Abstract
Although the complexity and circuitry of nervous systems undergo evolutionary change, we lack understanding of the general principles and specific mechanisms through which it occurs. The Drosophila larval neuromuscular junction (NMJ), which has been widely used for studies of synaptic development and function, is also an excellent system for studies of synaptic evolution because the genus spans >40 Myr of evolution and the same identified synapse can be examined across the entire phylogeny. We have now characterized morphology of the NMJ on muscle 4 (NMJ4) in >20 species of Drosophila. Although there is little variation within a species, NMJ morphology and complexity vary extensively between species. We find no significant correlation between NMJ phenotypes and phylogeny for the species examined, suggesting that drift alone cannot explain the phenotypic variation and that selection likely plays an important role. However, the nature of the selective pressure is still unclear because basic parameters of synaptic function remain uniform. Whatever the mechanism, NMJ morphology is evolving rapidly in comparison with other morphological features because NMJ phenotypes differ even between several sibling species pairs. The discovery of this unexpectedly extensive divergence in NMJ morphology among Drosophila species provides unique opportunities to investigate mechanisms that regulate synaptic growth; the interrelationships between synaptic morphology, neural function, and behavior; and the evolution of nervous systems and behavior in natural populations.
Over the course of evolution, the complex circuitry of nervous systems is modified both by the addition of new types of neurons and by alterations in the synaptic connectivity of preexisting neurons (1). These changes presumably provide the physical basis for acquisition of new behaviors that serve adaptive functions as new species evolve. Although the evolution of brains and behaviors is of fundamental biological importance, we lack comprehensive understanding of the general principles governing these processes or the specific mechanisms and molecules through which evolutionary changes are effected. Because synapses are the basic structural and functional units of nervous systems, one way to begin to address these problems is to investigate a defined synapse across evolutionary time, which allows us to correlate morphological variation with genetic differences.
The Drosophila larval neuromuscular junction (NMJ) is a powerful system for investigating molecular mechanisms of synaptic growth and development. The muscles are large, arranged in an invariant, segmentally repeating pattern, and each muscle is innervated by the same identified motor neurons and forms NMJs with stereotypic morphology in each animal. These NMJs are large, easily accessible for microscopic and electrophysiological analyses, and their morphological features, such as the number of synaptic boutons and branch points, can be readily observed and quantified. A number of investigators have taken advantage of these features to identify genes encoding positive and negative regulators of NMJ growth that have revealed the signaling pathways regulating NMJ development (2).
The same features of the larval NMJ that make it well suited for developmental studies also make it an ideal system for studying the evolution of synaptic morphology by analyzing NMJ phenotypes in different species of Drosophila. The overall body plan, musculature, and motor innervation pattern are precisely conserved among all species of Drosophila and are even shared with more distantly related groups of Diptera (3), despite enormous differences in size, habitat, food source, predation, and other aspects of their natural history and concomitant differences in behavior. Moreover, the subgenus Drosophila by itself encompasses at least 40 Myr of evolutionary history (4). The conservation of body plan and muscle innervation over the evolutionary history of Drosophila immediately raises the question of whether NMJ morphology is also conserved or has undergone modification in different species. Because the body plan is conserved, it is possible to compare exactly the same genetically determined synapse among all species of Drosophila, which is particularly advantageous for this type of analysis.
Drosophila has featured prominently in the molecular analysis of regulatory changes in gene expression and developmental mechanisms that underlie evolutionary changes in morphological features among different species such as wing spots, body coloration, and denticle pattern (5–8). Surprisingly, however, synaptic morphology, with its implications for evolution of brains and behavior, has not previously been examined as an evolutionary trait among different species of Drosophila. Thus, an investigation of variation in NMJ morphology among different species of Drosophila should provide a wealth of important new information with implications for the evolution of nervous systems.
