Abstract
The dicistrovirus intergenic region internal ribosome entry site (IRES) utilizes a unique mechanism, involving P-site tRNA mimicry, to directly assemble 80S ribosomes and initiate translation at a specific non-AUG codon in the ribosomal A site. A subgroup of dicistrovirus genomes contains an additional stem-loop 5′-adjacent to the IRES and a short open reading frame (ORFx) that overlaps the viral structural polyprotein ORF (ORF2) in the +1 reading frame. Using mass spectrometry and extensive mutagenesis, we show that, besides directing ORF2 translation, the Israeli acute paralysis dicistrovirus IRES also directs ORFx translation. The latter is mediated by a U∶G base pair adjacent to the P-site tRNA-mimicking domain. An ORFx peptide was detected in virus-infected honey bees by multiple reaction monitoring mass spectrometry. Finally, the 5′ stem-loop increases IRES activity and may couple translation of the two major ORFs of the virus. This study reveals a novel viral strategy in which a tRNA-like IRES directs precise, initiator Met-tRNA-independent translation of two overlapping ORFs.
Keywords: frameshifting, Israeli acute paralysis virus, protein synthesis, pseudoknot, genetic recoding
Dicistroviruses possess a positive-sense, monopartite single-stranded RNA genome that encodes nonstructural and structural polyproteins within two open reading frames (ORF1 and ORF2) that are separated by an intergenic region (IGR) (1). Translation of each ORF is directed by distinct internal ribosome entry site (IRES) mechanisms (2). Members of the family Dicistroviridae include cricket paralysis virus (CrPV), Taura syndrome virus (TSV), Plautia stali intestine virus, and Drosophila C virus. Within this family, the honey bee viruses acute bee paralysis virus (ABPV), Israeli acute paralysis virus (IAPV), and Kashmir bee virus (KBV) and the fire ant virus Solenopsis invicta virus (SINV-1) form a tight clade. Interestingly, IAPV has been associated with colony collapse disorder (3). Uniquely, the IGR IRES can directly assemble 80S ribosomes and direct translation initiation at a non-AUG codon from the ribosomal A site without the aid of initiation factors or initiator Met-tRNAi (2, 4–9). The IGR IRES achieves this function by folding into a structure containing three overlapping pseudoknots: PKI, PKII, and PKIII (Fig. 1). PKII and PKIII form one domain that folds into a compact core that is responsible for ribosome binding (5, 9–12). PKI mimics a tRNA anticodon stem-loop and P-site codon:anticodon duplex (11, 13, 14). This structure occupies the P site and sets the ribosome into the elongation phase in a specific reading frame, determined by the non-AUG codon in the A site (4, 6). This mechanism likely provides a selective advantage by allowing translation of the viral structural proteins during virus infection when host translation is shut off (2, 15).
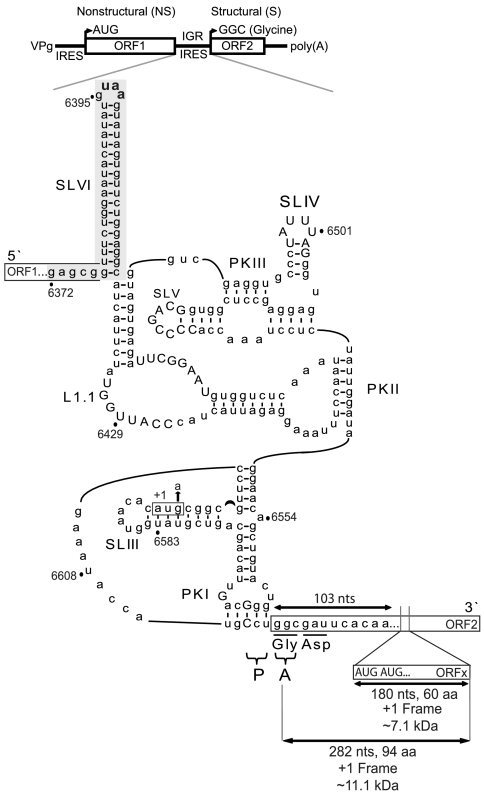
Fig. 1.
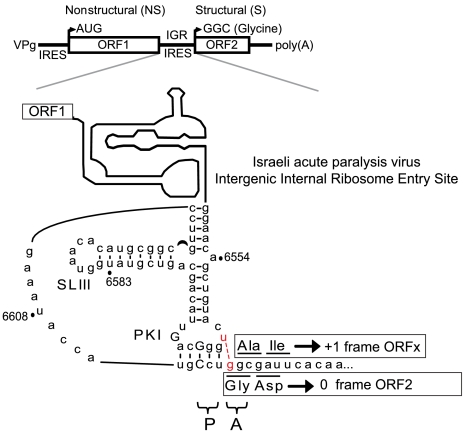
Secondary structure of IAPV IGR IRES. (Top) Schematic of IAPV genome. Distinct IRESs direct translation of nonstructural (ORF1) and structural (ORF2) polyproteins. Schematic of IAPV IGR IRES showing pseudoknots PKI, PKII, and PKIII, stem loops SLIII, SLIV, SLV, and SLVI (shaded gray), and loop L1.1. The UAA stop codon of ORF1 is shown in bold within the loop of SLVI. The overlapping +1 frame ORFx, within ORF2, is shown. Tandem ORFx-frame AUG codons, 103 nucleotides downstream from the IGR IRES, are indicated and, if utilized as the initiation site for ORFx translation, would lead to a 7.1-kDa protein. If, on the other hand, ORFx translation initiates at or near the IGR IRES, then an 11.1-kDa protein is expected. AUG6592-4 (boxed) is an alternative potential initiation site for +1 frame ORFx. G6594A tests whether the AUG6592-4 codon is used for ORFx translation. Conserved nucleotides among type II IGR IRESs are in capital letters. The CCU triplet mediates PKI base pairing and occupies the P site, whereas the first codon of the 0 frame ORF2 is the 3′-adjacent GGC codon in the A site.
Phylogenetic analysis has revealed that dicistrovirus IGR IRESs can be grouped into two types (16, 17). The CrPV IRES typifies the type I IGR IRES, and the TSV, SINV-1, and honey bee paralysis virus IRESs exemplify the type II IGR IRES. The main difference is that type II IRESs contain a longer L1.1 region and an extra stem loop (SLIII) within the PKI domain (Fig. 1). We and others have shown that the L1.1 region is responsible for 80S assembly on the type I and II IGR IRESs (11, 18). The role of SLIII is not clear but appears to be involved in ribosome positioning and binding (12, 19, 20). We have shown that the PKII/PKIII and PKI domains of type I and II IRESs are functionally interchangeable, which suggests that the IGR IRESs are modular (20). In general, both type I and II IRESs recruit ribosomes directly and set the ribosome within a specific reading frame, initiating at a non-AUG codon (12, 18–22).
Recently, we and others reported bioinformatic evidence for a hidden overlapping gene, called ORFx, located just downstream of the IGR IRES within a subset of dicistroviruses including the honey bee paralysis viruses and SINV-1 (23, 24) (Fig. 1). Conserved tandem AUG codons, which provide one potential initiation site for ORFx translation, are located approximately 100 nucleotides downstream of the PKI domain (Fig. 1). Moreover, a 14–18 bp stem-loop structure (SLVI) 5′-adjacent to the IGR IRES was found specifically in the honey bee viruses (23). In these viruses, the ORF1 stop codon is positioned within SLVI (Fig. 1). By using IAPV as a model, this study probes the role of these features and their relationship to IAPV IGR IRES-mediated translation.
