Abstract
The mechanisms underlying the biosynthesis of cellulose in plants are complex and still poorly understood. A central question concerns the mechanism of microfibril structure and how this is linked to the catalytic polymerization action of cellulose synthase (CESA). Furthermore, it remains unclear whether modification of cellulose microfibril structure can be achieved genetically, which could be transformative in a bio-based economy. To explore these processes in planta, we developed a chemical genetic toolbox of pharmacological inhibitors and corresponding resistance-conferring point mutations in the C-terminal transmembrane domain region of CESA1A903V and CESA3T942I in Arabidopsis thaliana. Using 13C solid-state nuclear magnetic resonance spectroscopy and X-ray diffraction, we show that the cellulose microfibrils displayed reduced width and an additional cellulose C4 peak indicative of a degree of crystallinity that is intermediate between the surface and interior glucans of wild type, suggesting a difference in glucan chain association during microfibril formation. Consistent with measurements of lower microfibril crystallinity, cellulose extracts from mutated CESA1A903V and CESA3T942I displayed greater saccharification efficiency than wild type. Using live-cell imaging to track fluorescently labeled CESA, we found that these mutants show increased CESA velocities in the plasma membrane, an indication of increased polymerization rate. Collectively, these data suggest that CESA1A903V and CESA3T942I have modified microfibril structure in terms of crystallinity and suggest that in plants, as in bacteria, crystallization biophysically limits polymerization.
Keywords: cell wall, polysaccharide, quinoxyphen
The fundamental unit comprising naturally produced cellulose is the para-crystalline microfibril, which consists of multiple parallel chains of unbranched β-1,4-glucan that aggregate via hydrogen bonds and van der Waals forces. Cellulose-synthesizing organisms typically produce microfibrils from large plasma membrane (PM)-localized cellulose-synthesizing complexes (CSCs) (1). The geometry of CSCs range from either linear arrays of particles with single rows, as seen in the bacterium Gluconacetobacter xylinus [formerly Acetobacter xylinus(m)], to multiple rows, as found in the green alga Oocystis apiculata, to hexagonal arrays of particles termed rosettes that occur in Charophycean algae and all land plants (2). The arrangement of particles within the CSCs of these and other organisms has been correlated with the size and shape of the cellulose microfibrils that they produce (2). A detailed analysis of microfibril structure has revealed the existence of two crystalline allomorphs of naturally occurring cellulose, cellulose Iα (triclinic unit cell) and Iβ (monoclinic unit cell) (3). The elucidation of the crystal structure of both forms has helped determine the hydrogen-bonding network within and between glucan chains as well as between sheets of chains so formed (4, 5). Analysis of higher plant cellulose suggests not only the presence of large numbers of ordered and disordered crystal-surface chains (6), but that the inner chains of the microfibril core are more crystalline, while more amorphous forms of cellulose compose the chains in the surrounding para-crystalline sheath (7). These analyses have raised the question that if the geometry of the CSC determines the lateral dimensions of the microfibril (8), then does it also determine microfibril structure or is this feature instead dictated by glucan chain self-assembly?
Pioneering studies with the bacterium A. xylinum have shown that the fluorescent dye Calcofluor disrupts the crystallization of cellulose by hydrogen bonding to individual glucan chains immediately upon their emergence from the cell, thereby preventing their assembly into a microfibril (9). The disruption of microfibril assembly caused an increase in the rate of glucan chain polymerization, suggesting that the two processes are biophysically coupled and that crystallization is rate limiting (10). However, during plant cellulose biosynthesis, it remains unclear whether a link exists between cellulose crystallization and the CSC function in terms of polymerization. This is in part due to a lack of low-crystallinity cellulose mutants available to explore this postulate. Moreover, in plant cells as opposed to bacteria, the requirement for cellulose production is not dispensable and is linked to the anisotropic expansion of the cell and assaying polymerization after pharmacological decrystallization is complicated by the inherent fluorescence of the β-glucan binding dye Calcofluor (11).
In Arabidopsis, 10 CESA genes have been identified (12). A combination of biochemical and genetic analyses has shown that at least three cellulose synthase (CESA) subunits directly interact to form a CSC during primary (13, 14) and secondary (15) cell wall cellulose formation. The temperature-sensitive mutation (or homozygous allele) rsw1-1 (CESA1) results in primary wall deficiencies accompanied by a disappearance of rosettes from the membrane at the nonpermissive temperature (16), and strong homozygous alleles are either male gametophyte or embryo lethal (14, 17). CESA3 is coexpressed with CESA1, and null homozygous cesa3 alleles are also male gametophyte lethal (14). The remaining primary cell wall CESA genes display a degree of redundancy (13, 14). In addition to CESA, several other genes have been linked to a role in cellulose biosynthesis by null alleles resulting in cellulose deficiency (18). Recent studies with CESA fused to fluorescent reporter proteins suggest that CSCs move at relatively stable trajectories at the PM via a microtubule (MT) guidance mechanism (19). However, when MTs are completely depolymerized, relative CESA velocity remains unchanged in otherwise wild-type (WT) plants, suggesting that energy provided by glucan chain polymerization is primarily responsible for movement rather than MT motor proteins (19, 20). The effect that genetic mutations have on cellulose polymerization rates can therefore be explored by examining the displacement velocity of fluorescently labeled CESA at the PM. Although previous analyses of the CESA velocity in cellulose deficient or MT mutants have suggested that polymerization rate can be altered, these studies have not addressed the influence that microfibril structure could impart on CESA velocity (21–24).