Here, we describe the results of such an investigation. We examined NMJ morphology in wild-caught Drosophila melanogaster for comparison with laboratory-bred wild-type stocks. We also examined NMJ morphology in >20 different species of Drosophila. Whereas variation in NMJ morphology within a species is limited, we discovered a surprisingly extensive degree of variation among different species. No phylogenetic structure is apparent among the NMJ phenotypes, suggesting that genetic drift alone cannot explain the observed phenotypic variation and that selection is likely to play a predominant role in sculpting the NMJ. Compared with evolution of other morphological traits, NMJ morphology appears to be evolving rapidly as revealed by the striking differences in phenotype we find even among sibling species. Despite the vast differences in NMJ structure, the basic electrophysiological features of synaptic function are relatively constant. Understanding why NMJ morphology should have undergone such dramatic divergence, the adaptive and behavioral consequences of the different phenotypes, and the genetic and molecular mechanisms that generate them are important challenges for the future.
Results
NMJ Morphology Shows Extensive Phenotypic Variation Among Different Species of Drosophila.
Despite differences in size, habitat, food preference, and natural history, all species of Drosophila share a common larval body plan with identical body wall musculature and motor neuron innervations. Fig. 1 shows the musculature of three Drosophila species: D. melanogaster, Drosophila punjabiensis (the smallest species examined), and Drosophila virilis (the largest and most distant from D. melanogaster of the species examined). However, the details of the morphological features of larval NMJs in these species have not previously been described. Is synaptic morphology also conserved or does it vary among species? If the phenotype is variable, does it show any predictable pattern of variation among different Drosophila species?
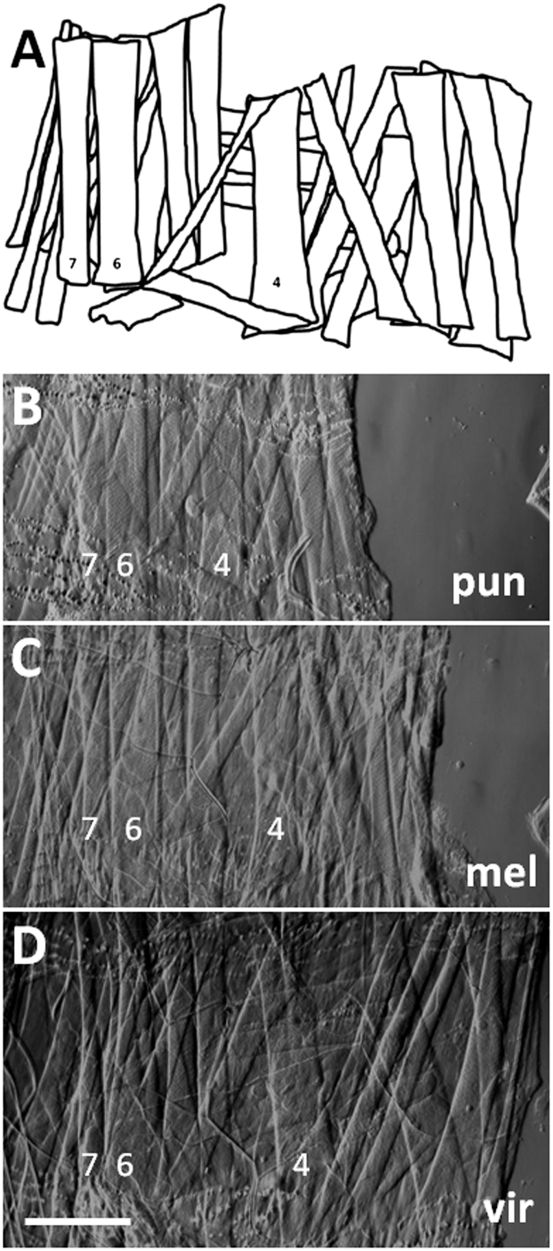
Fig. 1.
(A–D) Larval body wall musculature is precisely conserved among Drosophila species. Bright-field images of dissected third-instar larvae from D. punjabiensis (pun), D. melanogaster (mel), and D. virilis (vir) show musculature in right hemisegment 2. (A) Diagram illustrates the complete pattern of musculature in each of these species. All of these muscles cannot be seen in the microscopic images, which were focused on the most superficial muscle layers. For orientation, muscles 4, 6, and 7 are labeled. This analysis focused on the NMJs that form on muscle 4. (Scale bar, 200 μm.)
