Abstract
Myotonic dystrophy type 1 (DM1) is an RNA-dominant disease caused by abnormal transcripts containing expanded CUG repeats. The CUG transcripts aggregate in the nucleus to form RNA foci and lead to nuclear depletion of Muscleblind-like 1 (MBNL1) and stabilized expression of CUGBP Elav like family 1 (CELF1), both of which are splicing regulatory proteins. The imbalance of these proteins results in misregulation of alternative splicing and neuromuscular abnormalities. Here, we report the use of antisense oligonucleotides (ASOs) as a therapeutic approach to target the pathogenic RNA in DM1. We designed chimeric ASOs, termed gapmers, containing modified nucleic acid residues to induce RNase H-mediated degradation of CUG-repeat transcripts. The gapmers selectively knockdown expanded CUG transcripts and are sufficient to disrupt RNA foci both in cell culture and mouse models for DM1. Furthermore, combination of gapmers with morpholino ASOs that help release binding of MBNL1 to the toxic RNA can potentially enhance the knockdown effect. Additional optimization will be required for systemic delivery; however, our study provides an alternative strategy for the use of ASOs in DM1 therapy.
Keywords: microsatellite expansion, muscular dystrophy, phosphorothioate, gapmer
Myotonic dystrophy (DM) is the most common muscular dystrophy in adults, affecting 1 in 8,000 individuals. It is a multisystemic disease that affects mainly the skeletal muscle, heart, and central nervous system (1). DM1 patients have CTG trinucleotide repeat expansions (>50–3,000 repeats) in the 3′ untranslated region (3′ UTR) of the DMPK gene (2). The predominant cause of DM1 pathogenesis is the gain-of-function of mutant DMPK mRNA, which contains long CUG repeats that accumulate in the nuclei as RNA foci (3). Two known pathways contribute to DM1 pathogenesis. First, the CUG repeats sequester an RNA-binding protein, Muscleblind-like 1 (MBNL1), resulting in its depletion and loss of function (4). Second, the repeat RNA induces PKC-mediated phosphorylation of CUGBP Elav like family 1 (CELF1), resulting in increased stability and gain-of-function (5). MBNL1 and CELF1 are antagonistic regulators of alternative splicing and the imbalance of their activities results in abnormal expression of embryonic splice variants in adult tissue, some of which contribute to the pathogenesis of the disease (6).
Increased understanding of DM1 pathogenesis has led to therapeutic approaches including utilization of antisense oligonucleotides (ASOs), which can be used to block gene expression by steric hindrance or to elicit RNase H-mediated cleavage of the target RNA (7). RNase H is a non-sequence-specific enzyme that recognizes RNA–DNA heteroduplexes and specifically cleaves the RNA strand (8). By introducing ASOs complementary to a specific RNA sequence, RNase H can mediate cleavage and decay of the target RNA (9). The stability and efficiency of the ASO can be enhanced by substituting DNA with modified nucleotides with increased affinity for RNA and resistance to nucleases, including locked nucleic acids (LNA) or 2′-O-Methoxyethyl (MOE) nucleic acids. These modified nucleic acids are not recognized by RNase H; therefore, a center “gap” region with 7–10 nucleotides containing RNase H-compatible phosphorothioate (PS) DNA is required (10, 11).
There have been several reports applying ASOs for potential DM1 therapy. Wheeler et al. used morpholino ASOs that bind to the toxic CUG repeats, blocking sequestration of Mbnl1 and rescuing its loss-of-function (12). Another group used 2′-O-methyl (2’-OMe) phosphorothioate modified ASOs that reduced levels of the toxic CUG mRNA through unknown mechanisms that do not involve RNase H (13). Here we report a study to target degradation of toxic RNA in DM1 specifically through an RNase H-mediated mechanism. We generated gapmer ASOs with CAG repeat sequences that are sufficient to reduce expanded CUG transcripts and RNA foci in both cell culture and mouse models of DM1. Importantly, the gapmers preferentially target expanded CUG repeats compared with normal-size repeats. We also found that combined administration of gapmers with ASOs that displace proteins from the toxic RNA can enhance the knockdown effect. Our study provides an additional approach for DM1 therapy and may be applied to other RNA diseases.
Results
RNase H-Mediated Degradation of Expanded CUG Repeats in Cell Culture.
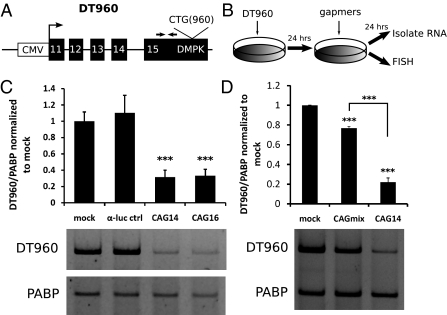
To determine whether ASOs can be used to induce RNase H-mediated degradation of CUG repeat RNA, we designed gapmer ASOs containing CAG sequences with 3–4 LNA or MOE nucleotides on the flanking ends and 8–10 PS nucleotides in the center region (Table 1). We first tested the gapmers in COSM6 cells transiently transfected with a plasmid (DT960) containing DMPK exons 11–15, with 960 interrupted CTG repeats in exon 15 (Fig. 1 A and B) (14). We found that both CAG gapmers with 14 nucleotides (LNA-CAG14) and 16 nucleotides (LNA-CAG16) resulted in a 70% decrease in DT960 mRNA by RT-PCR analysis (Fig. 1C).