Here, we identified a mutant, CESA1A903V (cesa1aegeus) by forward chemical genetics and created a double mutant with the previously identified CESA3T942I (cesa3ixr1-2) (25). X-ray diffraction (XRD) and 13C solid-state nuclear magnetic resonance (SSNMR) spectroscopy are employed to show that cesa1aegeus and cesa3ixr1-2 confer changes to the ordered crystallization of glucan chains in the interior of cellulose microfibrils. Lower crystallinity cellulose displays greater digestibility characteristics relative to WT and reveals a consistent coupled association between crystallization and polymerization that is conserved between bacterial (10) and plant cellulose biosynthetic processes.
Results
Identification and Characterization of the Cellulose Biosynthesis Inhibitor Quinoxyphen.
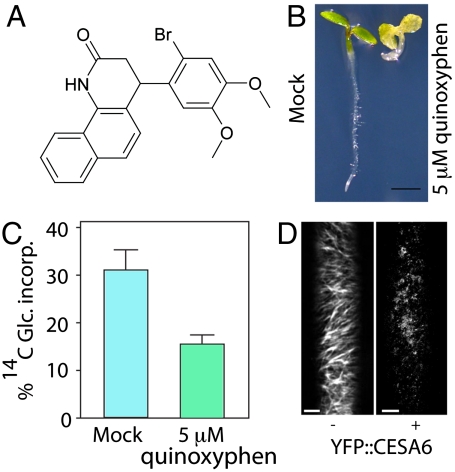
The herbicide quinoxyphen (Fig. 1A; Fig. S1) was identified in a screen for compounds that inhibit cell growth anisotropy. Germination on quinoxyphen above the IC50 value of 1.0 μM caused severe cell and tissue swelling of the hypocotyl and upper root in Arabidopsis seedlings (Fig. 1B). In addition, there was a 50% inhibition of 14C-glucose incorporation into crystalline cellulose in plants germinated and grown on a 5.0 μM concentration of quinoxyphen (Fig. 1C). Differences in cell wall composition based on FTIR spectra were detected in quinoxyphen treated seedlings (Fig. S2 A and B), as well as significant increases in the abundance of cell wall neutral sugars (Fig. S2C). Additional analysis of seedlings exposed to quinoxyphen showed other cell wall compositional alterations, including hyper-accumulation of callose and ectopic lignin production; both common phenotypes observed following chemical inhibition of cellulose biosynthesis (25, 26) (Fig. S3 A–D).
Fig. 1.
Identification of quinoxyphen as a cellulose biosynthesis inhibitor. (A) Structure of 4-(2-bromo-4,5-dimethoxy-phenyl)-3,4-dihydro-1H-benzo-quinolin-2-one (C21H18BrNO3 MW 412.28), named herein quinoxyphen. (B) Arabidopsis seedlings grown under continuous light for 5 d on MS agar containing 5 μM quinoxyphen (Right) showed reduce growth compared with control plant (Left). Scale bar, 10 mm. (C) Incorporation of 14C-glucose in the acid-insoluble fraction in WT treated and untreated with 5.0 μM quinoxyphen. (*P < 0.001, Student’s t test, n = 3 with 200 seedling per replicate). (D) Time-lapse confocal images of YFP∷CESA6 in hypocotyl cells of 3-d-old etiolated plants were compared between mock control treatment and 20 μM quinoxyphen treatment revealing clearance of the PM CSCs after 120 min. Each image in D is an average of 60 frames taken at 5-s intervals on the same Z plane (n = 3). Scale bar, 10 μm.
In expanding Arabidopsis hypocotyl tissue, cellulose biosynthesis can be visualized in living cells by labeling the CSC with fluorescently tagged CESA proteins. To establish a possible cause of the anisotropic growth disruption and cellulose content reduction, we used spinning disk confocal microscopy to examine the effect of quinoxyphen on the behavior of YFP-CESA6 particles in the focal plane of the PM. Consistent with previous observations, following treatment with the herbicide isoxaben (19), CSCs were depleted from the PM, a change visualized in time-projected images by the loss of tracks created by particle translocation (Fig. 1D). Concurrently, label accumulated in trafficking compartments at the cell cortex, most of which were immobilized (Fig. 1D, see ref. 27 for further analysis of this redistribution syndrome). In contrast, no marked differences in morphology or general dynamics were observed in actin or MT cytoskeletons after quinoxyphen treatment (Fig. S4). Hence, we hypothesized that quinoxyphen targets cellulose biosynthesis, rather than causing broad cell toxicity, and that its mechanism of action is similar to that of isoxaben.
Resistance to Quinoxyphen Is Conferred by a Semidominant CESA1 Allele.