As a basis for comparison, we first examined a number of isolates from natural populations of D. melanogaster to determine the extent of variation in synaptic morphology within a species. To describe the phenotype quantitatively, we counted the number of synaptic boutons and branch points, commonly used metrics for this type of analysis, focusing on the NMJ on muscle 4 (NMJ4) because of its relative simplicity. At least 20 isolates were examined both from local populations in Madison, Wisconsin and from more distant populations including African isolates. Although we did find an occasional variant, NMJ morphology in natural populations of D. melanogaster is generally uniform and comparable to the phenotype described for wild-type laboratory stocks (average number of boutons per NMJ4 ranged from 17 to 28 with a SE of 1.0 between groups, which is comparable to the error observed within a single population).
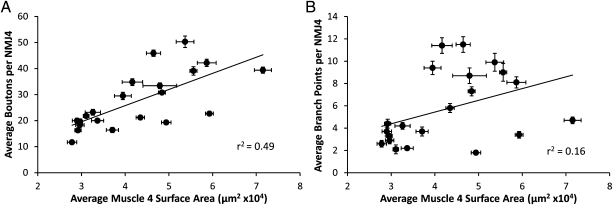
We performed a similar quantitative analysis of NMJ4 morphology on 20 additional species of Drosophila. Although we again found little variation within each species for the parameters we measured, we discovered surprisingly extensive variation in NMJ size and complexity between species (Fig. 2 and Table 1). Indeed, the range of phenotypes we observed in these species from D. punjabiensis to Drosophila funebris is comparable to the phenotypic differences observed in D. melanogaster between mutants with the most severe NMJ undergrowth (∼10 boutons per NMJ4) (9) to those with the most elaborate overgrowth (∼50 boutons per NMJ4) (10). A substantial fraction of this variation in bouton number can be attributed to differences in larval body size (as estimated by average surface area of muscle 4) (Table 1 and Fig. 3). This result is in accord with the correlation between bouton number and muscle size observed in D. melanogaster throughout larval development and reflects the fact that as muscles increase in size they require more neurotransmitter release from presynaptic motor terminals to drive their contraction (11). Nonetheless, the value of r2 for the correlation between bouton number and muscle size for NMJ4 among the different species (Fig. 3) indicates that only ∼50% of the variation in bouton number can be attributed to variation in muscle size, with the remaining 50% dependent on other factors. Moreover, variations in synaptic complexity and architecture are not strongly dependent on muscle size. For example, r2 for the correlation between NMJ branch points and muscle area for NMJ4 is only 0.16 (Table 1 and Fig. 3). We also note that the parameters we have measured capture only a portion of the observed variation in NMJ phenotypes as some species also exhibit novel morphological features that are difficult to quantify such as the unusual arrangement or appearance of boutons observed in Drosophila vulcana, with boutons growing off the main stem rather than the typical “beads-on-a-string” morphology, and Drosophila willistoni, with boutons appearing diffuse and not readily resolved from the rest of the axon terminal. These differences in NMJ morphology that we observe are not limited to NMJ4 as we observe similar phenotypic differences for NMJ6/7 and NMJ12/13. Thus, in contrast to the overall larval body plan, which is invariant among Drosophila species, NMJ morphology shows remarkably extensive evolutionary divergence.
Fig. 2.
Larval NMJs show extensive morphological variation in different species of Drosophila. Third-instar larvae of each species were dissected and stained with FITC-conjugated anti-HRP. Representative confocal images of NMJ4 in abdominal segment 2 are shown. There is substantial variation in NMJ morphology among these species with respect to overall geometry, number of boutons, and number of branch points. The 11 species enclosed by red boxes are those whose genome has been completely sequenced and whose precise phylogenetic relationships have been ascertained. (Scale bar, 20 μm.)
Table 1.