Table 1.
Antisense oligonucleotide sequences and modifications
| ASO name | Sequence | Modifications | Length (nt) |
| LNA-CAG14 | AGC AGCAGCAG CAG | LNA/PS | 14 |
| LNA-CAG16 | CAG CAGCAGCAGC AGC | LNA/PS | 16 |
| GAC14 | ACG ACGACGAC GAC | LNA/PS | 14 |
| α-luc | TTC CCGTCATCGT CTTT | LNA/PS | 17 |
| MOE-CAG14 | AGC AGCAGCAG CAG | MOE/PS | 14 |
| MOE-CTG14 | TGC TGCTGCTG CTG | MOE/PS | 16 |
| CAGmix | AG CAG CA G CA G CAG | MOE/PS | 14 |
| morCAG13 | AGCAGCAGCAGCA | morpholino | 13 |
| morCAG25 | AGCAGCAGCAGCAGCAGCAGCAGCA | morpholino | 25 |
Phosphorothioate (PS) nucleotides are in bold, locked nucleic acid (LNA) or 2′-O-Methoxyelthyl (MOE) nucleotides are underlined.
Fig. 1.
RNase H-mediated degradation of expanded DMPK transcript in cell culture. (A) Diagram of the DT960 minigene construct. The DT960 minigene contains the human DMPK genomic segment with exons 11–15 and 960 interrupted CTG repeats expressed by a CMV promoter/enhancer. Primer pairs for RT-PCR [E15upF (forward) and E15upR (reverse)] are located in exon 15 upstream of the repeats (indicated by arrows). (B) Schematic of the experimental strategy. DT960 plasmid was transiently transfected into COSM6 cells, followed by transfection of gapmers 24 h later. RNA isolation or fluorescence in situ hybridization (FISH) was performed the next day. (C) (Upper) Standard RT-PCR showed 70% reduction of DT960 RNA in cells treated with 50 nM of LNA-CAG14 or LNA-CAG16 gapmers. A gapmer complementary to luciferase (α-luc) served as control. Poly-A binding protein (PABP) was used as internal control. The results of four independent transfection experiments were averaged (***P < 0.001). (Lower) Representative gel image used for quantification. (D) (Upper) Standard RT-PCR results. MOE-CAGmix treatment resulted in 25% decrease of the DT960 RNA, whereas the MOE-CAG14 gapmer induced 80% reduction compared with mock. The data represent the average of three independent transfection experiments (***P < 0.001). (Lower) Representative gel image used for quantification.
We designed the gapmers for RNase H accessibility; however, previous studies have shown that ASOs can also initiate degradation through RNase H-independent pathways (12, 13). To determine whether the effect of the CAG gapmers is dependent on accessibility to RNase H, we tested a CAG “mixmer” (CAGmix) containing MOE and PS modifications that do not form a gap of more than 3 nucleotides, preventing binding of RNase H (15). We found that at the same concentration (50 nM), MOE-CAGmix decreased the DT960 transcript by 25%, but was significantly less efficient than the MOE-CAG14 gapmer that resulted in 80% reduction (Fig. 1D). Thus, part of the reduction may result from RNase H-independent mechanisms, but the majority of the knockdown effect involves RNase H.
CAG Gapmers Are Sufficient to Disrupt RNA Foci in Cell Culture.
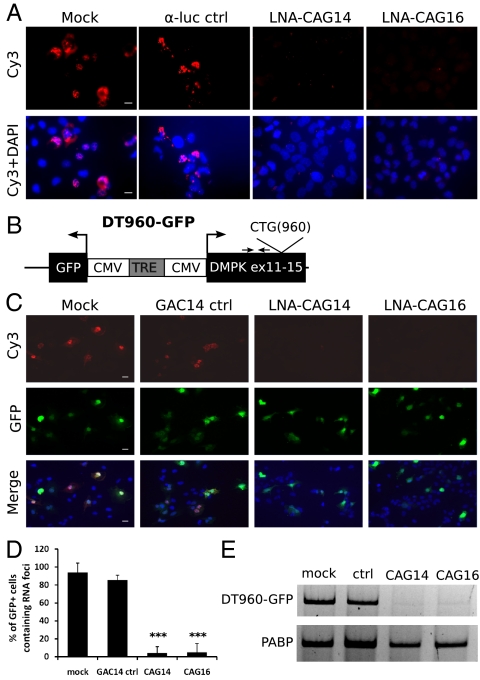
Mutant DMPK transcripts containing long CUG repeats bind specific proteins, including MBNL1, and accumulate in nuclear RNA foci. This is a main characteristic in DM1 patient cells and is also recapitulated in cell culture systems (3). To determine whether CAG gapmers are able to disrupt RNA foci in cells, we performed fluorescent in situ hybridization (FISH) using Cy3-labeled probes that recognize CUG-containing RNA foci. We found a significant reduction of RNA foci in cells treated with LNA-CAG14 or LNA-CAG16 gapmers, whereas a control LNA gapmer against luciferase (α-luc) had no effect (Fig. 2A). To confirm this result, we designed a DT960-GFP plasmid that uses a tet-inducible bidirectional promoter to express both the expanded DMPK transcript and GFP (Fig. 2B). By expressing both GFP and DT960 RNA from the same plasmid, we were able to examine whether GFP positive cells contain RNA foci with or without administration of CAG gapmers. In the absence of gapmers, 95% of GFP-positive cells were found to have RNA foci. In cells treated with the CAG gapmers, there was a significant decrease in the number of GFP positive cells containing RNA foci, which was verified by quantification (Fig. 2 C and D). RT-PCR confirmed that the expanded CUG transcripts were indeed degraded (Fig. 3E). Taken together, these results indicate that CAG gapmers are sufficient to disrupt CUG repeat-containing RNA foci.