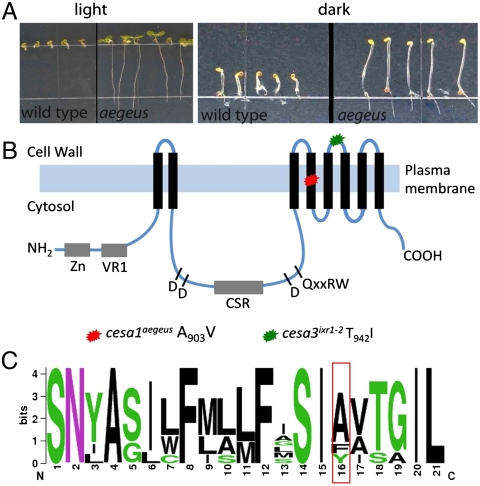
To identify possible targets of quinoxyphen, we performed a forward genetic screen and isolated an Arabidopsis mutant showing strong resistance to quinoxyphen (> 10 mM), which we named aegeus (Fig. 2A). Given the similar phenotypes induced by quinoxyphen and isoxaben, as observed with live-cell imaging, we tested whether the two drugs shared the same resistance locus. In an allelism test between aegeus and the isoxaben resistant mutant ixr1-2, the F1 progeny from crosses between the two homozygotes showed sensitivity when germinated and grown in media containing 5 μM of quinoxyphen or 10 nm of isoxaben (both lethal doses to WT), indicating that the two resistance loci were not allelic. In addition, homozygous aegeus plants displayed sensitivity to isoxaben, and homozygous ixr1-2 plants were sensitive to quinoxyphen. Using a positional cloning approach to identify the gene conferring resistance to quinoxyphen, we found that the aegeus mutation corresponds to the replacement of an alanine with a valine in the fourth transmembrane spanning domain at amino acid position 903 in the C-terminal end of the CESA1 protein (Fig. 2 B and C). The mutated alanine is not highly conserved among primary cell wall CESAs, but is flanked by several highly conserved residues (Fig. 2C). The aegeus mutant displayed semidominant inheritance similar to that observed for isoxaben resistance in the ixr mutants (25, 26). Both cesa1aegeus and cesa3ixr1-2 occur within the C-terminal region of CESA proteins comprising six transmembrane domains, and both occur in essential genes for cellulose biosynthesis in the primary wall (Fig. 2B (14, 16). Complementation of cesa1aegeus by transformation with WT CESA1 restored sensitivity to quinoxyphen.
Fig. 2.
The aegeus mutant is resistant to quinoxyphen. (A) The phenotype of the aegeus mutant compared to WT when grown for 7 d in the light (Left) or the dark (Right) on media containing 5 μM quinoxyphen (n = 5). (B) The gene corresponding to the aegeus mutation was cloned using a map-based approach (Fig. S3 and Table S2) and revealed an alanine to valine change at position 903 in the AtCESA1 protein (At4g3241). (C) Sequence logo assessment of residues in the fourth transmembrane domain of primary cell wall CESA proteins illustrates the location and conservation of the mutated alanine residue in CESA1 (red box). Amino acids are colored according to their chemical properties: Polar amino acids are green, basic are blue, acidic are red, and hydrophobic are black.
Generation of Double Drug-Resistant Mutants.
We generated the double mutant cesa1aegeus/cesa3ixr1-2, thereby creating a primary cell wall CSC containing two essential CESA subunits with mutations conferring drug resistance. The double mutant showed resistance to both quinoxyphen and isoxaben, but also a far more pronounced dwarf phenotype than either of the single mutants (Fig. S2 D–F). Significant differences were also apparent during the first 7 d of dark-grown hypocotyl elongation (Fig. S2F and Fig. S5b) and in light-grown root growth (Fig. S5 A and C). Examination of crystalline cellulose content of mature stem and dark-grown hypocotyls revealed a significant reduction in all mutants compared with WT (Table 1).
Table 1.
Effect of mutation on cellulose content and crystallite size
| Cellulose content, μg/mg AIR | |||
| Genotype | Hypocotyls | Stems | Crystallite size, Å ± SE |
| WT | 415 ± 20.8 | 217 ± 6.5 | 23.4 ± 0.26 |
| cesa1aegeus | 249 ± 13.4* | 146 ± 7.2* | 21.9 ± 0.35* |
| cesa3ixr1-2 | 297 ± 6.8* | 150 ± 13.7* | 22.0 ± 0.48* |
| cesa1aegeus/cesa3ixr1-2 | 161 ± .6.3* | 140 ± 12.0* | 22.5* ± 0.34* |
Asterisks in cellulose measurements of dark-grown hypocotyls and mature stems indicate P < 0.001 Student’s t test, n = 3. Cellulose content presented as μg cellulose per mg acetone insoluble residue (AIR). Mutant and WT plants were grown to maturity, and the first internode of the stem was exposed to synchrotron X-ray analysis to measure crystallite size (lateral dimensionality). Asterisks indicate P < 0.01 Student’s t test, n = 4.
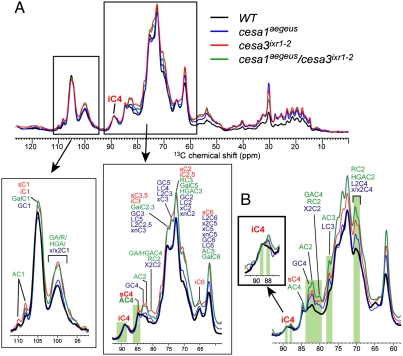
To dissect the structure of the cell wall in these mutants, we combined neutral sugar analysis, glycosidic linkage analysis (Tables S1 and S2) and 13C SSNMR (Fig. 3). Neutral sugar composition analysis revealed significant differences in neutral sugars as a proportion of total cell wall (Table S1). In addition, although there was little to no hyper-accumulation of callose in the mutant hypocotyls (Fig. S3 E–H), similar to quinoxyphen treatment, the double mutant cesa1aegeus/cesa3ixr1-2 showed increased ectopic lignin production (Fig. S3 I–L). Quantitative 13C SSNMR spectra of uniformly 13C labeled cell walls indicated increased intensities of pectin and hemicellulose peaks relative to the interior cellulose C4 peak at 89 ppm for all mutants (Fig. 3 A and B). For example, the arabinan (Ara) C1 signal at 108 ppm, the galacturonic acid (GalA) and rhamnose (Rha) C1 signal at 101 ppm, and the C2/C4 signals of xyloglucan (XG) Glc, GalA, Rha, and the Ara signals at 80–83 ppm, increased markedly, consistent with linkage analysis (Fig. S6 A and B and Table S2). The relative amounts of glycoprotein to interior cellulose also increased in the mutants by about twofold compared to WT. Hence, the cesa1aegeus, cesa3ixr1-2, and cesa1aegeus/cesa3ixr1-2 display quantitatively less cellulose and relative increases in pectin, hemicellulose and glycoprotein as a proportion of the cell wall.