Quantification of bouton number, branch points, and muscle 4 surface area in various species of Drosophila
| n | Boutons | Branch points | Muscle surface area (μm2 × 104) | |
| ananassae | 25 | 22.7 ± 0.8 | 3.4 ± 0.3 | 5.44 ± 0.23 |
| erecta | 25 | 29.5 ± 1.4 | 9.4 ± 0.6 | 3.95 ± 0.19 |
| funebris | 30 | 42.2 ± 1.4 | 8.1 ± 0.5 | 5.87 ± 0.22 |
| greeni | 25 | 33.4 ± 1.1 | 8.7 ± 0.7 | 4.79 ± 0.39 |
| jambulina | 25 | 19.5 ± 1.2 | 3.3 ± 0.4 | 2.95 ± 0.07 |
| malerkotliana | 30 | 18.3 ± 0.8 | 2.9 ± 0.3 | 2.96 ± 0.10 |
| mauritiana | 25 | 30.8 ± 0.7 | 7.3 ± 0.4 | 4.84 ± 0.18 |
| melanogaster | 45 | 19.3 ± 0.8 | 1.8 ± 0.2 | 4.94 ± 0.11 |
| mojavensis | 30 | 34.8 ± 1.3 | 11.4 ± 0.7 | 4.16 ± 0.23 |
| pallidosa | 25 | 21.8 ± 0.7 | 2.1 ± 0.4 | 3.11 ± 0.06 |
| persimilis | 25 | 39.1 ± 1.6 | 9.0 ± 0.8 | 5.56 ± 0.08 |
| pseudoobscura | 30 | 50.3 ± 2.2 | 9.9 ± 0.8 | 5.37 ± 0.20 |
| punjabiensis | 25 | 11.7 ± 0.5 | 2.6 ± 0.3 | 3.78 ± 0.11 |
| sechellia | 45 | 45.9 ± 1.2 | 11.5 ± 0.7 | 4.65 ± 0.16 |
| seguyi | 30 | 23.2 ± 1.1 | 4.2 ± 0.3 | 3.26 ± 0.17 |
| serrata | 25 | 20.0 ± 0.9 | 3.7 ± 0.4 | 2.90 ± 0.11 |
| simulans | 30 | 21.2 ± 0.7 | 5.8 ± 0.4 | 4.35 ± 0.10 |
| virilis | 30 | 39.4 ± 1.2 | 4.7 ± 0.3 | 7.15 ± 0.19 |
| vulcana | 35 | 16.3 ± 0.7 | 4.4 ± 0.4 | 2.92 ± 0.07 |
| willistoni | 30 | 20.0 ± 0.5 | 2.2 ± 0.2 | 3.97 ± 0.14 |
| yakuba | 25 | 16.4 ± 1.0 | 3.7 ± 0.4 | 3.71 ± 0.14 |
Mean ± SEM is shown. All analyses were performed on NMJ4 in abdominal segments 2–4 of each species.
Fig. 3.
Differences in muscle size cannot fully explain all of the variation observed in NMJ size. For each species the average numbers of boutons (A) or branch points (B) per NMJ4 are plotted against the average muscle 4 surface area with the linear regression line superimposed. (Scale bars, SEM for each parameter.)
NMJ Phenotypes Do Not Correlate with Phylogenetic Relationships.
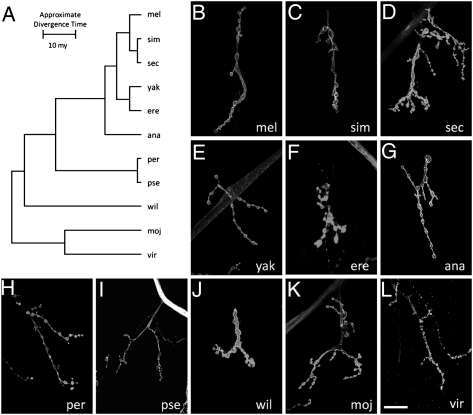
The discovery of this striking and extensive variation in NMJ morphology among Drosophila species immediately raises the question of what biological significance this variation has. One way to begin to address this question is to determine whether any consistent patterns emerge when we compare morphological phenotypes with the phylogenetic relationships among the various species. Are the NMJ phenotypes for two closely related species more similar than for two distantly related species? To minimize uncertainty about the phylogeny, we focused on 11 species of Drosophila whose genomes have been sequenced and whose evolutionary relationships are therefore known with a high degree of accuracy (12).