Fig. 2.
Disruption of RNA foci by CAG gapmers in cell culture. (A) RNA foci containing DT960 RNA were detected by FISH using Cy3-labeled probes. Nuclei were counterstained with DAPI (Lower). All images were taken at the same exposure time. (Scale bars: 20 μm.) (B) Diagram of the DT960-GFP construct. The expression of DMPK RNA and GFP are controlled by a bidirectional tetracycline responsive element (TRE), which is activated by the transactivator in the presence of doxycycline (TetON). Primers (E15upF and E15upR) for RT-PCR are indicated with arrows. (C) FISH was performed on cells transfected with transfection reagent only (mock), GAC14 control gapmer, LNA-CAG14 and LNA-CAG16. All images were taken at the same exposure time. (Scale bars: 20 μm.) (D) Bar graph represents average percent of GFP+ cells containing RNA foci. For the results, ≥7 microscopic fields and a total of >90 cells were counted from three independent transfection experiments. (***P < 0.001). (E) Reduction of DT960 transcript was confirmed by standard RT-PCR. Poly-A binding protein (PABP) was used as internal control.
Fig. 3.
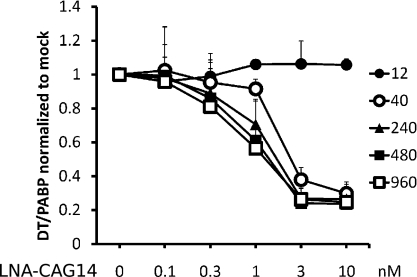
CAG gapmers preferentially degrade RNAs containing expanded CUG repeats. Cells expressing identical DMPK transcripts except containing 12, 40, 240, 480, or 960 CUG repeats were treated with increased dosage (0, 0.1, 0.3, 1, 3, 10 nM) of LNA-CAG14 gapmer. LNA-CAG14 had no effect on RNA containing 12 CUG repeats. DMPK transcripts containing longer repeats are affected at lower concentrations. Data represent average of three independent experiments.
CAG Gapmers Specifically Target Expanded CUG Repeats.
DM1 is an autosomal dominant disease in which only one allele contains the expanded CTG repeats, whereas the other allele contains nonpathogenic repeats of 5–38 CTGs. An ideal therapy will discriminate between RNA from mutant and wild-type alleles, and preferentially target the former. Targeting RNA from mutant alleles will prevent loss of DMPK protein, which could result in detrimental consequences (16). To determine whether CAG gapmers discriminate between expanded and nonexpanded RNA transcripts, we transiently transfected cells expressing DMPK mRNAs containing from 12 to 960 CUG repeats and administered increasing concentrations of the LNA-CAG14 gapmer. We found that up to the highest concentration of LNA-CAG14 gapmer tested (10 nM), there was no effect on levels of RNA containing 12 CUG repeats, which is the average number of repeats among non-DM1 individuals (Fig. 3) (17). For RNA containing 40 CUG repeats, the gapmer starts to have a significant knockdown at 3 nM (P < 0.001); whereas for 240 and 480 repeats, the knockdown is significant at 1 nM (P < 0.05). For RNA containing 960 repeats, significant knockdown is achieved at as low as 0.3 nM (P < 0.01) (Fig. 3). Thus, DMPK transcripts containing longer repeats are affected at lower concentrations. The data suggest that the CAG gapmers can potentially target expanded CUG repeats compared with normal length repeats.
CAG Gapmers Induce Degradation of Expanded DMPK Transcripts in Skeletal Muscle From a DM1 Mouse Model.
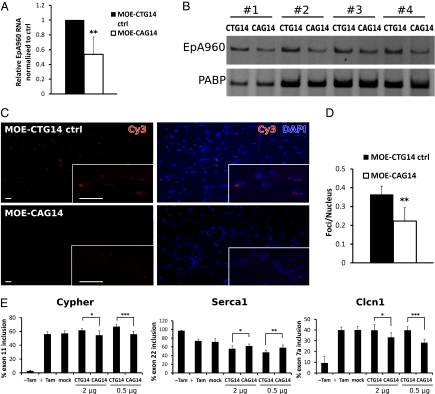
To assess the efficacy of CAG gapmers in vivo, we tested them in our EpA960/HSA-Cre DM1 mouse model (18). The EpA960 transgene contains the human DMPK exon 15 including 960 interrupted CTG repeats. To inducibly express the transgene specifically in skeletal muscle, EpA960 mice are crossed with HSA-CreERT2 mice (19) and bitransgenic animals were mated to generate double homozygous EpA960/HSA-Cre mice. Mice 2–3 mo of age were injected with tamoxifen to induce expression of the mRNA containing 960 CUG repeats. At least 2 wk postinduction, MOE-CAG14 gapmers (2 μg) were injected into the tibialis anterior (TA) muscle of one hind limb, followed by in vivo electroporation. The TA muscle of the contralateral hind limb was injected with MOE-CTG14 gapmer as control and electroporated following the same procedure. The level of EpA960 mRNA was determined by real-time RT-PCR 2 wk after gapmer administration. We observed a 50% decrease in the level of EpA960 transcript in the TA muscle treated with MOE-CAG14 compared with the CTG control (Fig. 4A). This result was confirmed by standard RT-PCR assays (Fig. 4B). Consistent with the EpA960 transcript level, we observed a 40% decrease in the average number of RNA foci per nuclei by FISH (Fig. 4 C and D). This result shows that CAG gapmers can induce degradation of expanded DMPK transcripts and disrupt RNA foci in DM1 mice.