Fig. 3.
13C magic-angle-spinning SSNMR analysis of cellulose and cell wall structure in cesa1aegeus, cesa3ixr1-2, and cesa1aegeus/cesa3ixr1-2. (A) Quantitative 13C magic-angle-spinning SSNMR spectra of WT and mutant cell walls. The intensity ratio of interior (88.5–91.5 ppm) to surface (84–86 ppm) cellulose C4 peaks was the lowest for the double mutant. The relative amount of hemicellulose and pectin signals to interior cellulose is significantly higher in all mutants compared to WT. The spectra were measured quantitatively by 13C direct polarization using a long recycle delay of 20 s. (B) 13C direct-polarization spectra measured with a short recycle delay of 2 s, which preferentially enhances the signals of mobile polysaccharides. The hemicellulose and pectin signals and the 88-ppm less-crystalline cellulose signal are increased for the mutants relative to the WT.
cesa1aegeus and cesa3ixr1-2 Mutations Cause Increased Cellulose Synthase Movement in the PM.
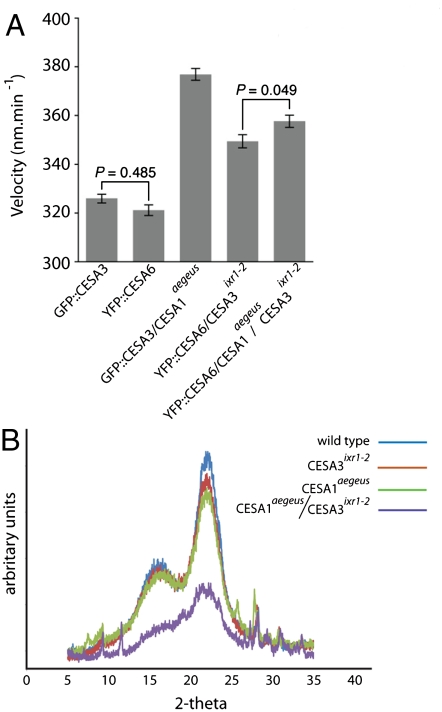
We examined the in vivo behavior of the CSC complexes in the aegeus mutant, with and without the ixr1-2 mutation, by creating double and triple mutants in stable transgenic plants expressing fluorescent reporters, and using spinning disk laser confocal microscopy to visualize the movement of the CSCs in living cells. Bidirectional trajectories of fluorescent CSCs were visualized at the PM focal plane of dark-grown transgenic seedlings in all mutant combinations (epidermal cells in the upper hypocotyl; Fig. 4A and Fig. S4). The relative mean velocity of labeled CESA complexes was significantly greater in cesa1aegeus, cesa3ixr1-2, and the double mutant than in transgenic lines that carried WT alleles of CESA1 and CESA3 (P < 0.0001, ANOVA with Bonferroni tests) (Fig. 4A). Previously, increased polymerization rates were observed in studies of bacterial cellulose synthesis, where the pharmacological disruption of crystallization prevented microfibril assembly and caused an increased polymerization rate (9, 10). Based on the difference in CESA behavior (Fig. 4A), and prior evidence from bacteria, we postulated that cellulose microfibril structure may be aberrant in cesa1aegeus and cesa3ixr1-2 mutants, and may reflect differences in cellulose crystallization.
Fig. 4.
CESA1aegeus and CESA3ixr1-2 mutants display increased CESA particle velocity at the PM focal plane and altered cellulose structure. (A) Epidermal cells of 3-d-old seedling expressing translational fusion reporters were imaged in the upper hypocotyl with spinning disk confocal microscopy (72 frames). CESA complex velocities from the indicated genotypes were compared using ANOVA with Bonferroni tests and significant difference indicated at P < 0.0001. Each result represents ≥720 CESA complexes from ≥10 cells from unique plants. Error bars represent SEM. The mean velocities, from left to right, were 377 ± 2.4, 349 ± 2.7, 358 ± 2.5, 326 ± 1.8, and 321 ± 2.2. (B) Analysis of cesa1aegeus, cesa3ixr1-2, and cesa1aegeus/cesa3ixr1-2 structural changes in the cellulose fingerprint compared between mutant plants and WT were initially screened by wide-angle XRD using Bragg–Brentano reflective geometries to obtain an RCI: WT = 73.1 ± 1.1, *cesA3ixr1-2 = 68.7 ± 1.4, *cesA1aegeus = 68.4 ± 2.0, *cesA1aegeus/cesA3ixr1-2 = 65.4 ± 1.8 (*P < 0.01 Student’s t test, n = 3).
cesa1aegeus, cesa3ixr1-2, and cesa1aegeus/cesa3ixr1-2 Mutants Produce Structurally Aberrant Cellulose Microfibrils.