Results
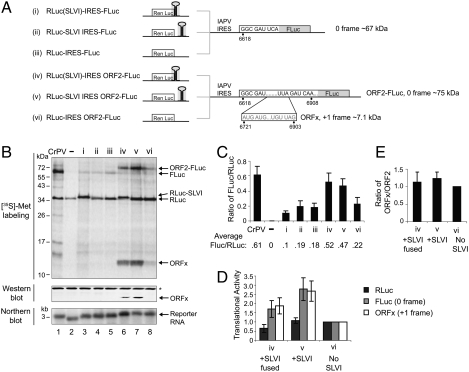
To determine the role of the 5'-adjacent stem-loop and ORFx of the IAPV IGR, dicistronic reporter constructs were engineered to contain different versions of the IGR IAPV IRES [Fig. 2A, constructs (i)–(vi)]. The upstream renilla (RLuc) and downstream firefly (FLuc) luciferase genes monitor scanning- and IRES-dependent translation, respectively. Specifically, reporter constructs contained either the IRES alone, the IRES with the upstream SLVI positioned downstream of the RLuc stop codon, or the IRES with SLVI positioned such that the UAA stop codon of ORF1 is fused in frame with the RLuc ORF and serves as the stop codon for RLuc translation [Fig. 2A, constructs (i)–(iii)]. In these constructs, the FLuc ORF is fused in frame with the first three codons of ORF2 so that ORFx is not present. Additional constructs were designed in which the FLuc ORF is fused in frame with a longer region of the IAPV ORF2 (nucleotides 6618–6908), which contains the overlapping +1 frame ORFx [Fig. 2A, constructs (iv)–(vi)].
Fig. 2.
Characterization of novel features within the IAPV IGR IRES. (A) Schematic of dicistronic reporter constructs containing the IAPV IGR IRES inserted in the intergenic region between two reporter genes: Renilla luciferase (RLuc), which monitors scanning-mediated translation, and firefly luciferase (FLuc), which monitors IRES-mediated translation. Constructs (i)–(iii) contain the IAPV IGR IRES alone. Constructs (iv)–(vi) also contain a part of the IAPV ORF2 (6618–6908) fused in the 0 frame with FLuc (ORF2-FLuc). This region also contains the predicted ORFx in the +1 frame. Dicistronic reporter constructs were engineered to contain no SLVI (iii, vi), SLVI positioned in the IGR (ii, v), or SLVI positioned such that the UAA stop codon within SLVI serves as the termination codon for RLuc (i, iv). (B) The presence of SLVI and part of ORF2 stimulates IAPV IGR IRES translation. Dicistronic reporter constructs were incubated in Sf21 extracts at 30 °C for 120 min in the presence of [35S]-methionine. CrPV and (−) denote dicistronic reporters containing the CrPV IGR IRES and an empty no-IRES control, respectively. In parallel, reactions were subjected to Western blotting, or to Northern blotting using a probe specific to the reporter RNA. (C) Quantitation of radiolabeled protein products calculated by taking the ratio FLuc/RLuc. Averages from at least three independent experiments ± SD are shown. (D) SLVI increases IGR IRES activity. Graph of translational activities determined via quantitation of radiolabeled proteins produced from constructs (iv), (v), and (vi). Values were normalized relative to (vi). (E) Ratio of ORFx/ORF2 protein products produced from the indicated constructs. All calculations are from at least three independent experiments ± SD.
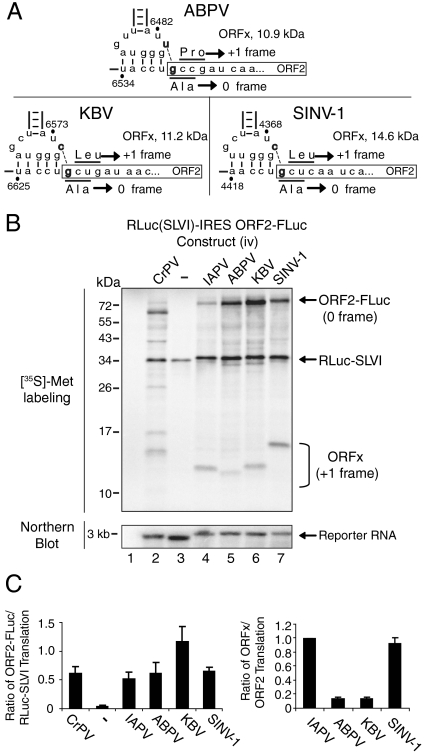
Translational activities were monitored by incubating dicistronic constructs in insect Sf21 extracts in the presence of [35S]-methionine, and the resulting protein products were detected by SDS-PAGE analysis. IRES activity was measured by calculating the expression of FLuc and RLuc individually as well as the ratio of FLuc/RLuc (Fig. 2 C and D). To compare translational activities between constructs, the number of methionines was taken into account in cases where the reporter gene lengths are different. As shown previously (20), IAPV IGR IRES translation was significantly weaker compared to that of the CrPV IGR IRES, and, as expected, an empty construct did not produce IRES activity (Fig. 2B, compare lanes 1, 2, and 5). Insertion of SLVI did not significantly affect IRES activity (Fig. 2B, compare lanes 3–5). Note the slower migration of RLuc derived from constructs where the UAA codon of SLVI is fused in frame with RLuc thus resulting in a longer RLuc protein [Fig. 2B, lane 3, construct (i)]. In constructs (iv)–(vi), insertion of the ORF2 region (6618–6908) downstream of the IAPV IGR IRES resulted in a larger ORF2-FLuc protein (Fig. 2B, lanes 6–8). Interestingly, in these constructs, the presence of SLVI had an approximately twofold stimulatory effect on IRES activity (Fig. 2 B, lanes 6–8, and C). Similarly, when compared to constructs that contained only SLVI and the IAPV IGR IRES [Fig. 2B, lanes 3 and 4, constructs (ii) and (iii)], the addition of the ORF2 (6618–6908) region increased IRES translation by approximately 2.5- to 5-fold (Fig. 2B, compare lanes 3 and 6 and lanes 4 and 7), which is similar to previous reports that sequences downstream of the IGR IRES contribute to IRES activity (7, 25). In contrast, the presence of the ORF2 (6618–6908) region did not stimulate IRES translation in constructs containing the IGR IRES but not SLVI [Fig. 2B, compare lanes 5 and 8, constructs (iii) and (vi)]. Importantly, Northern blotting showed similar levels of reporter RNAs in each reaction, which indicates that the differences in reporter gene expression are due to differences in translation (Fig. 2B). Thus, these results demonstrate that the presence of both SLVI and a part of ORF2 act synergistically to stimulate IAPV IGR IRES translation in a context-dependent manner.
The UAA stop codon of ORF1 is located within the loop of SLVI (Fig. 1). We next investigated whether fusing the UAA of SLVI in frame with RLuc affects translation of either the upstream RLuc or the downstream FLuc. We compared the translation of scanning-dependent RLuc and IRES-dependent FLuc in constructs where SLVI is inserted in the IGR [construct (v), +SLVI] or where the UAA of SLVI is fused in frame with RLuc [construct (iv), +SLVI fused]. Both RLuc and FLuc expression were consistently lower by approximately 35% when the UAA of SLVI is fused in frame with RLuc than when SLVI is within the intergenic region (Fig. 2 B, compare lanes 6 and 7, and D). These results indicate that translation of the upstream ORF may affect translation of the downstream ORF when the UAA stop codon of SLVI is fused in frame with the upstream ORF.
Interestingly, reporter constructs that contain the ORF2 (6618–6908) region also produced an approximately 12-kDa protein (Fig. 2B, lanes 6–8). To determine whether this protein is an ORFx product, we performed a Western blot analysis by using an antibody that was raised against a peptide encoded within the predicted ORFx. The ORFx antibody reacted with the approximately 12-kDa protein specifically in those reactions that had reporter constructs containing the ORF2 region (Fig. 2B, compare lanes 3–5 and 6–8). Similar to the expression observed for the 0 frame ORF2-FLuc, the presence of SLVI within the intergenic region of the dicistronic construct increased ORFx translation by approximately 2.5-fold (Fig. 2B, compare lanes 7 and 8, and D). Furthermore, fusing the UAA codon of SLVI in frame with RLuc decreased ORFx translation by approximately 35%, similar to the reduction observed for the 0 frame ORF2-FLuc (Fig. 2D). The presence of SLVI, regardless of whether the UAA is fused in frame with RLuc, did not, however, affect the ratio of ORFx/ORF2 translation (Fig. 2E). In summary, insertion of SLVI with the IAPV IGR IRES within the intergenic region of the reporter constructs increased both 0 frame ORF2 and +1 frame ORFx translation, but fusing the UAA stop codon of SLVI with the upstream RLuc decreased scanning-dependent RLuc, IRES-dependent 0 frame FLuc, and ORFx translation.