Fig. 4.
CAG gapmer administration reduces expanded CUG RNA levels and aberrant splicing in a DM1 mouse model. (A) Real-time RT-PCR indicates a 50% decrease of the relative EpA960 transcript level in the TA muscle 2 wk after administration of MOE-CAG14 compared with the MOE-CTG14 control (n = 6 mice, triplicate assays per sample, **P < 0.01). β-actin was used as the internal control. (B) Standard RT-PCR shows a decrease in EpA960 transcript in muscle treated with MOE-CAG14 compared with the MOE-CTG14 control. Representative results from four treated mice (numbered) are shown. (C) Fewer RNA foci were detected in muscle treated with MOE-CAG14 gapmer compared MOE-CTG14 control. Nuclei were stained using DAPI (Right). All images were taken at the same exposure time. (Inset) Higher magnification of foci. (Scale bars: 20 μm.) (D) Quantification reveals 40% reduction of the number of foci per nucleus in MOE-CAG14 muscle compared with control. Five microscopic fields were counted per muscle. Bar graph represents the average of [number of foci/number of nuclei] each field (n = 5 mice). (E) RT-PCR quantification of inclusion of alternative exons from three misregulated splicing events in DM1. Lanes are as follows: EpA960/HSA-Cre mice not injected with tamoxifen, n = 3 (-Tam); untreated mice 2 wk post tam, n = 4 (+Tam); mock-treated mice 2 wk post tam, n = 4 (mock); tamoxifen-induced EpA960/HSA-Cre mouse muscle injected with MOE-CTG14 control (2 μg), n = 6 ; MOE-CAG14 (2 μg), n = 6; MOE-CTG14 control (0.5 μg), n = 7; MOE-CAG14 (0.5 μg), n = 7 (*P < 0.05, **P < 0.01, ***P < 0.001).
To assess the downstream effect of expanded CUG transcript degradation on aberrant splicing, we examined the alternative splicing of three DM1-associated splicing events: Cypher, Serca1, and Clcn1 (20–22). At time points more than 2 wk after tamoxifen injection, all three events reverted toward the embryonic splicing pattern as expected (Fig. 4E, +Tam). MOE-CAG14 administration (2 μg) partially reversed the embryonic splicing isoforms toward adult splicing isoforms compared with the MOE-CTG14 control, consistent with the reduction of EpA960 transcripts. Interestingly, however, treatment with a lower dose (0.5 μg) of MOE gapmers had a slightly stronger effect on splicing (Fig. 4E) without reduction of EpA960 RNA (Fig. S1), implicating other mechanisms besides reduction of repeat RNA that could contribute to the splicing change. Muscle injected with PBS and electroporated with the same procedures (mock) did not show splicing changes compared with untreated posttamoxifen mice (+Tam). Surprisingly, the splicing of Cypher and Serca1 shifted more toward the embryonic pattern in the muscle injected with the MOE-CTG14 control. This result suggested that electroporation of the MOE gapmers induced muscle damage and regeneration (see below), which causes a splicing switch toward embryonic patterns (23).
CAG Gapmers Induce Histological Abnormalities in Mouse Skeletal Muscle.
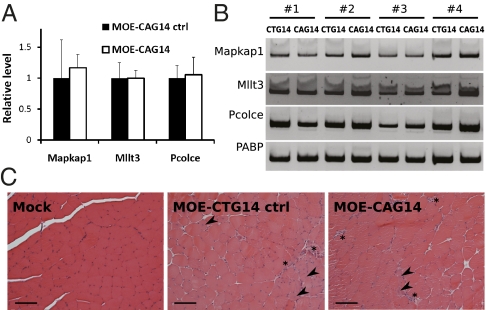
We have shown that CAG gapmers reduce the level of pathogenic CUG RNA in cell culture and mouse skeletal muscle. We next tested whether there are off-target effects on endogenous transcripts containing CUG repeats. There are at least eleven genes in mice that express mRNAs containing ≥ 6 CUG repeats (12, 13). To determine the nontarget effect of the gapmers, we examined the mRNA levels of three genes: Mapkap1, Mllt3 and Pcolce, which contain 26, 8 and 12 CUG repeats respectively. Real-time and standard RT-PCR showed no significant difference in the level of these transcripts in muscle treated with MOE-CTG14 control or MOE-CAG14 (Fig. 5 A and B). This result is consistent with our cell culture experiments, where the CAG gapmers preferentially target the longer repeats.
Fig. 5.