As an initial experiment, we measured the relative crystallinity index (RCI) for each sample using XRD based on the Segal method (28). This assay is incapable of determining any accurate structural information for the cellulose crystallites and instead reflects the proportion of crystalline to amorphous cell wall components (29, 30). cesa1aegeus, cesa3ixr1-2, and cesa1aegeus/cesa3ixr1-2 all show significantly reduced RCI values compared to WT (Fig. 4B). However, as indicated by the 13C SSNMR analysis, the hemicellulose fraction relative to interior cellulose also increased in the mutants, which will contribute to the amorphous peak present in the XRD data and skew the RCI determination toward lower crystallinity. Therefore, to obtain a more accurate estimation of the microfibril structure, we examined the first internode of mature stem using high-energy synchrotron radiation XRD and obtained an estimation of the crystallite width using the Scherrer equation (31). This method allows the examination of intact plant samples, and so disruptive grinding or chemical treatments are unnecessary. Synchrotron XRD data indicated a significant reduction in the crystallite width for cesa1aegeus, cesa3ixr1-2, and cesa1aegeus/cesa3ixr1-2 compared to WT (Table 1). However, within biomass samples the use of 13C SSNMR is required to delineate accurate structural information about cellulose (32, 33).
Further analysis of the actual crystallite structure was obtained from quantitative 13C SSNMR spectra of uniformly 13C-labeled cell wall (Fig. 3A). Based on the intensity ratio of interior C4 (iC4) (88.5–91.5 ppm) to surface C4 (sC4) (84–86 ppm) peak intensities, we found that the double mutant had 8% lower crystallinity relative to WT (Table S3), while the single mutants had smaller differences from the WT. The mutants also displayed additional intensities in the regions of the iC4 peak, with a new 88-ppm peak that is largely absent in the WT spectrum (Fig. S6). This 88-ppm peak is closer to the 85-ppm C4 peak of the more amorphous surface cellulose, suggesting that it corresponds to less crystalline cellulose (Fig. 3B, Inset). Further, the 88-ppm intensity is preferentially enhanced in the mutant samples relative to the WT when the 13C spectra were measured using short recycle delays to increase the intensities of dynamic polysaccharides. This suggests that the less crystalline glucan chains in the mutants are more dynamic, which is consistent with weakened hydrogen bonding and the amorphous nature of the chains. In 2D double-quantum correlation spectra, we also observed two types of interior cellulose, which were best resolved in the CESA3ixr1-2 mutant but were also present in the other mutants (Fig. S6). Therefore, in the mutants, the microfibrils displayed reduced width and an additional cellulose C4 peak indicative of a degree of crystallinity that is intermediate between the surface and interior cellulose of WT, suggesting a difference in glucan chain association during microfibril formation.
We tested whether cellulose with more mobile amorphous iC4 intensity might show differences in saccharification efficiency when hydrolyzed with cellulase enzymes, as cellulose crystallinity has been shown to be an important predictor of enzymatic hydrolysis rate (34). The catalytic efficiency (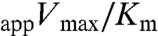 ) value showed a 32% and 49% increase in hydrolytic efficiency when using the cesa1aegeus and cesa1aegeus/cesa3ixr1-2 cellulose extracts, respectively (Fig. S5), consistent with our prior examination of cesa3ixr1-2 (35). These saccharification data support the quantitative 13C SSNMR spectra, indicating increased mobile iC4 assignments, from which we deduce that the amino acid sequence of the cesa1aegeus and cesa3ixr1-2 has an important influence on cellulose microfibril architecture.
) value showed a 32% and 49% increase in hydrolytic efficiency when using the cesa1aegeus and cesa1aegeus/cesa3ixr1-2 cellulose extracts, respectively (Fig. S5), consistent with our prior examination of cesa3ixr1-2 (35). These saccharification data support the quantitative 13C SSNMR spectra, indicating increased mobile iC4 assignments, from which we deduce that the amino acid sequence of the cesa1aegeus and cesa3ixr1-2 has an important influence on cellulose microfibril architecture.
Discussion
It is evident from these studies that cellulose microfibrils displayed inherent structural changes in the cesa1aegeus and cesa3ixr1-2 mutants. There seem to be two possible hypotheses to explain how cesa1aegeus and cesa3ixr1-2 mutants create a more amorphous and mobile cellulose-like structure (Fig. 3B). First, the mutated residues may occur in a region of the CESA protein involved in associations with other proteins in the CSC (not CESAs), which in turn play a role in microfibril assembly. However, this is only weakly supported by the nature of the cesa1aegeus mutation, which is predicted to lie within the fourth transmembrane spanning domain; it would be necessary to invoke a change in conformation state for the interaction with such protein cofactors. Alternatively, the transmembrane region containing mutated residues in CESA1aegeus and CESA3ixr1-2, while distal to the catalytic site, has been proposed to form a pore in the PM, which facilitates unidirectional glucan passage into the cell wall (36). The mutated residues may be necessary to organize the individual glucan chains into the composite microfibril and thus promote crystallinity (H-bonding) by facilitating association with other glucan chains. This model is consistent with the only reported crystal structure of a CSC, an octameric structure of the periplasmic AxCeSD subunit in the CSC of the bacterium A. xylinum in complex with a 5-glucose-cellulose product (37). Here, a dimer interface was necessary to facilitate passage and proper alignment of glucan chains into a crystalline microfibril. If applied to the proposed pore forming transmembrane regions of the plant CESA, this model would suggest that subtle alterations in the transmembrane helix structure via cesa1aegeus (A-V) and cesa3ixr1-2 (T-I) would induce changes in the organization of glucan chains via the inner side of a multimeric subunit association.