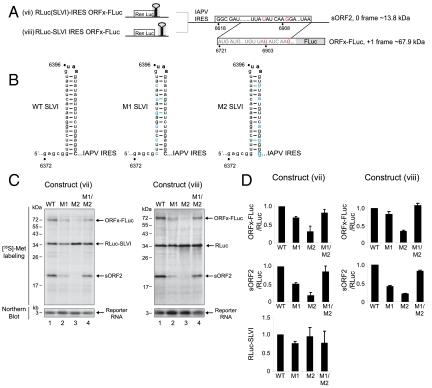
To further confirm that ORFx is expressed, mutations within ORF2 were made such that ORFx is fused in frame with FLuc to produce an ORFx-FLuc fusion protein [Fig. 3A, construct (vii)]. Specifically, we mutated the stop codon of ORFx and inserted an extra G nucleotide at position 6910 to shift FLuc into the ORFx frame (Fig. 3A, nucleotides shown in red). In doing so, a smaller truncated 0 frame ORF2 (sORF2) is created. Thus both 0 and +1 frame translation products can be simultaneously monitored by SDS-PAGE. Incubating construct (vii) in Sf21 extracts produced the +1 frame ORFx-FLuc and 0 frame truncated sORF2 proteins (Fig. 3C, lane 1). We note that the sORF2 protein (approximately 20 kDa) migrates more slowly than expected from the predicted molecular mass (approximately 13.8 kDa). Small proteins often migrate at unusual positions, depending on their amino acid composition. However, from results below, we are confident that the product migrating at approximately 20 kDa represents 0 frame sORF2 translation mediated by the IGR IRES.
Fig. 3.
The integrity of SLVI affects IAPV IGR IRES translation. (A) Schematic of dicistronic constructs containing the IAPV IGR IRES and ORFx fused in frame with FLuc. Red nucleotides denote mutations inserted to fuse the +1 frame ORFx in frame with FLuc. These mutations also create a short 0 frame ORF2 protein (sORF2). (B) Schematic of WT and mutant (M1 and M2) SLVI. Mutations that disrupt the helical base pairing within SLVI (M1 and M2) are indicated in blue. Compensatory mutations (M1/M2 combined) restore base pairing within SLVI. Bold nucleotides denote the UAA stop codon of ORF1. (C) Dicistronic constructs containing IGR SLVI (viii) or SLVI fused with RLuc (vii) were incubated in Sf21 translation extracts. Northern blotting using a probe specific to the reporter RNA is shown. (D) Quantitation of translation products normalized relative to the dicistronic construct containing the wild-type SLVI. Averages ± SD are from at least three independent experiments.
To address whether SLVI is important for IGR IRES translation, we created mutations on either the 5′ or the 3′ side of SLVI to disrupt the helical stem (Fig. 3B, mutants M1 and M2, nucleotide changes shown in blue). The SLVI 5′ mutations were designed to be silent with respect to the amino acid coding sequence up to the UAA stop codon. These mutations ensure that the effects observed are due to disruption of SLVI and not the amino acid sequence. The 3′ mutations were designed to complement the 5′ mutations. The SLVI mutants were inserted into the IGR of the dicistronic construct (viii), or such that the UAA stop codon of SLVI is fused in frame with the RLuc ORF [construct (vii)] (Fig. 3 A). Disruption of the helical stem of SLVI on the 5′ side (M1) in both constructs (vii) and (viii) moderately inhibited IRES translation, whereas mutating the 3′ side (M2) inhibited 0 and +1 frame translation significantly (Fig. 3 C and D). Given that fusing the UAA stop codon of SLVI in frame with the RLuc ORF inhibited RLuc translation, it was surprising that mutating the helical stem did not significantly affect RLuc translation (Fig. 3 C and D). Because only every third nucleotide was mutated in mutants M1 and M2 and, further, because of the desire to maintain the encoded amino acid sequence some mutated sites could still form U∶G base pairings (Fig. 3B), it is possible that the helical stem is still partly intact in these constructs. In contrast to M1 and M2 mutant SLVI, combining the mutations to restore the helical stem rescued 0 and +1 frame translation (Fig. 3 C and D, M1/M2). In summary, these data show that SLVI contributes to IAPV IRES translation.
From our reporter analysis, fusing the UAA stop codon of SLVI in frame with RLuc inhibited RLuc, ORFx, and ORF2 translation (Fig. 2D). One possibility is that ribosomes translating the upstream RLuc may disrupt SLVI and thereby inhibit translation of the downstream 0 and +1 frame ORFs. We reasoned that if translation of the upstream RLuc was abolished, this inhibition should increase IRES-mediated 0 and +1 frame translation. To knock out translation of RLuc, we inserted a strong hairpin upstream of the RLuc ORF, which inhibits scanning (Fig. S1A) (26). As expected, insertion of the hairpin abolished scanning-dependent RLuc translation (Fig. S1 B and C). In contrast, the addition of the hairpin resulted in a 40% increase in 0 and +1 frame translation in construct (vii), in which the UAA of SLVI is fused in frame with RLuc, as compared to constructs where the hairpin is absent (Fig. S1 B and C). When SLVI is inserted within the intergenic region of construct (viii), 0 and +1 frame translation was not significantly affected by the insertion of the hairpin upstream of RLuc (Fig. S1 B and C). These results are consistent with the idea that translation of the upstream ORF affects the translation of the downstream 0 and +1 frame ORFs only when the UAA stop codon of SLVI is fused in frame with the upstream ORF.
Mechanism of ORFx Translation.
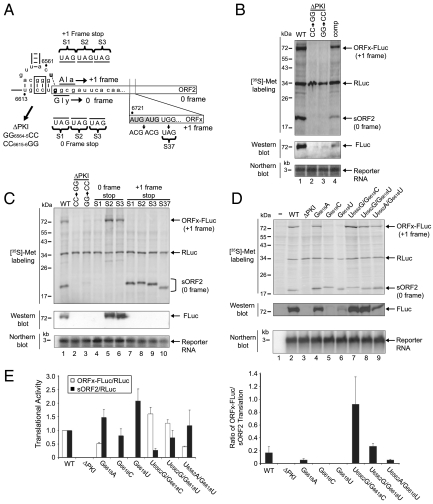
We next addressed the mechanism by which ORFx is translated. We first asked whether ORFx translation is dependent on the IAPV IGR IRES. Disruption of PKI within dicistrovirus IGR IRESs abolishes IRES-dependent translation by inhibiting ribosome positioning on the IRES (2, 5, 8, 9). As expected, disrupting the base pairing within the IAPV PKI by mutating GG6564-5CC or CC6615-6GG inhibited 0 frame translation (Fig. 4 B, lanes 2 and 3, and C, lanes 2 and 3). Furthermore, disruption of PKI also inhibited +1 frame ORFx expression. Compensatory mutations restoring the PKI base pairing partially rescued both 0 and +1 frame translation, which indicates that +1 frame ORFx translation is dependent on the integrity of the IAPV IGR IRES (Fig. 4B, lane 4).
Fig. 4.
U6562/G6618 base pairing directs +1 frame ORFx translation. (A) Schematic of the IAPV PKI domain. Black dashes and bold denote the predicted U6562/G6618 base pairing within PKI which directs +1 frame ORFx translation. The first amino acid of ORF2 is encoded by a glycine GGC codon in the 0 frame. The predicted first amino acid of ORFx—if ORFx is initiated at the IGR IRES—is encoded by an alanine GCG codon in the +1 frame. Tandem AUG codons within ORFx are shown. Mutations that replace codons with a stop codon in the 0 or +1 frame are denoted by S1-3 and S37. Mutations that disrupt PKI base pairing are shown (ΔPKI). Mutations were inserted in reporter constructs (viii). (B–D) Reporter constructs were incubated in Sf21 extracts. In parallel, reactions were subjected to Western blotting or Northern blotting. “comp” denotes compensatory mutations to restore PKI base pairing. (E) Quantitation of radiolabeled protein products normalized relative to the wild-type IAPV IGR IRES dicistronic reporter is shown (Left). The ratio of ORFx-FLuc (+1 frame)/sORF2 (0 frame) translation, taking into account the number of methionines within each translated protein, is shown (right). Averages ± SD are from at least three independent experiments.