Secondary effects of CAG gapmers. (A) Real-time RT-PCR of three gene transcripts containing ≥8 CUG repeats. RNA levels are expressed as arbitrary units relative to β-actin, then normalized to the mean expression of control muscle. (n = 4 mice, triplicate assays per sample, P > 0.5 for each). (B) No consistent difference in expression level was seen by standard RT-PCR. Data from four mice is shown. (C) Hematoxylin and eosin staining of TA muscle cross-sections. Central nuclei (arrowheads) and regions with multiple nuclei (asterisks) are present in muscle treated with MOE-CTG14 control and MOE-CAG14 gapmers. (Scale bar = 100 μm.)
To further investigate secondary effects of the CAG gapmers in mouse skeletal muscle, we examined the histology of the TA muscle 2 wk postgapmer administration. PBS-treated muscle (mock) appeared essentially normal indicating that neither tamoxifen nor the injection/electroporation procedure induced persistent histopathology. However, muscles treated with either the MOE-CTG14 control gapmer or MOE-CAG14 showed focal regions with central nuclei and small myofibers containing multiple nuclei (Fig. 5C). The histological abnormalities appear to reflect muscle damage and regeneration induced by gapmer administration following electroporation, which explains the enhanced embryonic splicing patterns induced by the CTG control gapmer (Fig. 4E). The fact that both the MOE-CTG and MOE-CAG14 gapmers induce regeneration suggests that it is the gapmer chemistry that is toxic rather than the specific sequence. In addition, the MOE-CAG14 gapmer also induces regeneration, strongly suggesting that their effects to reverse the embryonic pattern by degradation of expanded CUG RNA is stronger than reflected in Fig. 4 because a component of the embryonic pattern is due to regeneration in addition to expression of expanded CUG RNA. We conclude that CAG gapmers do not predominantly target endogenous CUG transcripts; however, both CTG and CAG gapmers induce some level of muscle damage when electroporated into muscle tissue.
Combined Administration of CAG Morpholinos and Morpholinos Enhance CUG RNA Knockdown.
A previous study demonstrated that CAG-containing morpholinos (CAG25) bind to expanded CUG RNA, block sequestration of MBNL1, and reverse molecular features caused by MBNL1 loss-of-function. Morpholinos are not recognized by RNase H; however, CAG25 caused a 50% reduction of CUG repeat RNA presumably by releasing RNA from foci and enhancing its degradation (12).
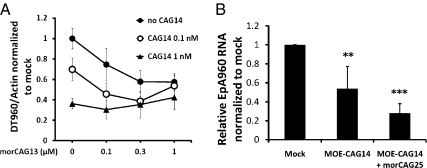
We wanted to determine whether morpholinos could enhance the effect of the CAG gapmers through release of the expanded CUG RNA from nuclear foci. DT960-transfected cells were exposed to mixtures of LNA-CAG14 gapmer combined with increasing concentrations of a CAG morpholino containing 13 nucleotides (morCAG13). morCAG13 alone was able to induce 50% decrease in the level of DT960 RNA at 0.3 μM, consistent with what has been reported (12). An enhanced knockdown effect was achieved when 0.1 and 0.3 μM of morpholino was added to 0.1 nM of gapmer compared with cells treated with gapmer only. However, this enhancement was compromised when the morpholino concentration increased to 1 μM (Fig. 6A). One explanation is that the CAG morpholino competes with the CAG gapmers for the same CUG binding sites, therefore reducing the RNase H-mediated knockdown by gapmers at higher concentrations. When the gapmer concentration increases to 1 nM, addition of morpholino no longer enhances degradation. Note that the concentration of morpholino tested is roughly 1,000 times that of the gapmer, indicating that much lower amount of gapmer is required for significant knockdown.
Fig. 6.
Combined effect of CAG gapmers and morpholinos. (A) Following expression of DT960 RNA in COSM6 cells, LNA-CAG14 gapmer (0, 0.1, 1 nM) was transfected into COSM6 cells with increasing doses (0, 0.1, 0.3, 1 μM) of CAG morpholino containing 13 nucleotides (morCAG13). β-actin was used as internal control. (B) Real-time RT-PCR revealed enhanced decrease of EpA960 transcript level when MOE-CAG14 (2 μg) was combined with morCAG25 (20 μg). EpA960 transcript levels from PBS-treated muscle served as control (mock). n = 4–6 mice per group, triplicates were done per sample. β-actin was used as internal control. (**P < 0.01, ***P < 0.001).
We then treated EpA960/HSA-Cre mice with a combination of MOE-CAG14 (2 μg) and morCAG25 (20 μg) to determine whether a similar synergistic effect occurs in vivo. Although MOE-CAG14 by itself induced 50% decrease in the level of EpA960 transcript, addition of morCAG25 further enhanced the reduction to 75% (Fig. 6B). These results indicate that combining ASOs that help release CUG transcripts from nuclear foci, with gapmers that target the CUGs for degradation, can have a synergistic effect at certain concentrations.
Discussion
The main goal of this study was to target degradation of expanded CUG repeats in DM1 by using gapmer ASOs. We designed gapmers with CAG repeat sequences containing LNA and MOE modifications, both of which are resistant to nucleases and have increased affinity to RNA (24, 25). The gapmers were sufficient to knockdown the mutant CUG transcripts both in cell culture and DM1 mice, providing proof of principle that the RNase H pathway may be exploited for use in DM1 therapy.