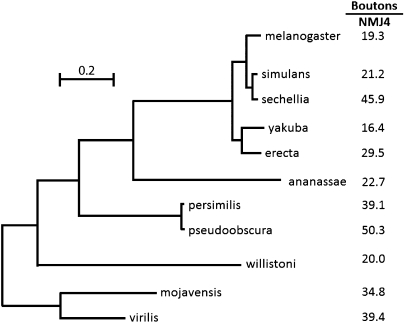
The phylogenetic tree for these 11 species is shown in Fig. 4. The number of boutons, which is the parameter of NMJ morphology we used for this analysis, is presented for each species on the same tree. Examination of these data reveals no discernible pattern between phenotypic similarity and phylogenetic distance. Some closely related species such as D. melanogaster and Drosophila simulans do share similar NMJ phenotypes but other pairs of sibling species such as D. simulans and Drosophila sechellia have very distinct NMJ phenotypes. Conversely, some distantly related species have morphologically similar NMJs. For example, the NMJ morphology of D. virilis is quite distinct from that of the related Drosophila mojavensis but the overall morphology is very similar to that of D. melanogaster from which it has been diverging for >40 Myr (4) (Figs. 2 and 4). We see a similar absence of a correlation between NMJ morphologies and phylogenetic relationships among the larger sample of 21 species even though the exact phylogenetic tree for all these species has not been determined from genome sequences. The lack of any apparent correlation between phylogeny and phenotype means that the NMJ phenotype of any given species of Drosophila can be determined only by direct examination; it cannot be predicted on the basis of NMJ phenotypes of any related species. Furthermore, as explored in more detail below, these results constrain the possible explanations for the evolutionary basis of the variation in NMJ phenotypes among different Drosophila species.
Fig. 4.
NMJ phenotypes among different species of Drosophila lack phylogenetic structure. The phylogenetic tree is derived from the complete genomic sequences of the species shown on the basis of rates of neutral substitutions at 12,000 sites (12). (Scale bar, number of substitutions per site.) The total evolutionary distance spanned by this phylogeny is >40 Myr with the distance from D. melanogaster to D. mojavensis being estimated at 40 Mya (4). Shown to the right of each species is the average number of boutons per NMJ4 as a representative quantitative measure of NMJ phenotype for that species (Table 1). Examination of the data reveals no obvious relationship between phylogenetic distance between the various species and their phenotypic similarity. The same is true if NMJ phenotypes are represented instead by number of synaptic branches.
NMJ Morphology Can Evolve Rapidly.
Regardless of the mechanism driving the evolution of NMJ morphology in nature, it is apparent that it can occur over a rapid timescale. Even pairs of sibling species such as D. simulans and D. sechellia, which diverged ∼0.5 Mya (4), or Drosophila yakuba and Drosophila erecta, which diverged ∼2 Mya (4), exhibit striking phenotypic differences in NMJ morphology (Figs. 2 and 4). The contrast between the conserved external morphological appearance of the adults and the extensive phenotypic variation in NMJ morphology in these species is especially striking. Adult females in the melanogaster clade are almost indistinguishable and adult males can be distinguished only by differences in their genitalia (13). In fact, we are unaware of any morphological trait other than NMJ morphology that varies as extensively among these closely related species or that distinguishes them so readily. Thus, NMJ morphology appears to be evolving rapidly in comparison with other morphological traits.
Genetic Drift Is Unlikely to Explain Evolution of NMJ Morphology.