Previous sequence analysis provided several hypotheses for the mechanism of ORFx translation, including (i) a portion of ribosomes recruited to the IGR IRES somehow start scanning and initiate at one of a conserved pair of AUG codons (nucleotides 6721-6) 103 nucleotides downstream of the IAPV IGR IRES (Fig. 1); (ii) the IGR IRES places a portion of ribosomes directly into the +1 reading frame at or near to the normal 0 frame IGR IRES initiation site; or (iii) IGR IRES initiation occurs only in the 0 frame, but a portion of ribosomes subsequently shift into the +1 reading frame at some specific slippery site (23). On the basis of the molecular mass of ORFx in reactions using our reporter constructs, we hypothesized that ORFx is initiated near the IAPV IGR IRES which would lead to an approximately 11.1-kDa ORFx protein.
To determine the initiation site of ORFx, stop codons were systematically introduced at different positions in the +1 frame downstream of the IGR IRES (Fig. 4A). Replacing the first, second, third, or 37th codon downstream from the IGR IRES with a UAG codon in the +1 frame abolished ORFx expression (Fig. 4C, lanes 7–10). Importantly, introducing these stop codons in the +1 frame did not significantly affect 0 frame IGR IRES-mediated sORF2 translation (Fig. 4C, lanes 7–10). The migration differences of the 0 frame sORF2 products is likely due to amino acid changes from introducing stop codons in the +1 frame. These results suggest that +1 frame ORFx translation initiates either close to or upstream of the 0 frame IGR IRES initiation site.
To determine whether IGR IRES-mediated 0 frame translation is required for +1 frame ORFx translation (e.g., if ORFx is translated by ribosomes frameshifting from the 0 frame), stop codons were introduced at different positions in the 0 frame (Fig. 4A). As expected, insertion of UAG codons in the 0 frame in any of the positions downstream of the IGR IRES abolished 0 frame sORF2 translation (Fig. 4C, lanes 4–6). Insertion of a UAG codon as the second or third codon in the 0 frame did not significantly affect +1 frame ORFx-FLuc translation, which suggests that +1 frame ORFx translation is independent of IGR IRES-mediated 0 frame ORF2 translation. Western blotting of the reactions using an anti-FLuc antibody confirmed the specific expression of the +1 frame ORFx-FLuc protein (Fig. 4C). However, replacing the initial 0 frame codon, Gly GGC, with a UAG stop codon eliminated not only 0 frame translation but also +1 frame ORFx-FLuc translation (Fig. 4C, lane 4).
U6562 can potentially base pair with G6618 and perhaps set the ribosome into the +1 frame to initiate from an Ala GCG6619-21 triplet (Fig. 4A, bold nucleotides). To test this hypothesis, G6618 was mutated to A, C, or U, and 0 and +1 frame translational activities were monitored by using dicistronic constructs (viii). We predicted that mutant IRESs that allow base pairing between nucleotides 6618 and 6562 should result in +1 frame ORFx translation. Mutating G6618 to C or U abolished +1 frame ORFx-FLuc expression, consistent with the idea that the U6562/G6618 base pair directs translation in the +1 frame (Fig. 4D, lanes 5 and 6). In contrast, mutating G6618 to A, which is predicted to maintain a U6562/A6618 base pair, retained +1 frame ORFx-FLuc translation, albeit at a level lower than wild type (Fig. 4D, lane 4). Mutants in which the base pairing was restored by compensatory mutations—U6562G/G6618C, U6562G/G6618U, and U6562A/G6618U—again regained ORFx-FLuc expression (Fig. 4D, lanes 7–9), with once again a lower expression for the A∶U base pairing than for the G∶C or G∶U base pairings. Quantitation of +1 and 0 frame translation is shown in Fig. 4E. In all cases, 0 frame expression was still detected in these mutant constructs. In confirmation of the expression of ORFx-FLuc, Western blotting using an anti-FLuc antibody detected the specific expression of the +1 frame ORFx-FLuc protein (Fig. 4D). Furthermore, Northern blotting confirmed that reporter RNAs were expressed similarly between experiments (Fig. 4D). +1 frame ORFx translation was 20% and 5% of the 0 frame translational activity in reporter constructs containing the wild-type and the G6618A mutant IAPV IGR IRESs, respectively (Fig. 4E). In contrast, the U6562G/G6618C mutations resulted in a large increase in the ORFx-FLuc/sORF2 ratio, which is likely due to a combination of higher +1 frame ORFx and decreased 0 frame sORF2 translation as compared to the wild-type IRES (Fig. 4E, left graph). One possibility is that a strong G∶C base pairing between nucleotides 6562 and 6618 is responsible for the higher +1 frame translation compared to that of the wild-type IRES where a weaker U∶G base pair exists. In summary, these results clearly demonstrate that base pairing between U6562 and G6618 directs +1 frame ORFx translation.
ORFx Translation from a Full-Length IAPV cDNA.
We next investigated whether ORFx can be expressed by using a full-length IAPV genome. We obtained a full-length IAPV cDNA and inserted an A at nucleotide 7348 to create a stop codon near the C terminus of VP2. From 5′ to 3′, the ORF2 encodes the VP2, VP4, VP3, and VP1 structural proteins that have predicted molecular masses of 35.5, 7.2, 33.4, and 23.8 kDa, respectively (27, 28). Insertion of the stop codon allows detection of 0 frame (VP2) and +1 frame (ORFx) translation without the effects of processing events of the ORF2 polyprotein. We incubated the full-length IAPV cDNA in Sf21 extracts in the presence of [35S]-methionine. The full-length IAPV cDNA produced two major proteins (approximately 34 and approximately 11 kDa) and one minor protein (24 kDa) in vitro (Fig. S2A, lane 2). To determine whether these proteins are expressed from the upstream nonstructural ORF1 or downstream structural ORF2, we disrupted the base pairing within PKI (ΔPKI) to inhibit expression of the downstream structural proteins. Disruption of PKI abolished expression of all of the proteins, which suggests that they are encoded within ORF2/ORFx (Fig. S2A, lane 3). Compensatory mutations that restore PKI rescued expression, which indicates that the expression of these proteins is IGR IRES-dependent (Fig. S2A, lanes 3 and 4). Furthermore, replacing the glycine GGC start codon of ORF2 with a UAG stop codon eliminated expression of the proteins, confirming that these proteins are likely encoded downstream of the IGR IRES (Fig. S2A, lane 6). To determine whether the approximately 34-kDa protein represents VP2, we raised a VP2 antibody against a peptide within the N terminus of VP2. The antibody detected the approximately 34-kDa protein in reactions that showed expression of the radiolabeled 34-kDa protein (Fig. S2A). From these findings, we conclude that the 34-kDa protein is encoded by the 5′ region of ORF2 and its presence is indicative of IAPV IGR IRES-mediated 0 frame translation. The lack of expression of the ORF1 proteins from the IAPV cDNA may be due to the weak 5′UTR IAPV IRES activity in this extract.
The expression of an approximately 11-kDa radiolabeled protein may represent ORFx expression. Like VP2, the expression of the 11-kDa protein is dependent on the integrity of the IGR IRES (Fig. S2A, lanes 2–4). Importantly, introduction of stop codons within the +1 frame downstream of the IGR IRES abolished expression of the 11-kDa protein but not 0 frame VP2 production, which demonstrates that the 11-kDa protein corresponds to the ORFx product (Fig. S2A, lanes 8–10). Furthermore, Western blotting using the anti-ORFx antibody confirmed the specific expression of ORFx (Fig. S2A). Northern blotting confirmed the expression of a full-length IAPV RNA. In summary, by using the IAPV full-length cDNA, both 0 frame VP2 and +1 frame ORFx proteins are expressed in Sf21 extracts.