We showed that the CAG gapmers are able to preferentially target long CUG repeats, which is likely to be due to the increased availability of hybridization sites with longer expansions as well as preferential localization of expanded transcripts in the nucleus where RNase H is localized. 2′-OMe ASOs have also been shown to selectively target expanded CUG repeats in DM1 myoblasts (13). Another study showed that ASOs used to inhibit translation of CAG expansions in Huntington disease only affected the expanded huntingtin allele but not the wild-type allele (26). This provides evidence that the ability to preferentially target expanded repeat sequence may be common to other ASO therapies.
We found that CAG gapmers reduce expanded DMPK transcripts by 80% and almost eliminate RNA foci in cell culture. However, a significant but smaller reduction (50%) was obtained in mouse skeletal muscle. Possible reasons for the decreased performance in mice include lower efficiency of delivery and variable localization of gapmers in muscle tissue. We found that fluorescently tagged ASOs transfected into cultured cells localize predominantly in the nucleus, consistent with previous results (27). Moreover, 2′-OMe ASOs injected in mouse muscle are concentrated in nuclei (13). It seems likely that a large fraction of the gapmers retained in muscle enter the nucleus given the 50% reduction of CUG RNA. In addition, CAG oligos can form self-structures and it is likely ASOs designed to have decreased propensity to form structure will increase availability to bind to target RNA.
Although the CAG gapmers reduced the level of toxic CUG RNA in skeletal muscle tissue by half, we observed only a slight reversal of Cypher, Serca1, and Clcn1 splicing when comparing CAG and CTG control gapmers. It is possible that 50% reduction does not surpass a threshold needed to fully rescue these splicing events. However, it is also likely that the regenerative response induced by the gapmers obfuscated the reversal of the embryonic splicing pattern. We have recently shown that muscle regeneration produces an adult-to-embryonic switch in splicing (23). Therefore, the rescue of splicing might have been mitigated by production of the embryonic splicing pattern in regenerating fibers. On the other hand, we found that a lower dose of CAG gapmer treatment can result in reversal of splicing even without reduction of toxic RNA. This finding suggests that the gapmers may work through other RNase-H independent mechanisms to affect downstream splicing events, perhaps through displacement of MBNL1 (15).
There are at least two possible explanations of how ASO treatment may result in muscle damage: (i) Sequence-specific off-target effects, which seem unlikely because both CTG and CAG gapmers resulted in similar histological changes. (ii) Sequence-dependent aptameric properties: ASOs with CAG and CTG repeats are predicted to form higher order structures which may be more immunostimulatory or induce aptameric effects (28, 29). Our CAG gapmers contain MOE modifications which are known to decrease the immunostimulatory effects (30), but the central region is not MOE-modified. On the other hand, MOE modified nucleotides prevent the ASO from degradation, which may enhance the toxic properties when electroporated into cells. There is limited knowledge of adverse effects of chemically modified ASOs in muscle tissue and side effects can be minimized with future development of antisense technology.
We also tested an approach of combined administration of RNase H-active gapmers with RNase H-inactive morpholinos in cell culture and mouse models for DM1. We found that CAG morpholinos could enhance the knockdown by CAG gapmers at specific concentration ratios. However, once the morpholino concentration exceeds a threshold, degradation is inhibited. This effect is expected because the morpholinos and gapmers compete for the CUG repeats and the high concentrations of morpholino are likely to displace the gapmers, inhibiting RNase H accessibility. A synergistic effect of morpholinos and gapmers was also observed in mice. A potential alternative approach is to use combined administration of CAG morpholinos with gapmers targeting different sequences within the DMPK mRNA to avoid binding competition. In addition, several reports have identified small molecules and peptides with the ability to disrupt RNA foci formation (31, 32). This opens up the possibility of combining ASOs with other nonantisense strategies for DM1 therapy.
We did not test systemic delivery in this study due to toxicity concerns; however, it is possible that the toxicity we observed was secondary to an inflammatory response stimulated by the ASO combined with tissue damage due to the method of delivery. Different routes of administration as well as different chemistries will need to be tested to lower toxicity and improve efficacy. There are several systemically administered RNase H-based antisense drugs in clinical trials, suggesting bright prospects in the future (10). The growing interest on therapeutic ASOs will accelerate development of novel nucleotide modifications and delivery methods addressing issues of toxicity and bio-distribution.
Materials and Methods
Oligonucleotides.
LNA gapmers were purchased from Exiqon. MOE gapmers were obtained from ISIS Pharmaceuticals. CAG morpholinos were purchased from Gene Tools.
Cell Culture and Transfection.
COSM6 cells were plated in six-well plates containing DMEM supplemented with 10% certified FBS and 1% l-glutamine (all from Gibco). Twenty-four h after plating, 1 μg of CTG repeat plasmid was transfected per well using Fugene6 (Roche). The next day, indicated concentrations of gapmers were transfected using Lipofectamine 2000 (Invitrogen). Morpholinos were transfected using the Neon transfection system (Invitrogen).
Transgenic Mice and ASO Injection.
Animal experiments were approved by the Institutional Animal Care and Use Committee of Baylor College of Medicine. EpA960 and HSA-Cre-ERT2 mice were used to generate double homozygous EpA960/HSA-Cre mice (18). Mice were injected daily for 5 consecutive days with 1 mg of tamoxifen. At least 2–4 wk after tamoxifen injection, mice were anesthetized by i.p. injection of Avertin (0.5 mg/g weight). The tibialis anterior (TA) muscle was pretreated with bovine hyaluronidase (Sigma-Aldrich) for 1–2 h and the indicated amount of ASO was injected intramuscularly followed by in vivo electoporation (100 V/cm, 10 pulses at 1 Hz, 20-ms duration per pulse).
