 1987 Jul;6(7):2145–2155. doi: 10.1002/j.1460-2075.1987.tb02482.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeppesen C., Stebbins-Boaz B., Gerbi S. A. Nucleotide sequence determination and secondary structure of Xenopus U3 snRNA. Nucleic Acids Res. 1988 Mar 25;16(5):2127–2148. doi: 10.1093/nar/16.5.2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss T., Antal M., Solymosy F. Plant small nuclear RNAs. II. U6 RNA and a 4.5SI-like RNA are present in plant nuclei. Nucleic Acids Res. 1987 Jan 26;15(2):543–560. doi: 10.1093/nar/15.2.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss T., Antal M., Solymosy F. Plant small nuclear RNAs. III. The complete primary and secondary structure of broad bean U2 RNA: phylogenetic and functional implications. Nucleic Acids Res. 1987 Feb 11;15(3):1332–1332. doi: 10.1093/nar/15.3.1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss T., Jakab G., Antal M., Pálfi Z., Hegyi H., Kis M., Solymosy F. Plant small nuclear RNAs. V. U4 RNA is present in broad bean plants in the form of sequence variants and is base-paired with U6 RNA. Nucleic Acids Res. 1988 Jun 24;16(12):5407–5426. doi: 10.1093/nar/16.12.5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss T., Solymosy F. Isolation of high molecular weight plant nuclear DNA suitable for use in recombinant DNA technology. Acta Biochim Biophys Hung. 1987;22(1):1–5. [PubMed] [Google Scholar]
- Kiss T., Tóth M., Solymosy F. Plant small nuclear RNAs. Nucleolar U3 snRNA is present in plants: partial characterization. Eur J Biochem. 1985 Oct 15;152(2):259–266. doi: 10.1111/j.1432-1033.1985.tb09192.x. [DOI] [PubMed] [Google Scholar]
- Konings D. A., Hogeweg P. Pattern analysis of RNA secondary structure similarity and consensus of minimal-energy folding. J Mol Biol. 1989 Jun 5;207(3):597–614. doi: 10.1016/0022-2836(89)90468-3. [DOI] [PubMed] [Google Scholar]
- Krol A., Ebel J. P., Rinke J., Luhrmann R. U1, U2 and U5 small nuclear RNAs are found in plants cells. Complete nucleotide sequence of the U5 RNA family from pea nuclei. Nucleic Acids Res. 1983 Dec 20;11(24):8583–8594. doi: 10.1093/nar/11.24.8583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel G. R., Maser R. L., Calvet J. P., Pederson T. U6 small nuclear RNA is transcribed by RNA polymerase III. Proc Natl Acad Sci U S A. 1986 Nov;83(22):8575–8579. doi: 10.1073/pnas.83.22.8575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maser R. L., Calvet J. P. U3 small nuclear RNA can be psoralen-cross-linked in vivo to the 5' external transcribed spacer of pre-ribosomal-RNA. Proc Natl Acad Sci U S A. 1989 Sep;86(17):6523–6527. doi: 10.1073/pnas.86.17.6523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazan S., Bachellerie J. P. Structure and organization of mouse U3B RNA functional genes. J Biol Chem. 1988 Dec 25;263(36):19461–19467. [PubMed] [Google Scholar]
- Melton D. A., Krieg P. A., Rebagliati M. R., Maniatis T., Zinn K., Green M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984 Sep 25;12(18):7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker K. A., Steitz J. A. Structural analysis of the human U3 ribonucleoprotein particle reveal a conserved sequence available for base pairing with pre-rRNA. Mol Cell Biol. 1987 Aug;7(8):2899–2913. doi: 10.1128/mcb.7.8.2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter G. L., Brennwald P. J., Holm K. A., Wise J. A. The sequence of U3 from Schizosaccharomyces pombe suggests structural divergence of this snRNA between metazoans and unicellular eukaryotes. Nucleic Acids Res. 1988 Nov 11;16(21):10131–10152. doi: 10.1093/nar/16.21.10131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prestayko A. W., Tonato M., Busch H. Low molecular weight RNA associated with 28 s nucleolar RNA. J Mol Biol. 1970 Feb 14;47(3):505–515. doi: 10.1016/0022-2836(70)90318-9. [DOI] [PubMed] [Google Scholar]
- Reddy R. Compilation of small RNA sequences. Nucleic Acids Res. 1988;16 (Suppl):r71–r85. doi: 10.1093/nar/16.suppl.r71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy R., Henning D., Das G., Harless M., Wright D. The capped U6 small nuclear RNA is transcribed by RNA polymerase III. J Biol Chem. 1987 Jan 5;262(1):75–81. [PubMed] [Google Scholar]
- Russel M., Kidd S., Kelley M. R. An improved filamentous helper phage for generating single-stranded plasmid DNA. Gene. 1986;45(3):333–338. doi: 10.1016/0378-1119(86)90032-6. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R., Reddy R. Gamma-monomethyl phosphate: a cap structure in spliceosomal U6 small nuclear RNA. Proc Natl Acad Sci U S A. 1989 Nov;86(21):8280–8283. doi: 10.1073/pnas.86.21.8280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroke I. L., Weiner A. M. The 5' end of U3 snRNA can be crosslinked in vivo to the external transcribed spacer of rat ribosomal RNA precursors. J Mol Biol. 1989 Dec 5;210(3):497–512. doi: 10.1016/0022-2836(89)90126-5. [DOI] [PubMed] [Google Scholar]
- Vankan P., Edoh D., Filipowicz W. Structure and expression of the U5 snRNA gene of Arabidopsis thaliana. Conserved upstream sequence elements in plant U-RNA genes. Nucleic Acids Res. 1988 Nov 25;16(22):10425–10440. doi: 10.1093/nar/16.22.10425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vankan P., Filipowicz W. A U-snRNA gene-specific upstream element and a -30 'TATA box' are required for transcription of the U2 snRNA gene of Arabidopsis thaliana. EMBO J. 1989 Dec 1;8(12):3875–3882. doi: 10.1002/j.1460-2075.1989.tb08566.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vankan P., Filipowicz W. Structure of U2 snRNA genes of Arabidopsis thaliana and their expression in electroporated plant protoplasts. EMBO J. 1988 Mar;7(3):791–799. doi: 10.1002/j.1460-2075.1988.tb02877.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y., Reddy R. Genes for human U3 small nucleolar RNA contain highly conserved flanking sequences. Biochim Biophys Acta. 1989 Jun 1;1008(1):14–22. doi: 10.1016/0167-4781(89)90164-4. [DOI] [PubMed] [Google Scholar]
- Zuker M., Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981 Jan 10;9(1):133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Santen V. L., Spritz R. A. Nucleotide sequence of a bean (Phaseolus vulgaris) U1 small nuclear RNA gene: implications for plant pre-mRNA splicing. Proc Natl Acad Sci U S A. 1987 Dec;84(24):9094–9098. doi: 10.1073/pnas.84.24.9094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Santen V. L., Swain W., Spritz R. A. Nucleotide sequences of two soybean U1 snRNA genes. Nucleic Acids Res. 1988 May 11;16(9):4176–4176. doi: 10.1093/nar/16.9.4176. [DOI] [PMC free article] [PubMed] [Google Scholar]