The IAPV genome contains an AUG6592-4 codon in the +1 frame within the IGR IRES structure and tandem AUG codons at nucleotides 6721-26 (103 nt 3′ of the IGR IRES; Fig. 1). To check that ORFx initiation does not occur at these sites, we created mutations that substituted the AUG codons by mutating G6594A and AUGAUG6721-26 to ACGACG. Neither mutation abrogated expression of the 0 frame VP2 or +1 frame ORFx, which indicates that these AUG codons do not play a role in +1 frame translation initiation (Fig. S2A, lanes 5 and 11).
Our findings using dicistronic reporter constructs indicated that the U6562/G6618 base pairing directs +1 frame translation. To confirm these findings in the full-length IAPV cDNA, we created the same mutations and compensatory mutations at nucleotides 6562 and 6618. In summary, disruption of the base pairing between these two nucleotides eliminated, and compensatory mutations rescued, ORFx expression (Fig. S2 B and C). As expected, these mutations did not abolish 0 frame VP2 translation. Furthermore, specific VP2 and ORFx expression was confirmed by Western blotting (Fig. S2B). Similar to observations with the reporter constructs, a G∶C base pairing between nucleotides 6562 and 6618 directed higher +1 frame ORFx translation compared to an A∶U base pairing or the wild-type U∶G base pairing (Fig. S2C). Interestingly, the ratio of +1 frame ORFx to 0 frame VP2 expression was higher (approximately 0.5) with the full-length IAPV genome compared to the reporter RNA (Fig. S2C). One possibility is that sequences within the full-length IAPV genome missing from the dicistronic reporter RNA contribute to ORFx expression.
Our results showed that replacing the 0 frame GGC start codon with a UAG stop codon eliminated 0 and +1 frame translation (Fig. 4C and Fig. S3). To confirm that the loss of +1 frame ORFx translation is due to disruption of the base pairing between nucleotides 6562 and 6618, we created a compensatory mutation that restores the base pairing with the 0 frame stop codon. As shown previously, replacing the GGC start codon with a UAG stop codon abolished 0 and +1 frame translation (Fig. S3, lane 5). In contrast, mutating U6562G—which restores the G∶U base pairing between nucleotides 6562 and 6618—rescued +1 frame but not 0 frame translation (Fig. S3, lane 6). In summary, for both reporter constructs and the full-length IAPV genomic cDNA, ORFx is translated in vitro, and translation is mediated by base pairing between nucleotides 6562 and 6618 adjacent to the IGR IRES.
ORFx Translation Mediated by a Subset of Dicistroviruses.
Other viruses in the subset of dicistroviruses that contain an overlapping ORFx, apart from IAPV, include ABPV, KBV, and SINV-1 (23, 24). For these dicistroviruses, a predicted U∶G or C∶G base pairing adjacent to the IGR IRES PKI domain can potentially direct +1 frame ORFx translation similar to that mediated by the IAPV IRES (Fig. 5A). To test this hypothesis, we subcloned the ABPV, KBV, and SINV-1 IGRs, and part of the downstream ORF2, within the intergenic region of constructs (iv) (Fig. 5) and (vii) (Fig. S4). The ABPV, KBV, and SINV-1 IGR IRESs directed 0 frame ORF2-FLuc translation (Fig. 5B and Fig. S4). The 0 frame translation mediated by the KBV IGR IRES was approximately twofold higher compared to the other IRESs. Furthermore, similar to the IAPV IGR IRES, the predicted +1 frame ORFx product was detected from all IGR IRESs (Fig. 5B and Fig. S4). Note that the migration of the ORFx protein in the SDS-PAGE correlated with the predicted molecular masses for ABPV, KBV, and SINV-1. Interestingly, ORFx is more highly expressed from the IAPV and SINV-1 IGR IRESs than from the ABPV and KBV IGR IRESs (Fig. 5C). In summary, like IAPV, the KBV, ABPV, and SINV-1 IGR IRESs can direct ORFx translation in vitro.
Fig. 5.
A subset of dicistrovirus IGR IRESs direct ORFx translation. (A) Schematic of the PKI domains of the subset of dicistroviruses that contain the +1 frame ORFx. Black dashes denote the predicted additional base pairing within PKI which may direct +1 frame ORFx translation. (B) Dicistronic reporter constructs (iv) containing the KBV, ABPV, or SINV-1 IGR IRESs were incubated in Sf21 extracts. A representative SDS-PAGE of radiolabeled protein products detected by autoradiography is shown. In parallel, reactions were subjected to Northern blotting. (C) Quantitation of radiolabeled protein products normalized relative to the wild-type IAPV IGR IRES dicistronic reporter is shown at top. +1 frame ORFx or 0 frame sORF2-FLuc translated proteins were normalized to RLuc. The ratio of ORFx/sORF2-FLuc translation, taking into account the number of methionines within each translated protein, is shown. Averages ± SD are from at least three independent experiments.
Detection of ORFx in Vitro and in Vivo.
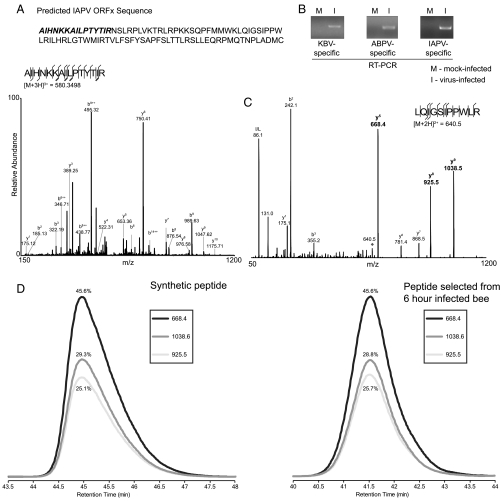
Our findings indicate that base pairing of nucleotides 6562 and 6618 can direct +1 frame ORFx translation initiating (in IAPV) at an alanine GCG6619-21 codon (Fig. 3A). To further confirm this result, in vitro synthesized ORFx was subjected to in-gel digestion with endopeptidase ArgC (which cleaves specifically after arginines), and the resulting peptides were analyzed by liquid chromatography-tandem mass spectrometry. A peptide with the sequence AIHNKKAILPTYTIR was detected, which corresponds to the predicted N terminus of ORFx on the basis of initiation at the GCG alanine codon (Fig. 6A). This result confirms that the in vitro translated ORFx initiates at GCG6619-21, 3′-adjacent to the IGR IRES.
Fig. 6.
Detection of ORFx in vitro and in virus-infected honey bee pupae. (A) Identification of the start site of ORFx. Fragment spectra from the 580.3498 Th, triply charged precursor ion of AIHNKKAILPTYTIR from ArgC-digested ORFx indicating that ORFx translation starts at this Ala residue. Individual fragment ions are annotated in the spectrum and in the sequence representation. (B) Detection of KBV, ABPV, and IAPV from virus-infected honey bee pupae by RT-PCR using virus-specific primers. RNA was extracted from honey bee pupae 96 h after injection with PBS (mock-infected) or virus particles. (C) Fragment spectra from a shared peptide of KBV and IAPV ORFx with the three y ions selected for transitions labeled in bold. (D) MRM traces for the pure synthetic peptide and the same three transitions detected in virally infected bees at 6 h after injection. The relative percentages of area under the curve for each transition are labeled.
We next examined whether ORFx is expressed in IAPV-infected honey bees by first obtaining and harvesting IAPV from infected honey bees. IAPV virions were injected into approximately 17-d-old honey bee pupae, and after 96 h, RNA and protein were extracted. By using virus-specific primers, RT-PCR analysis detected KBV, ABPV, and IAPV RNAs in virus-infected samples (Fig. 6B), which thus indicates that the virus sample we obtained contained a mixed population of viruses. Western blotting analysis detected the expression of IAPV VP2 but not IAPV ORFx, which suggests that ORFx may be expressed in low abundance during virus infection or may have more rapid turnover than VP2. However, using a more sensitive and specific multiple reaction monitoring-MS analysis, we detected the expression of a peptide that is found in both IAPV and KBV ORFx, in virally infected bees at 6 and 24 h after injection (Fig. 6 C and D). We monitored three different transitions, and the relative peak areas of those transitions in the infected samples were exactly the same as measured for the synthetic peptide. Importantly, no signal was seen in uninfected samples. These data demonstrate unequivocal evidence that ORFx is expressed during virus infection.