Fluorescence in Situ Hybridization (FISH).
Cells were fixed in 4% paraformaldehyde and permeabilized with 0.02% Triton X-100 in PBS. Mouse muscle tissues were fixed overnight in 10% formalin. Tissues were then paraffin-embedded and cut in cross-section at 10 μm. CUG transcripts were detected using (CAG)5-Cy3-labeled LNA probes (Exiqon) as described (18). The nuclei were stained with DAPI using Vectashield (Vector).
RT-PCR.
Total RNA was isolated from skeletal muscle using TRIzol (Invitrogen). cDNA was generated from 4 μg of RNA using oligo dT and AMV reverse transcriptase (Life Science). To assay alternative splicing events, flanking primer pairs were designed for Clcn1, Serca1, and Cypher. See Table S1 for oligo sequences. PCR products were separated on 5% nondenaturing polyacrylamide gels and quantified using the Kodak Gel Logic 2200 and Molecular Imaging software.
Real-Time RT-PCR.
Taqman primers were used to quantify EpA960 repeat recombined allele mRNA products as described (18). Primer pairs for Mapkap1, Mllt3 and Pcolce were purchased from Applied Biosystems. Real-time RT-PCR was performed on the ABI Prism 7000 sequence detection system. All samples were normalized to β-actin (Applied Biosystems part no. 4352341E).
Statistics.
All data are expressed as mean ± SD. Statistical significance was determined using two-tailed Student t test. A P value of less than 0.05 was considered significant.
Supplementary Material
Acknowledgments
We thank Dr. C. Thornton and Dr. T. Wheeler (University of Rochester) for their expertise with the in vivo electroporation experiments, M. Koshelev for initial help with the project, and D. Bundman for technical assistance. All histology was performed by the Center for Comparative Medicine pathology core facility at Baylor College of Medicine, with a special thanks to B. Bhatti. This work was supported by the National Institutes of Health Grant R01AR45653 (to T.A.C.), the Muscular Dystrophy Association (T.A.C.), and the Shanna and Andrew Linbeck Family Charitable Fund.
Footnotes
Conflict of interest statement: C.F.B. is an employee of Isis Pharmaceuticals, Inc., and may materially benefit either directly or indirectly through stock options.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1117019109/-/DCSupplemental.
References
- 1.Harper P.S. Myotonic Dystrophy. London: W.B. Saunders; 2001. [Google Scholar]
- 2.Brook JD, et al. Molecular basis of myotonic dystrophy: expansion of a trinucleotide (CTG) repeat at the 3′ end of a transcript encoding a protein kinase family member. Cell. 1992;68:799–808. doi: 10.1016/0092-8674(92)90154-5. [DOI] [PubMed] [Google Scholar]
- 3.Davis BM, McCurrach ME, Taneja KL, Singer RH, Housman DE. Expansion of a CUG trinucleotide repeat in the 3′ untranslated region of myotonic dystrophy protein kinase transcripts results in nuclear retention of transcripts. Proc Natl Acad Sci USA. 1997;94:7388–7393. doi: 10.1073/pnas.94.14.7388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miller JW, et al. Recruitment of human muscleblind proteins to (CUG)(n) expansions associated with myotonic dystrophy. EMBO J. 2000;19:4439–4448. doi: 10.1093/emboj/19.17.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuyumcu-Martinez NM, Wang GS, Cooper TA. Increased steady-state levels of CUGBP1 in myotonic dystrophy 1 are due to PKC-mediated hyperphosphorylation. Mol Cell. 2007;28:68–78. doi: 10.1016/j.molcel.2007.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee JE, Cooper TA. Pathogenic mechanisms of myotonic dystrophy. Biochem Soc Trans. 2009;37:1281–1286. doi: 10.1042/BST0371281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bennett CF, Swayze EE. RNA targeting therapeutics: molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annu Rev Pharmacol Toxicol. 2010;50:259–293. doi: 10.1146/annurev.pharmtox.010909.105654. [DOI] [PubMed] [Google Scholar]
- 8.Stein H, Hausen P. Enzyme from calf thymus degrading the RNA moiety of DNA-RNA Hybrids: effect on DNA-dependent RNA polymerase. Science. 1969;166:393–395. doi: 10.1126/science.166.3903.393. [DOI] [PubMed] [Google Scholar]
- 9.Walder RY, Walder JA. Role of RNase H in hybrid-arrested translation by antisense oligonucleotides. Proc Natl Acad Sci USA. 1988;85:5011–5015. doi: 10.1073/pnas.85.14.5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wahlestedt C, et al. Potent and nontoxic antisense oligonucleotides containing locked nucleic acids. Proc Natl Acad Sci USA. 2000;97:5633–5638. doi: 10.1073/pnas.97.10.5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lima WF, et al. Structural requirements at the catalytic site of the heteroduplex substrate for human RNase H1 catalysis. J Biol Chem. 2004;279:36317–36326. doi: 10.1074/jbc.M405035200. [DOI] [PubMed] [Google Scholar]
