Abstract
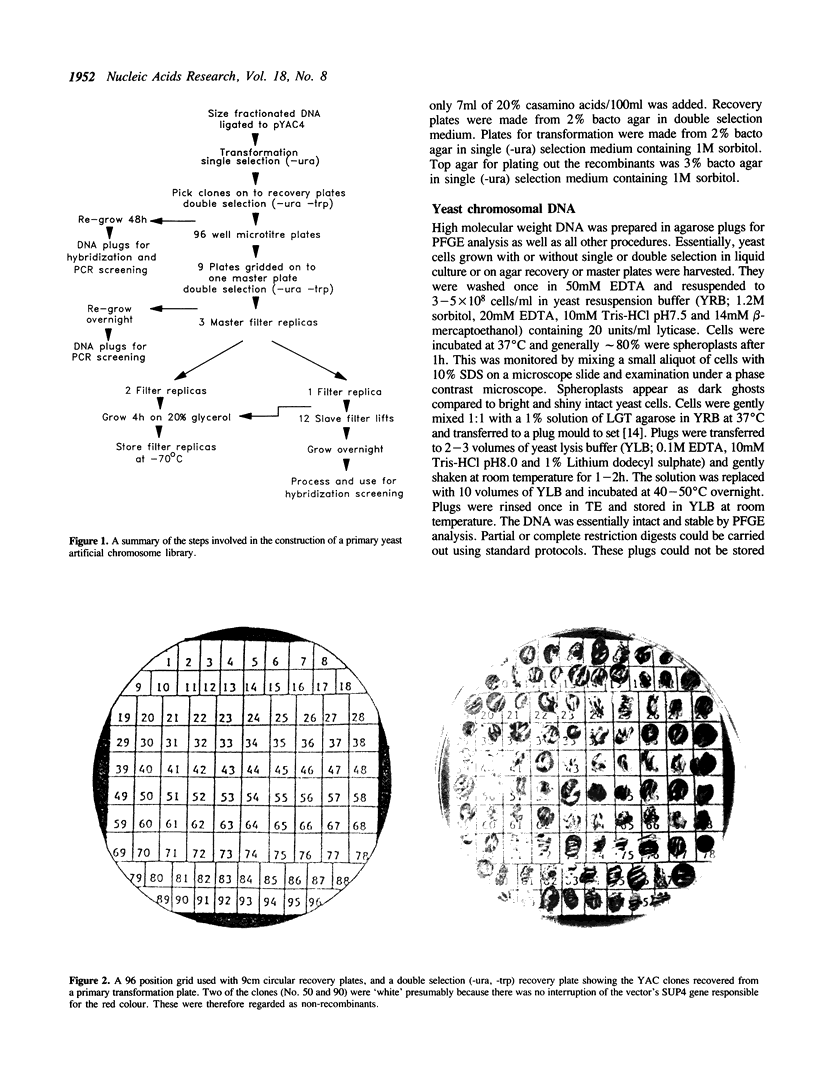
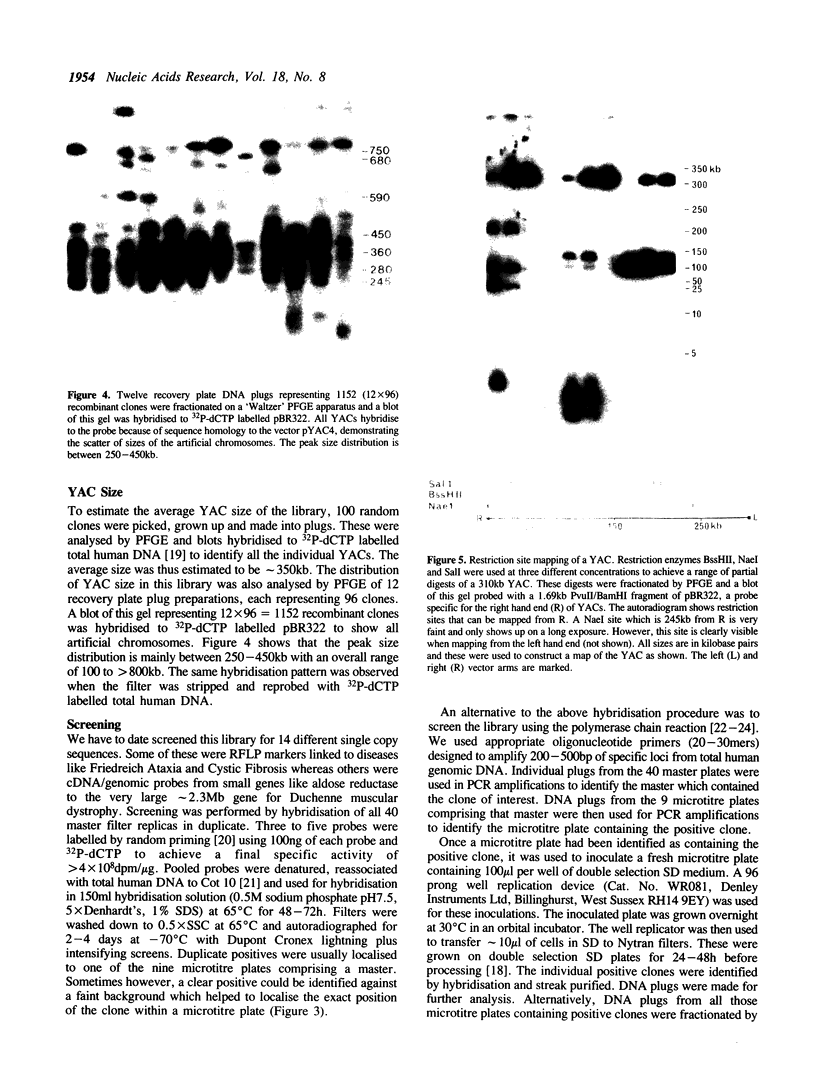
The construction of a yeast artificial chromosome (YAC) primary gridded library of 35,000 clones from human lymphoblastoid (48,XXXX) cell line DNA is described. The average YAC size is approximately 350kb representing a greater than 3.5 times coverage of the genome. The library is stored at -70 degrees C as gridded clones on nylon filters impregnated with 20% glycerol and as glycerol suspensions of individual clones in microtitre plates providing a prolonged multi-user potential. To date we have used 14 single copy probes to screen this library by colony hybridisation as well as PCR and have isolated between 1 and 5 YAC clones for every probe.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anand R., Villasante A., Tyler-Smith C. Construction of yeast artificial chromosome libraries with large inserts using fractionation by pulsed-field gel electrophoresis. Nucleic Acids Res. 1989 May 11;17(9):3425–3433. doi: 10.1093/nar/17.9.3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownstein B. H., Silverman G. A., Little R. D., Burke D. T., Korsmeyer S. J., Schlessinger D., Olson M. V. Isolation of single-copy human genes from a library of yeast artificial chromosome clones. Science. 1989 Jun 16;244(4910):1348–1351. doi: 10.1126/science.2544027. [DOI] [PubMed] [Google Scholar]
- Burgers P. M., Percival K. J. Transformation of yeast spheroplasts without cell fusion. Anal Biochem. 1987 Jun;163(2):391–397. doi: 10.1016/0003-2697(87)90240-5. [DOI] [PubMed] [Google Scholar]
- Burke D. T., Carle G. F., Olson M. V. Cloning of large segments of exogenous DNA into yeast by means of artificial chromosome vectors. Science. 1987 May 15;236(4803):806–812. doi: 10.1126/science.3033825. [DOI] [PubMed] [Google Scholar]
- Collins F. S., Weissman S. M. Directional cloning of DNA fragments at a large distance from an initial probe: a circularization method. Proc Natl Acad Sci U S A. 1984 Nov;81(21):6812–6816. doi: 10.1073/pnas.81.21.6812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulson A., Waterston R., Kiff J., Sulston J., Kohara Y. Genome linking with yeast artificial chromosomes. Nature. 1988 Sep 8;335(6186):184–186. doi: 10.1038/335184a0. [DOI] [PubMed] [Google Scholar]