Is NMJ morphology for each species being shaped primarily by natural selection or is it mainly due to genetic drift with random accumulation of adaptively neutral mutations? If genetic drift is the major force affecting the evolution of a trait (as it is for a noncoding DNA sequence), the similarity in phenotype between two species will generally be primarily dependent on their time of divergence; closely related species should be more similar in phenotype than more distantly related species. Similar reasoning has been used to develop computational models that statistically evaluate the evolution of more complex quantitative traits (14). The models partition the phenotypic value, z, for a quantitative trait (such as bouton number) into three components: zi = u + ai + ei, where u is the underlying phenotypic component shared by all members of a phylogeny; ai is the heritable additive effect for a particular species; and ei is the residual error, which includes measurement error, phylogenetic uncertainty, fluctuating selection, etc. Phylogenetic heritability, 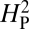 , is then defined as
, is then defined as 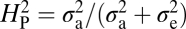 , where
, where 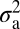 is the variance associated with the heritable additive effect and
is the variance associated with the heritable additive effect and 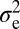 is the variance associated with the residual error. Because
is the variance associated with the residual error. Because 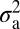 is dependent primarily on phylogenetic relatedness,
is dependent primarily on phylogenetic relatedness, 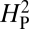 estimates the proportion of the phenotypic variation that is attributable to the underlying phylogeny. Thus,
estimates the proportion of the phenotypic variation that is attributable to the underlying phylogeny. Thus, 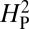 provides some measure of genetic drift.
provides some measure of genetic drift.
An underlying assumption of this model is that the phenotypic values are normally distributed. We confirmed this assumption for our data by using a Q-Q plot and did not observe any deviation from normality within our dataset (P = 0.03). Furthermore, a log transformation of the data to force normality yielded a similar 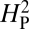 value (4.63 × 10−3, P = 0.24). This model also assumes that the error associated with phenotypic measurement is randomly distributed across the phylogenetic tree. In this case, all measurements were conducted in the same manner and the error associated with these measurements is quite small. Regardless, there is no discernible pattern of error across the phylogeny (r2 = 0.1). A third assumption of the model is that the phylogeny is known without error. We used a phylogeny based on complete sequenced genomes; therefore the error associated with the phylogeny is minimal. Thus, it appears that the necessary conditions have been met to apply this model to our data.
value (4.63 × 10−3, P = 0.24). This model also assumes that the error associated with phenotypic measurement is randomly distributed across the phylogenetic tree. In this case, all measurements were conducted in the same manner and the error associated with these measurements is quite small. Regardless, there is no discernible pattern of error across the phylogeny (r2 = 0.1). A third assumption of the model is that the phylogeny is known without error. We used a phylogeny based on complete sequenced genomes; therefore the error associated with the phylogeny is minimal. Thus, it appears that the necessary conditions have been met to apply this model to our data.
Using average bouton number as the quantitative measure of NMJ morphology, we can compute the value of 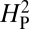 for the data presented in Fig. 4. We apply this analysis to the 11 species whose genomes have been completely sequenced, thereby minimizing any sources of residual error due to inaccuracies in presumed phylogenetic relationships. Under this model, if a trait is evolving primarily under the influence of genetic drift, most of the phenotypic variation should be attributable to the underlying phylogeny and the value of
for the data presented in Fig. 4. We apply this analysis to the 11 species whose genomes have been completely sequenced, thereby minimizing any sources of residual error due to inaccuracies in presumed phylogenetic relationships. Under this model, if a trait is evolving primarily under the influence of genetic drift, most of the phenotypic variation should be attributable to the underlying phylogeny and the value of 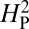 should approach 1. Conversely, if the trait is under selective pressure, phylogeny alone is relatively less important in determining the phenotypic values and
should approach 1. Conversely, if the trait is under selective pressure, phylogeny alone is relatively less important in determining the phenotypic values and 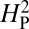 should approach 0. To increase statistical power in determining whether the computed value for