- 12.Wheeler TM, et al. Reversal of RNA dominance by displacement of protein sequestered on triplet repeat RNA. Science. 2009;325:336–339. doi: 10.1126/science.1173110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mulders SA, et al. Triplet-repeat oligonucleotide-mediated reversal of RNA toxicity in myotonic dystrophy. Proc Natl Acad Sci USA. 2009;106:13915–13920. doi: 10.1073/pnas.0905780106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Philips AV, Timchenko LT, Cooper TA. Disruption of splicing regulated by a CUG-binding protein in myotonic dystrophy. Science. 1998;280:737–741. doi: 10.1126/science.280.5364.737. [DOI] [PubMed] [Google Scholar]
- 15.Monia BP, et al. Evaluation of 2′-modified oligonucleotides containing 2′-deoxy gaps as antisense inhibitors of gene expression. J Biol Chem. 1993;268:14514–14522. [PubMed] [Google Scholar]
- 16.Jansen G, et al. Abnormal myotonic dystrophy protein kinase levels produce only mild myopathy in mice. Nat Genet. 1996;13:316–324. doi: 10.1038/ng0796-316. [DOI] [PubMed] [Google Scholar]
- 17.Ansved T, Edström L, Grandell U, Hedberg B, Anvret M. Variation of CTG-repeat number of the DMPK gene in muscle tissue. Neuromuscul Disord. 1997;7:152–155. doi: 10.1016/s0960-8966(97)00443-4. [DOI] [PubMed] [Google Scholar]
- 18.Orengo JP, et al. Expanded CTG repeats within the DMPK 3′ UTR causes severe skeletal muscle wasting in an inducible mouse model for myotonic dystrophy. Proc Natl Acad Sci USA. 2008;105:2646–2651. doi: 10.1073/pnas.0708519105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schuler M, Ali F, Metzger E, Chambon P, Metzger D. Temporally controlled targeted somatic mutagenesis in skeletal muscles of the mouse. Genesis. 2005;41:165–170. doi: 10.1002/gene.20107. [DOI] [PubMed] [Google Scholar]
- 20.Kanadia RN, et al. Reversal of RNA missplicing and myotonia after muscleblind overexpression in a mouse poly(CUG) model for myotonic dystrophy. Proc Natl Acad Sci USA. 2006;103:11748–11753. doi: 10.1073/pnas.0604970103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kimura T, et al. Altered mRNA splicing of the skeletal muscle ryanodine receptor and sarcoplasmic/endoplasmic reticulum Ca2+-ATPase in myotonic dystrophy type 1. Hum Mol Genet. 2005;14:2189–2200. doi: 10.1093/hmg/ddi223. [DOI] [PubMed] [Google Scholar]
- 22.Mankodi A, et al. Expanded CUG repeats trigger aberrant splicing of ClC-1 chloride channel pre-mRNA and hyperexcitability of skeletal muscle in myotonic dystrophy. Mol Cell. 2002;10:35–44. doi: 10.1016/s1097-2765(02)00563-4. [DOI] [PubMed] [Google Scholar]
- 23.Orengo JP, Ward AJ, Cooper TA. Alternative splicing dysregulation secondary to skeletal muscle regeneration. Ann Neurol. 2011;69:681–690. doi: 10.1002/ana.22278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kurreck J, Wyszko E, Gillen C, Erdmann VA. Design of antisense oligonucleotides stabilized by locked nucleic acids. Nucleic Acids Res. 2002;30:1911–1918. doi: 10.1093/nar/30.9.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Monia BP, Johnston JF, Sasmor H, Cummins LL. Nuclease resistance and antisense activity of modified oligonucleotides targeted to Ha-ras. J Biol Chem. 1996;271:14533–14540. doi: 10.1074/jbc.271.24.14533. [DOI] [PubMed] [Google Scholar]
- 26.Hu J, et al. Allele-specific silencing of mutant huntingtin and ataxin-3 genes by targeting expanded CAG repeats in mRNAs. Nat Biotechnol. 2009;27:478–484. doi: 10.1038/nbt.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leonetti JP, Mechti N, Degols G, Gagnor C, Lebleu B. Intracellular distribution of microinjected antisense oligonucleotides. Proc Natl Acad Sci USA. 1991;88:2702–2706. doi: 10.1073/pnas.88.7.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gürsel M, Verthelyi D, Gürsel I, Ishii KJ, Klinman DM. Differential and competitive activation of human immune cells by distinct classes of CpG oligodeoxynucleotide. J Leukoc Biol. 2002;71:813–820. [PubMed] [Google Scholar]
- 29.Vollmer J, et al. Characterization of three CpG oligodeoxynucleotide classes with distinct immunostimulatory activities. Eur J Immunol. 2004;34:251–262. doi: 10.1002/eji.200324032. [DOI] [PubMed] [Google Scholar]
- 30.Henry S, et al. Chemically modified oligonucleotides exhibit decreased immune stimulation in mice. J Pharmacol Exp Ther. 2000;292:468–479. [PubMed] [Google Scholar]
- 31.Warf MB, Nakamori M, Matthys CM, Thornton CA, Berglund JA. Pentamidine reverses the splicing defects associated with myotonic dystrophy. Proc Natl Acad Sci USA. 2009;106:18551–18556. doi: 10.1073/pnas.0903234106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.García-López A, Llamusí B, Orzáez M, Pérez-Payá E, Artero RD. In vivo discovery of a peptide that prevents CUG-RNA hairpin formation and reverses RNA toxicity in myotonic dystrophy models. Proc Natl Acad Sci USA. 2011;108:11866–11871. doi: 10.1073/pnas.1018213108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.