Disruption of microfibril assembly induced pharmacologically in bacteria caused an increase in the rate of glucan chain polymerization, illustrating that the two processes are coupled and that crystallization is rate limiting (9, 10). Our results show that cesa1aegeus and cesa3ixr1-2 single mutations displayed higher relative CESA velocity and reduced crystallinity, but were not additive in their effect on velocity in the double mutant. By contrast, more severe defects on cellular expansion were seen in the double mutant. One explanation is that mutated subunits impart influence on the polymerization rates on a single enzyme (CESA) and its catalytic process (β-1,4 glucan synthesis), but that additively the polymerization rates for individuals do not double in combination (as a CSC) and therefore appear as an intermediate velocity (polymerization) rate. Alternatively, the intermediate CSC velocity in the double mutant could be explained if a direct inhibitory effect of reduced crystallization counteracts some of the indirect (via enhanced rate of polymerization) stimulatory effect of reduced crystallization. Collectively, data from plants appear consistent with bacterial cellulose synthesis (9, 10) and illustrate a biophysical link between crystallinity and polymerization.
The identification of quinoxyphen represents a unique cellulose biosynthesis inhibitor (CBI) (Fig. 1 B and C) that induces rapid clearance of YFP∷CESA6 from the PM (Fig. 1D), reminiscent of isoxaben (19). While the identification of quinoxyphen represents a useful research tool, the resistance loci provide intriguing similarities and differences to isoxaben. Resistance to isoxaben was conferred by mutations in CESA3T942I and CESA6R1064W (26). By contrast, CESA3T942I and CESA6R1064W do not confer quinoxyphen resistance, which was mapped to CESA1A903V. Despite subunit specificity, quinoxyphen and isoxaben resistance was similarly conferred by mutations in the C-terminal transmembrane spanning region. It is possible that for both CBIs the site of inhibition occurs in this region. The C-terminal transmembrane spanning region has been proposed to mediate subunit association in the CSC (38), and therefore a CBI targeting this region could inhibit CSC formation. Supporting this postulate, live-cell imaging studies revealed clearance of YFP∷CESA6 from the PM after quinoxyphen treatment, although resistance was conferred by CESA1A903V. Precedence for the observed resistance mechanism whereby drug-binding affinity is displaced via mutation of residues in a transmembrane spanning region has been observed in the case of hydrophobic sliding for drug resistance in HIV-type 1 protease (39).
In terms of application, if cellulose crystallinity influences the rate of enzymatic hydrolysis (34), cellulose from CESA1aegeus and CESA3ixr1-2 (Fig. 3) should be more digestible by a cellulase cocktail. Our results supported this postulate and revealed that mutant cellulose was more readily enzymatically cleaved to individual glucan components. Hence, mutations identified herein could provide insights about how to modify cellulose synthesis in planta for improved bioconversion.
Materials and Methods
Chemical libraries used and screening conditions have been described in ref. 20 and supplemental on-line material. Transgenic plants expressing various florescent protein tagged genes or gene knockouts were developed by reciprocal crossing and have been described elsewhere (13, 19, 20).
Seedlings expressing GFP∷CESA3 or YFP∷CESA6 were grown in the dark for 3 d and imaged using spinning disk confocal microscopy, as described previously (26) except a DMI6000 B inverted microscope with Adaptive Focus Control (Leica) was used.
Cell Wall Structural Analysis by XRD.
Arabidopsis material was prepared and relative crystallinity index (RCI) determined as previously described (35) with a control of synthetic crystalline cellulose (Avicel®, FMC-Biopolymer). Three experimental replicates for mutant and WT biomass samples were performed. Synchrotron X-ray scattering experiments were performed at the macromolecular crystallography beamline F1 of the Cornell High Energy Synchrotron Source. A high-flux wiggler X-ray beam with a photon energy of 13 keV was focused to a spot size of 20 microns using an X-ray focusing capillary with 4 mrad acceptance. Plant samples were mounted in a helium-filled enclosure to reduce air scattering. Due to the high intensity of the microbeam, no further processing was necessary. A Quantum 210 area detector was placed at a distance of 120 mm from the sample, and the full 2D cellulose fiber diffraction image was recorded. Data were integrated using the Fit2D software. Scherrer equation analysis was performed by fitting a Gauss peak to the 200 reflection.
Cell Wall Structural Analysis by 13C Magic Angle Spinning SSNMR.
For 13C solid-state magic angle NMR plants were grown in sterile media supplemented with U-13C glucose and 15N NH4Cl (100 mM), and cell walls were extracted, desalted, and examined by NMR as previously described (40). 13C SSNMR spectra were measured on a Bruker Avance 600 (14.1 T) spectrometer operating at resonance frequencies of 600.13 MHz for 1H and 150.9 MHz for 13C. A double-resonance 4-mm magic-angle-spinning (MAS°) probe was used to measure all spectra. Typical radio-frequency field strengths were 62–70 kHz for 1H decoupling and 50 kHz for 13C pulses. The samples were spun under 7- to 12-kHz MAS. 13C chemical shifts were referenced to the 13CO signal of α-glycine at 176.49 ppm on the TMS scale. For quantitative 13C 1D experiments, the 13C magnetization was created by direct excitation using a 90° pulse and using a long recycle delay of 20 s. To preferentially detect mobile polysaccharides, a short recycle delay of 2 s was used in the 13C direct polarization experiment. 13C chemical shifts were assigned by reference with the recently published result (40) and were confirmed by 2D 13C-13C spin diffusion correlation experiments with a 40-ms mixing time and by 2D J-INADEQUATE experiments (40). The latter correlates 13C double-quantum chemical shifts with single-quantum chemical shifts, and gives relatively high spectral resolution.
Additional Details.
See SI Materials and Methods for additional details.