ORFx Translation when Specific Initiation Factors Are Compromised.
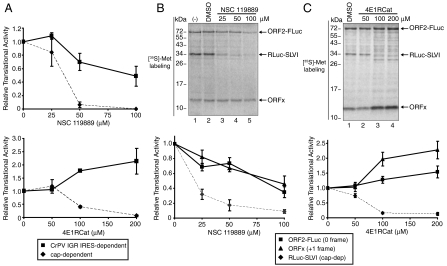
The IGR IRES can mediate translation under conditions in which cap-dependent translation is inhibited, in particular when specific initiation factors are compromised such as during endoplasmic reticulum stress (9, 29). Because ORFx translation depends on the IGR IRES, we predicted that ORFx would still be translated when cap-dependent translation is inhibited. Thus, we examined whether ORFx translation occurs when eIF2 activity or the cap-binding complex eIF4F are inhibited (eIF2 interacts with and recruits initiator Met-tRNAi; eIF4F is a complex that includes eIF4E and eIF4G). To inhibit these factors, we used two compounds discovered by Pelletier and colleagues: NSC119889, which inhibits eIF2 and Met-tRNAi interactions, and 4E1RCat, which binds to eIF4E and inhibits eIF4E and eIF4G interactions (30, 31). We monitored cap-dependent, CrPV IRES-dependent or IAPV IGR IRES-dependent 0 and +1 frame translation by incubating capped dicistronic reporter RNAs [construct (iv)] in Sf21 extract which was preincubated with increasing amounts of either NSC119889 or 4E1RCat. As shown previously, addition of NSC119889 or 4E1RCat resulted in a significant decrease in cap-dependent RLuc translation (> 90% decrease at the highest concentration of compound) (30, 31) (Fig. 7). In contrast, CrPV IGR IRES-mediated translation is relatively resistant to treatment with these compounds (Fig. 7A). At 50 μM NSC119889, cap-dependent translation is inhibited by approximately 95%, whereas CrPV IGR IRES-mediated translation remained relatively active (approximately 75% compared to untreated) (Fig. 7A). This result is similar to that observed in rabbit reticulocyte lysates (RRL) treated with NSC119889 (20). Like the CrPV IGR IRES, IAPV IGR IRES-mediated 0 frame ORF2-FLuc and +1 frame ORFx translation also remained relatively resistant to treatment with these compounds (Fig. 7 B and C). Interestingly, 4E1RCat treatment stimulated CrPV IGR IRES-mediated and IAPV IGR IRES-mediated 0 and +1 frame translation by approximately 1.5- to 2.5-fold (Fig. 7 A and C). In summary, like ORF2, ORFx can be efficiently translated when cap-dependent translation initiation is inhibited.
Fig. 7.
The 0 and +1 frame translation when eIF2 or eIF4F activity is compromised in vitro. Capped in vitro transcribed dicistronic reporter RNAs containing the (A) CrPV IGR IRES or (B and C) IAPV IGR IRES (construct iv) were incubated in Sf21 extracts alone, with DMSO or with NSC119889 or 4E1RCat, which inhibit eIF2 activity or eIF4E-eIF4G interactions, respectively. Extracts were preincubated for 5 min with the compound prior to adding the reporter RNA. Shown are the relative quantitation of the cap-dependent (RLuc), 0 frame (ORF2-FLuc), and +1 frame (ORFx) translation radiolabeled protein products. All calculations are from at least three independent experiments ± SD.
Discussion
The dicistrovirus IGR IRES has the remarkable ability to directly recruit ribosomes and set them into a specific reading frame through a tRNA-like anticodon-codon interaction occupying the ribosomal P site and making the ribosomal A site available for delivery of aminoacyl-tRNA. Here, we describe a mechanism used by a subset of dicistrovirus IGR IRESs that can selectively place ribosomes and initiate translation into either of two distinct open reading frames. The consequence of this mechanism is that, on top of initiating translation in the 0 frame to produce the viral ORF2-encoded structural proteins, an overlapping ORFx in the +1 frame is also translated. Translation of ORFx is directed by a U∶G base pairing adjacent to the tRNA-like anticodon-codon interaction of the PKI domain. Several lines of evidence support this result: (i) Disruption of the PKI domain or the U6562/G6618 base pairing abolishes ORFx translation, whereas compensatory mutations that restore base pairing rescues ORFx translation. The U∶G base pairing directs the ribosome to initiate translation at a +1 frame GCG alanine codon occupying the ribosomal A site. (ii) Detection of a peptide from an in vitro synthesized ORFx by mass spectrometric analysis confirmed that ORFx is initiated at this GCG codon. (iii) Detection of an IAPV and KBV ORFx peptide in virally infected honey bees by a sensitive selective multiple reaction monitoring (MRM)-MS approach demonstrated that ORFx is expressed during virus infection. (iv) Selective pressure to maintain the overlapping ORFx within a subset of dicistroviruses strongly suggests a mechanism to direct translation of ORFx (23, 24).
The tRNA-like anticodon-codon interaction is apparent from structural studies of the PKI domain of the type I IGR IRES, and, although the structure of the type II IGR IRES has not yet been solved, it is presumed to be similar (12–14, 32). In our in vitro assays, +1 frame translation occurs 20% of the time as compared to 0 frame translation (Fig. 4E). In general, disruption of +1 frame translation variably affects 0 frame translation (Fig. 4E and Fig. S2), but there does not appear to be a strict correlation to suggest competition for ribosome site selection in the 0 or +1 frame (i.e., the extra U∶G base pairing does not appear to occlude the 0 frame A site such that the A-site tRNA can be delivered only into the +1 frame). However, mutations that disrupt +1 frame translation also change the 0 frame initiation codon and thus make it difficult to interpret whether alterations in 0 frame translation are due to effects by disruption of +1 frame translation or due to delivery of a different aminoacyl-tRNA to the ribosomal A site. Currently, it is not clear if two distinct conformations exist that direct 0 and +1 frame translation or if initiation of 0 and +1 frame translation relies on the same tRNA-like structure of the PKI domain and that the extra U∶G base pairing directs +1 frame translation. Consistent with the latter hypothesis is that, similar to 0 frame translation, +1 frame translation requires base pairing of the PKI region (Fig. 4B and Fig. S2). This scenario is reminiscent of the base pairing that has been postulated by some for a subset of tRNA frameshift mutant suppressors that shift the reading frame by +1 nucleotide (33). The difference between the two mechanisms is that tRNA frameshift mutant suppressors shift the reading frame of elongating ribosomes, whereas the IAPV IGR IRES shifts the reading frame of an initiating ribosome, albeit a ribosome that is initiating directly into the elongation phase from the A site.
Although the in vitro translation system allows relatively high +1 frame translation compared to 0 frame translation, we had to resort to a very sensitive MRM-MS approach to detect ORFx in virus-infected honey bees (Fig. 6). It is possible that ORFx is translated only during a specific window of virus infection or that ORFx is inherently unstable (relative to the structural proteins) and is rapidly degraded. However, we show that, like IGR IRES-mediated 0 frame translation, +1 frame ORFx translation is relatively resistant to conditions in which eIF2 or eIF4F activities are compromised in in vitro experiments (Fig. 7). The stimulatory effect of IRES activity under 4E1RCat treatment is similar to that observed previously (30) and supports the idea that inhibition of cap-dependent translation frees ribosomes for IRES translation. In summary, because both 0 and +1 frame translation are IGR IRES-dependent, it is likely that both frames are regulated similarly during the viral life cycle, especially when certain initiation factors are compromised. We note that in the insect lysate NSC119889 treatment does not stimulate IRES activity in contrast to that shown in a study using Krebs extract (31). In RRL, we have also shown that translation mediated by the CrPV IRES and the type II TSV IRES is not stimulated but remains relatively resistant to NSC119889 treatment (20). Thus, the difference in NSC119889 sensitivity to IRES translation is probably dependent on the experimental system used. It will be of interest to examine these discrepancies on IRES activity in more detail.