- Dawkins H. J. Large DNA separation using field alternation agar gel electrophoresis. J Chromatogr. 1989 Aug 11;492:615–639. doi: 10.1016/s0378-4347(00)84481-3. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Guzmán P., Ecker J. R. Development of large DNA methods for plants: molecular cloning of large segments of Arabidopsis and carrot DNA into yeast. Nucleic Acids Res. 1988 Dec 9;16(23):11091–11105. doi: 10.1093/nar/16.23.11091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleppe K., Ohtsuka E., Kleppe R., Molineux I., Khorana H. G. Studies on polynucleotides. XCVI. Repair replications of short synthetic DNA's as catalyzed by DNA polymerases. J Mol Biol. 1971 Mar 14;56(2):341–361. doi: 10.1016/0022-2836(71)90469-4. [DOI] [PubMed] [Google Scholar]
- Nelson D. L., Ledbetter S. A., Corbo L., Victoria M. F., Ramírez-Solis R., Webster T. D., Ledbetter D. H., Caskey C. T. Alu polymerase chain reaction: a method for rapid isolation of human-specific sequences from complex DNA sources. Proc Natl Acad Sci U S A. 1989 Sep;86(17):6686–6690. doi: 10.1073/pnas.86.17.6686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiki R. K., Gelfand D. H., Stoffel S., Scharf S. J., Higuchi R., Horn G. T., Mullis K. B., Erlich H. A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988 Jan 29;239(4839):487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Schwartz D. C., Cantor C. R. Separation of yeast chromosome-sized DNAs by pulsed field gradient gel electrophoresis. Cell. 1984 May;37(1):67–75. doi: 10.1016/0092-8674(84)90301-5. [DOI] [PubMed] [Google Scholar]
- Sealey P. G., Whittaker P. A., Southern E. M. Removal of repeated sequences from hybridisation probes. Nucleic Acids Res. 1985 Mar 25;13(6):1905–1922. doi: 10.1093/nar/13.6.1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M., Anand R., Brown W. R., Fletcher D. S. A model for the separation of large DNA molecules by crossed field gel electrophoresis. Nucleic Acids Res. 1987 Aug 11;15(15):5925–5943. doi: 10.1093/nar/15.15.5925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Traver C. N., Klapholz S., Hyman R. W., Davis R. W. Rapid screening of a human genomic library in yeast artificial chromosomes for single-copy sequences. Proc Natl Acad Sci U S A. 1989 Aug;86(15):5898–5902. doi: 10.1073/pnas.86.15.5898. [DOI] [PMC free article] [PubMed] [Google Scholar]