Supplementary Material
Acknowledgments.
The authors extend thanks to Herman Hofte, Samantha Vernettes (Institut National de la Recherche Agronomique, France), and Richard Williamson (Australian National University, Canberra, Australia) for supplying genetic material and to Parastoo Azadi (University of Georgia, Athens, GA) and Ulrich Englich (Macromolecular Diffraction at Cornell High Energy Synchrotron Source, Ithaca, NY) for technical assistance. This work was supported by the National Science Foundation Grants NSF-IOS-0922947 (S.D.), NSF-REU (S.D. and K.C.), PICT2010-0658 (J.M.E.), Department of Energy DOE-FOA 10-0000368 (S.D., D.-M.S., and J.K.C.R.), DE-AC02-07CH11358 (M.H.), and DOE-FGO2-03ER20133 (C.R.S.). The Cornell High Energy Synchrotron Source is a national user facility supported by the National Science Foundation and National Institutes of Health/National Institute of General Medical Sciences via National Science Foundation award DMR-0936384.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1200352109/-/DCSupplemental.
References
- 1.Brown R, Jr, Montezinos D. Cellulose microfibrils: Visualization of biosynthetic and orienting complexes in association with the plasma membrane. Proc Natl Acad Sci USA. 1976;73:143–147. doi: 10.1073/pnas.73.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tsekos I. The sites of cellulose synthesis in algae: Diversity and evolution of cellulose-synthesizing enzyme complexes. J Phycol. 1999;35:635–655. [Google Scholar]
- 3.Atalla RH, VanderHart DL. Native cellulose: A composite of two distinct crystalline forms. Science. 1984;223:283–285. doi: 10.1126/science.223.4633.283. [DOI] [PubMed] [Google Scholar]
- 4.Nishiyama Y, Langan P, Chanzy H. Crystal structure and hydrogen-bonding system in cellulose 1b from synchrotron X-ray and neutron fiber diffraction. J Am Chem Soc. 2002;124:9074–9082. doi: 10.1021/ja0257319. [DOI] [PubMed] [Google Scholar]
- 5.Nishiyama Y, Sugiyama J, Chanzy H, Langan P. Crystal structure and hydrogen bonding system in cellulose 1a from synchrotron X-ray and neutron fibre diffraction. J Am Chem Soc. 2003;125:14300–14306. doi: 10.1021/ja037055w. [DOI] [PubMed] [Google Scholar]
- 6.Viëtor RJ, Newman RH, Ha M-A, Apperley DC, Jarvis MC. Conformational features of crystal-surface cellulose from higher plants. Plant J. 2002;30:721–731. doi: 10.1046/j.1365-313x.2002.01327.x. [DOI] [PubMed] [Google Scholar]
- 7.Šturcová A, His I, Apperley DC, Sugiyama J, Jarvis MC. Structural details of crystalline cellulose from higher plants. Biomacromolecules. 2004;5:1333–1339. doi: 10.1021/bm034517p. [DOI] [PubMed] [Google Scholar]
- 8.Jarvis M. Chemistry: Cellulose stacks up. Nature. 2003;426:611–612. doi: 10.1038/426611a. [DOI] [PubMed] [Google Scholar]
- 9.Haigler C, Brown R, Benziman M. Calcofluor white ST Alters the in vivo assembly of cellulose microfibrils. Science. 1980;210:903–906. doi: 10.1126/science.7434003. [DOI] [PubMed] [Google Scholar]
- 10.Benziman M, Haigler CH, Brown RM, White AR, Cooper KM. Cellulose biogenesis: Polymerization and crystallization are coupled processes in Acetobacter xylinum. Proc Natl Acad Sci USA. 1980;77:6678–6682. doi: 10.1073/pnas.77.11.6678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anderson CT, Carroll A, Akhmetova L, Somerville C. Real-time imaging of cellulose reorientation during cell wall expansion in Arabidopsis roots. Plant Physiol. 2010;152:787–796. doi: 10.1104/pp.109.150128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Richmond T. Higher plant cellulose synthases. Genome Biol. 2000;1:30011–30016. doi: 10.1186/gb-2000-1-4-reviews3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Desprez T, et al. Organization of cellulose synthase complexes involved in primary cell wall synthesis in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2007;104:15572–15577. doi: 10.1073/pnas.0706569104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Persson S, et al. Genetic evidence for three unique components in primary cell-wall cellulose synthase complexes in Arabidopsis. Proc Natl Acad Sci USA. 2007;104:15566–15571. doi: 10.1073/pnas.0706592104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taylor NG, Howells RM, Huttly AK, Vickers K, Turner SR. Interactions among three distinct CesA proteins essential for cellulose synthesis. Proc Natl Acad Sci USA. 2003;100:1450–1455. doi: 10.1073/pnas.0337628100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arioli T, et al. Molecular analysis of cellulose biosynthesis in Arabidopsis. Science. 1998;279:717–720. doi: 10.1126/science.279.5351.717. [DOI] [PubMed] [Google Scholar]
- 17.Gillmor CS, Poindexter P, Lorieau J, Palcic MM, Somerville C. α-Glucosidase I is required for cellulose biosynthesis and morphogenesis in Arabidopsis. J Cell Biol. 2002;156:1003–1013. doi: 10.1083/jcb.200111093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Somerville C. Cellulose synthesis in higher plants. Annu Rev Cell Dev Biol. 2006;22:53–78. doi: 10.1146/annurev.cellbio.22.022206.160206. [DOI] [PubMed] [Google Scholar]