IAPV, KBV, ABPV, and SINV-1 IGR IRESs can all mediate ORFx translation, and an ORFx is present in all four viruses; however, SLVI is present only in IAPV, KBV, and ABPV (23) (Fig. 5). The in vitro experiments showed that the presence of SLVI does not affect the ratio of +1 frame ORFx and 0 frame ORF2 translation (Fig. 2). Instead, we show that the integrity and presence of SLVI and part of the downstream ORF2 region moderately increases IGR IRES activity in general (Figs. 2 and 3). Previous reports have shown that unstructured regions outside of the IGR IRES may help the IRES fold properly and stimulate IRES activity (7, 25). Similarly, SLVI and the downstream ORF2 region may have a role in assisting in the proper folding of the IRES. The precise role of SLVI in IRES structure and activity are currently being investigated. Mutations (M1 and M2; Fig. 3) that are predicted to disrupt the helical stem of SLVI decrease IRES activity. We are currently investigating whether these mutations have an effect on the structure of SLVI specifically or on the IRES structure itself. Interestingly, fusing the UAA stop codon in SLVI in frame with the upstream ORF moderately decreased translation of both upstream and downstream reporter genes (Fig. 2). This result suggests that translation of both ORFs is negatively coupled. Consistent with this model, when a strong hairpin is inserted in the 5′UTR to inhibit scanning-dependent initiation of the upstream ORF in the reporter construct, translation of the downstream ORF driven by the IGR IRES increases but only when the UAA stop codon is fused in frame with the upstream ORF (Fig. S1). One model is that ribosomes translating the upstream ORF must unwind SLVI in order to reach the UAA stop codon, which effectively disrupts SLVI, and thus decreases IGR IRES-mediated 0 and +1 frame translation. Ribosomes encountering SLVI have to unwind a strong helical region—which may induce sufficient pausing to create a ribosome “logjam,” thus also leading to a modest down-regulation of translation of the upstream ORF. It is possible that SLVI plays a role in temporal regulation of the translation of the nonstructural and structural proteins during honey bee virus infection. The dicistrovirus ORF1 encodes the nonstructural proteins such as the replicase and protease, which would presumably have to be expressed early during infection for viral replication and protein processing. An interesting scenario would be that translation of ORF1 during honey bee virus infection down-regulates translation of the downstream ORF2, which encodes the structural proteins, as production of the capsid proteins is not needed until virion packaging is required. At later stages of infection, translation of ORF1 may be down-regulated because of factor dependencies of the 5′UTR IRES, which results in the reformation of SLVI and stimulation of IGR IRES-mediated translation of the structural proteins. It has been well-documented that the structural proteins are produced in molar excess over the nonstructural proteins (2, 15, 34). During CrPV infection in Drosophila S2 cells, translation of the two ORFs does not appear to be temporally regulated (15). The presence of SLVI in the honey bee viruses may play a role in regulating translation of the different ORFs for optimal total and/or temporal expression of the viral proteins. Alternatively, SLVI may have roles besides translation such as viral replication or RNA stability.
The selective pressure to maintain ORFx in the honey bee viruses strongly indicates a prominent biological function, possibly a role in combating the innate immune response or a direct role in viral translation or replication. ORFx does not have any obvious domains that would hint at its function (23, 24). It will be of considerable interest to obtain an infectious honey bee dicistrovirus cDNA to determine the role of ORFx during virus infection. RNA viruses have evolved many elegant strategies to optimize the coding capacity of compact genomes via the utilization of overlapping genes whereby one nucleotide sequence codes for two (or even three) different proteins in different reading frames. The controlled expression of multiple overlapping genes may be directed at the pretranslational stage (e.g., via subgenomic RNAs in the nodaviruses), at the stage of translation initiation (e.g., via leaky ribosomal scanning in a number of RNA viruses), or at the stage of translation elongation (e.g., via ribosomal frameshifting as in alphaviruses). The discovery that a subset of IGR IRESs can precisely select alternative reading frames for translation initiation highlights the remarkable strategies that viruses utilize to increase their coding capacity.
Materials and Methods
In Vitro Translation.
Dicistronic reporter constructs (1 μg), linearized with XbaI, were incubated in Sf21 extract (Promega) for 2 h at 30 °C in the presence of [35S]-methionine (PerkinElmer). Plasmid constructs are described in more detail in SI Materials and Methods. For optimal IRES activity, a final concentration of 0.5 mM MgCl2/40 mM KOAc was used. Proteins were analyzed by 16% SDS-PAGE and by using a Typhoon imager (Amersham).
MRM Assay.
The synthesized peptide was diluted 100-fold by using 0.5% acetic acid, and multiple injections of 1 μL each were used to optimize the fragmentor voltage (0–180 V, 20-V increments) and subsequently the collision energy (5–25 V, 5-V increments; 13–18 V, 1-V increments). Fragment ions with a mass-to-charge ratio (m/z) greater than the precursor ion m/z were prioritized, leading to three transitions that were selected for the MRM assay. A fragmentor voltage of 40 V was selected for all transitions, and a collision energy of 15 V was selected for transitions 640.6 → 1038.6 and 640.6 → 668.4 and of 14 V was selected for transition 640.6 → 925.5.
Virus Infection in Honey Bee.
Honey bee pupae (approximately 17 d old) injected with a mixture of IAPV, KBV, and ABPV were dissected on ice to separate the head and body. Samples were bead-homogenized in 1% sodium deoxycholate and 50 mM NH4HCO3 at three pulses of 20 s and 6.5 m/s, followed by incubation at 99 °C for 5 min. The lysate was centrifuged at 8,000 × g for 10 min, and the resulting supernatant was further centrifuged at 5,000 × g for 5 min, all at 4 °C. The final supernatant was subjected to ethanol precipitation. Proteins precipitated after 1.5 h incubation at RT were collected by centrifugation at 16,100 × g. The protein pellet was resolubilized in 6 M urea, 2 M thiourea, and 100 mM Tris-Cl (pH 8.0), and any insoluble material was subsequently removed by centrifugation at 16,100 × g. Resolubilized proteins were resolved on a 16% SDS gel, and a band corresponding to the molecular mass of ORFx was excised and in-gel digested with trypsin. The resulting peptides were stored on stop and go extraction tips until MS analysis (35).
Supplementary Material
Acknowledgments.
We thank Paul Van Westendorp (British Columbia Ministry of Agriculture and Lands Diagnostic Laboratory) for providing virus-infected bees. The work was supported by a Canadian Institute of Health Research (CIHR) grant [MOP-81244] (to E.J.), a Natural Sciences and Engineering Research Council Discovery grant (L.J.F.), Science Foundation Ireland [08/IN.1/B1889] and National Institutes of Health grants [R01 GM079523] (to J.F.A), Wellcome Trust [088789] (to A.E.F), and China Scholarship Council (Q.R.). L.J.F. is the Canada Research Chair in Quantitative Proteomics. The mass spectrometry infrastructure was supported by the Canada Foundation for Innovation, the BC Knowledge Development Fund, and the BC Proteomics Network. E.J. is a Michael Smith Foundation for Health Research Scholar and CIHR New Investigator scholar.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. P.S. is a guest editor invited by the Editorial Board.
See Author Summary on page 4033 (volume 109, number 11).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1111303109/-/DCSupplemental.