- 19.Paredez AR, Somerville CR, Ehrhardt DW. Visualization of cellulose synthase demonstrates functional association with microtubules. Science. 2006;312:1491–1495. doi: 10.1126/science.1126551. [DOI] [PubMed] [Google Scholar]
- 20.DeBolt S, et al. Morlin, an inhibitor of cortical microtubule dynamics and cellulose synthase movement. Proc Natl Acad Sci USA. 2007;104:5854–5859. doi: 10.1073/pnas.0700789104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gu Y, et al. Identification of a cellulose synthase-associated protein required for cellulose biosynthesis. Proc Natl Acad Sci USA. 2010;107:12866–12871. doi: 10.1073/pnas.1007092107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fujita M, et al. Cortical microtubules optimize cell-wall crystallinity to drive unidirectional growth in Arabidopsis. Plant J. 2011;66:915–928. doi: 10.1111/j.1365-313X.2011.04552.x. [DOI] [PubMed] [Google Scholar]
- 23.Bischoff V, et al. Phytochrome regulation of cellulose synthesis in Arabidopsis. Curr Biol. 2011;21:1822–1827. doi: 10.1016/j.cub.2011.09.026. [DOI] [PubMed] [Google Scholar]
- 24.Chen S, Ehrhardt DW, Somerville C. Mutations of cellulose synthase (CESA1) phosphorylation sites modulate anisotropic cell expansion and bidirectional mobility of cellulose synthase. Proc Natl Acad Sci USA. 2010;107:17188–17193. doi: 10.1073/pnas.1012348107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scheible WR, Eshed R, Richmond T, Delmer D, Somerville CR. Modifications of cellulose synthase confer resistance to isoxaben and thiazolidinone herbicides in Arabidopsis Ixr1 mutants. Proc Natl Acad Sci USA. 2001;98:10079–10084. doi: 10.1073/pnas.191361598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Desprez T, et al. Resistance against herbicide isoxaben and cellulose deficiency caused by distinct mutations in same cellulose synthase isoform CESA6. Plant Physiol. 2002;128:482–490. doi: 10.1104/pp.010822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gutierrez R, Lindeboom JJ, Paredez AR, Emons AMC, Ehrhardt DW. Arabidopsis cortical microtubules position cellulose synthase delivery to the plasma membrane and interact with cellulose synthase trafficking compartments. Nat Cell Biol. 2009;11:797–806. doi: 10.1038/ncb1886. [DOI] [PubMed] [Google Scholar]
- 28.Segal L, Creely JJ, Martin AE, Conrad CM. An empirical method for estimating the degree of crystallinity of native cellulose using the X-ray diffractometer. Text Res J. 1959;29:786–794. [Google Scholar]
- 29.Andersson S, Serimaa R, Paakkari T, SaranpÄÄ P, Pesonen E. Crystallinity of wood and the size of cellulose crystallites in Norway spruce (Picea abies) J Wood Sci. 2003;49:531–537. [Google Scholar]
- 30.Thygesen A, Oddershede J, Lilholt H, Thomsen AB, Ståhl K. On the determination of crystallinity and cellulose content in plant fibres. Cellulose. 2005;12:563–576. [Google Scholar]
- 31.Patterson AL. The Scherrer formula for X-ray particle size determination. Phys Rev. 1939;56:978–982. [Google Scholar]
- 32.Fernandes AN, et al. Nanstructure of cellulose microfibrils in spruce wood. Proc Natl Acad Sci USA. 2011;108:1195–1203. doi: 10.1073/pnas.1108942108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Newman RH. Estimation of the lateral dimensions of cellulose crystallites using 13C NMR signal strengths. Solid State Nucl Magn Reson. 1999;15:21–29. doi: 10.1016/s0926-2040(99)00043-0. [DOI] [PubMed] [Google Scholar]
- 34.Hall M, Bansal P, Lee JH, Realff MJ, Bommarius AS. Cellulose crystallinity—A key predictor of the enzymatic hydrolysis rate. FEBS J. 2010;277:1571–1582. doi: 10.1111/j.1742-4658.2010.07585.x. [DOI] [PubMed] [Google Scholar]
- 35.Harris D, Stork J, DeBolt S. Genetic modification in cellulose-synthase reduces crystallinity and improves biochemical conversion to fermentable sugar. Glob Change Biol Bioenergy. 2009;1:51–61. [Google Scholar]
- 36.Carpita NC. Update on mechanisms of plant cell wall biosynthesis: How plants make cellulose and other (1 → 4)-β-D-glycans. Plant Physiol. 2011;155:171–184. doi: 10.1104/pp.110.163360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hu S-Q, et al. Structure of bacterial cellulose synthase subunit D octamer with four inner passageways. Proc Natl Acad Sci USA. 2010;107:17957–17961. doi: 10.1073/pnas.1000601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang J, Howles PA, Cork A, Birch R, Williamson RE. Chimeric proteins suggest that the catalytic and/or C-terminal domains give CesA1 and CesA3 access to their specific sites in the cellulose synthase of primary walls. Plant Physiol. 2006;142:685–695. doi: 10.1104/pp.106.084004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Foulkes-Murzycki JE, Scott WRP, Schiffer CA. Hydrophobic sliding: A possible mechanism for drug resistance in human immunodeficiency virus type 1 protease. Structure. 2007;15:225–233. doi: 10.1016/j.str.2007.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dick-Pérez M, et al. Structure and interactions of plant cell-wall polysaccharides by two-and three-dimensional magic-angle-spinning solid-state NMR. Biochemistry. 2011;50:989–1000. doi: 10.1021/bi101795q. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.