References
- 1.Bonning BC, Miller WA. Dicistroviruses. Annu Rev Entomol. 2010;55:129–150. doi: 10.1146/annurev-ento-112408-085457. [DOI] [PubMed] [Google Scholar]
- 2.Wilson JE, Powell MJ, Hoover SE, Sarnow P. Naturally occurring dicistronic cricket paralysis virus RNA is regulated by two internal ribosome entry sites. Mol Cell Biol. 2000;20:4990–4999. doi: 10.1128/mcb.20.14.4990-4999.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cox-Foster DL, et al. A metagenomic survey of microbes in honey bee colony collapse disorder. Science. 2007;318:283–287. doi: 10.1126/science.1146498. [DOI] [PubMed] [Google Scholar]
- 4.Jan E, Kinzy TG, Sarnow P. Divergent tRNA-like element supports initiation, elongation, and termination of protein biosynthesis. Proc Natl Acad Sci USA. 2003;100:15410–15415. doi: 10.1073/pnas.2535183100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jan E, Sarnow P. Factorless ribosome assembly on the internal ribosome entry site of cricket paralysis virus. J Mol Biol. 2002;324:889–902. doi: 10.1016/s0022-2836(02)01099-9. [DOI] [PubMed] [Google Scholar]
- 6.Pestova TV, Hellen CU. Translation elongation after assembly of ribosomes on the cricket paralysis virus internal ribosomal entry site without initiation factors or initiator tRNA. Genes Dev. 2003;17:181–186. doi: 10.1101/gad.1040803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sasaki J, Nakashima N. Translation initiation at the CUU codon is mediated by the internal ribosome entry site of an insect picorna-like virus in vitro. J Virol. 1999;73:1219–1226. doi: 10.1128/jvi.73.2.1219-1226.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sasaki J, Nakashima N. Methionine-independent initiation of translation in the capsid protein of an insect RNA virus. Proc Natl Acad Sci USA. 2000;97:1512–1515. doi: 10.1073/pnas.010426997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilson JE, Pestova TV, Hellen CU, Sarnow P. Initiation of protein synthesis from the A site of the ribosome. Cell. 2000;102:511–520. doi: 10.1016/s0092-8674(00)00055-6. [DOI] [PubMed] [Google Scholar]
- 10.Nishiyama T, et al. Structural elements in the internal ribosome entry site of Plautia stali intestine virus responsible for binding with ribosomes. Nucleic Acids Res. 2003;31:2434–2442. doi: 10.1093/nar/gkg336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pfingsten JS, Costantino DA, Kieft JS. Structural basis for ribosome recruitment and manipulation by a viral IRES RNA. Science. 2006;314:1450–1454. doi: 10.1126/science.1133281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pfingsten JS, Costantino DA, Kieft JS. Conservation and diversity among the three-dimensional folds of the Dicistroviridae intergenic region IRESes. J Mol Biol. 2007;370:856–869. doi: 10.1016/j.jmb.2007.04.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schuler M, et al. Structure of the ribosome-bound cricket paralysis virus IRES RNA. Nat Struct Mol Biol. 2006;13:1092–1096. doi: 10.1038/nsmb1177. [DOI] [PubMed] [Google Scholar]
- 14.Zhu J, et al. Crystal structures of complexes containing domains from two viral internal ribosome entry site (IRES) RNAs bound to the 70S ribosome. Proc Natl Acad Sci USA. 2011;108:1839–1844. doi: 10.1073/pnas.1018582108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garrey JL, Lee YY, Au HH, Bushell M, Jan E. Host and viral translational mechanisms during cricket paralysis virus infection. J Virol. 2010;84:1124–1138. doi: 10.1128/JVI.02006-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jan E. Divergent IRES elements in invertebrates. Virus Res. 2006;119:16–28. doi: 10.1016/j.virusres.2005.10.011. [DOI] [PubMed] [Google Scholar]
- 17.Nakashima N, Uchiumi T. Functional analysis of structural motifs in dicistroviruses. Virus Res. 2009;139:137–147. doi: 10.1016/j.virusres.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 18.Jang CJ, Lo MC, Jan E. Conserved element of the dicistrovirus IGR IRES that mimics an E-site tRNA/ribosome interaction mediates multiple functions. J Mol Biol. 2009;387:42–58. doi: 10.1016/j.jmb.2009.01.042. [DOI] [PubMed] [Google Scholar]
- 19.Hertz MI, Thompson SR. In vivo functional analysis of the Dicistroviridae intergenic region internal ribosome entry sites. Nucleic Acids Res. 2011;39:7276–7288. doi: 10.1093/nar/gkr427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jang CJ, Jan E. Modular domains of the Dicistroviridae intergenic internal ribosome entry site. RNA. 2010;16:1182–1195. doi: 10.1261/rna.2044610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cevallos RC, Sarnow P. Factor-independent assembly of elongation-competent ribosomes by an internal ribosome entry site located in an RNA virus that infects penaeid shrimp. J Virol. 2005;79:677–683. doi: 10.1128/JVI.79.2.677-683.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hatakeyama Y, Shibuya N, Nishiyama T, Nakashima N. Structural variant of the intergenic internal ribosome entry site elements in dicistroviruses and computational search for their counterparts. RNA. 2004;10:779–786. doi: 10.1261/rna.5208104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Firth AE, Wang QS, Jan E, Atkins JF. Bioinformatic evidence for a stem-loop structure 5′-adjacent to the IGR-IRES and for an overlapping gene in the bee paralysis dicistroviruses. Virol J. 2009;6:193–200. doi: 10.1186/1743-422X-6-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sabath N, Price N, Graur D. A potentially novel overlapping gene in the genomes of Israeli acute paralysis virus and its relatives. Virol J. 2009;6:144–150. doi: 10.1186/1743-422X-6-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shibuya N, Nishiyama T, Kanamori Y, Saito H, Nakashima N. Conditional rather than absolute requirements of the capsid coding sequence for initiation of methionine-independent translation in Plautia stali intestine virus. J Virol. 2003;77:12002–12010. doi: 10.1128/JVI.77.22.12002-12010.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vattem KM, Wek RC. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc Natl Acad Sci USA. 2004;101:11269–11274. doi: 10.1073/pnas.0400541101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maori E, et al. Isolation and characterization of Israeli acute paralysis virus, a dicistrovirus affecting honeybees in Israel: Evidence for diversity due to intra- and inter-species recombination. J Gen Virol. 2007;88:3428–3438. doi: 10.1099/vir.0.83284-0. [DOI] [PubMed] [Google Scholar]
- 28.Agirre J, et al. Capsid protein identification and analysis of mature Triatoma virus (TrV) virions and naturally occurring empty particles. Virology. 2011;409:91–101. doi: 10.1016/j.virol.2010.09.034. [DOI] [PubMed] [Google Scholar]
- 29.Fernandez J, Yaman I, Sarnow P, Snider MD, Hatzoglou M. Regulation of internal ribosomal entry site-mediated translation by phosphorylation of the translation initiation factor eIF2alpha. J Biol Chem. 2002;277:19198–19205. doi: 10.1074/jbc.M201052200. [DOI] [PubMed] [Google Scholar]
- 30.Cencic R, et al. Reversing chemoresistance by small molecule inhibition of the translation initiation complex eIF4F. Proc Natl Acad Sci USA. 2011;108:1046–1051. doi: 10.1073/pnas.1011477108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robert F, et al. Initiation of protein synthesis by hepatitis C virus is refractory to reduced eIF2 GTP.Met-tRNA(i)(Met) ternary complex availability. Mol Biol Cell. 2006;17:4632–4644. doi: 10.1091/mbc.E06-06-0478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Costantino DA, Pfingsten JS, Rambo RP, Kieft JS. tRNA-mRNA mimicry drives translation initiation from a viral IRES. Nat Struct Mol Biol. 2008;15:57–64. doi: 10.1038/nsmb1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Atkins JF, Bjork GR. A gripping tale of ribosomal frameshifting: Extragenic suppressors of frameshift mutations spotlight P-site realignment. Microbiol Mol Biol Rev. 2009;73:178–210. doi: 10.1128/MMBR.00010-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moore NF, Kearns A, Pullin JSK. Characterization of cricket paralysis virus-induced polypeptides in Drosophila cells. J Virology. 1980;33:1–9. doi: 10.1128/jvi.33.1.1-9.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chan QW, Howes CG, Foster LJ. Quantitative comparison of caste differences in honeybee hemolymph. Mol Cell Proteomics. 2006;5:2252–2262. doi: 10.1074/mcp.M600197-MCP200. [DOI] [PubMed] [Google Scholar]