should approach 0. To increase statistical power in determining whether the computed value for 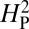 is consistent with genetic drift, the topology of the phylogenetic tree is held constant and the phenotypic values are randomly shuffled and reassigned to the tree. The resulting value of
is consistent with genetic drift, the topology of the phylogenetic tree is held constant and the phenotypic values are randomly shuffled and reassigned to the tree. The resulting value of 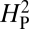 for this reassignment is then recomputed and the process repeated 10,000 times. Because the random reassignment of phenotypic values is independent of actual phylogenic relationship, the value of
for this reassignment is then recomputed and the process repeated 10,000 times. Because the random reassignment of phenotypic values is independent of actual phylogenic relationship, the value of 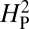 is expected to be low for most permutations. Genetic drift is presumed to be acting when the actual observed value of
is expected to be low for most permutations. Genetic drift is presumed to be acting when the actual observed value of 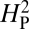 is significantly greater than the value of
is significantly greater than the value of 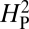 calculated from the random permutations. The value of
calculated from the random permutations. The value of 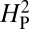 calculated from our data, 5.01 × 10−4, is not significantly different from the values of
calculated from our data, 5.01 × 10−4, is not significantly different from the values of 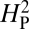 generated by random shuffling of phenotypic values (P = 0.27) (Fig. 5). Note that although we used bouton number for this analysis, equivalent conclusions are reached if we use other parameters such as branch number
generated by random shuffling of phenotypic values (P = 0.27) (Fig. 5). Note that although we used bouton number for this analysis, equivalent conclusions are reached if we use other parameters such as branch number 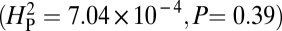 as a quantitative metric of NMJ phenotypes. The data for these parameters are more consistent with the hypothesis that genetic drift alone is insufficient to account for the observed variation and that selection is likely to be a significant factor in shaping NMJ morphology. Although we favor this interpretation, it is appropriate to emphasize that our analysis does not enable us to exclude other more complicated scenarios or to conclude that NMJ morphology is being shaped exclusively by selection.
as a quantitative metric of NMJ phenotypes. The data for these parameters are more consistent with the hypothesis that genetic drift alone is insufficient to account for the observed variation and that selection is likely to be a significant factor in shaping NMJ morphology. Although we favor this interpretation, it is appropriate to emphasize that our analysis does not enable us to exclude other more complicated scenarios or to conclude that NMJ morphology is being shaped exclusively by selection.
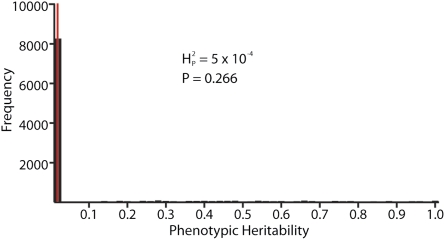
Fig. 5.
Genetic drift does not seem to account for variation in NMJ phenotypes among different species of Drosophila. Computational analysis was performed for the 11 species shown in Fig. 2 whose genomes have been sequenced and whose phylogenetic tree has been precisely established. The average number of boutons per NMJ4 for each species was used as the quantitative character analyzed. Comparable results were obtained using number of branch points 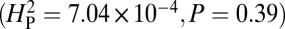 instead of number of boutons. The value of
instead of number of boutons. The value of 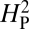 for bouton number was calculated as described in the text and Materials and Methods. According to the model, if the variation in bouton number among the different species is primarily determined by their phylogenetic relatedness, the value of
for bouton number was calculated as described in the text and Materials and Methods. According to the model, if the variation in bouton number among the different species is primarily determined by their phylogenetic relatedness, the value of 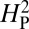 will approach 1. A high value of
will approach 1. A high value of 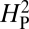 is therefore consistent with genetic drift playing a major role in the evolution of NMJ morphology. The computed value for
is therefore consistent with genetic drift playing a major role in the evolution of NMJ morphology. The computed value for 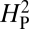 for our dataset is 5.01 × 10−4 (thin red line). To determine whether this outcome is consistent with a strong phylogenetic effect on NMJ morphology, bouton numbers shown in Fig. 2 were randomly shuffled among species on the phylogenetic tree and the value of
for our dataset is 5.01 × 10−4 (thin red line). To determine whether this outcome is consistent with a strong phylogenetic effect on NMJ morphology, bouton numbers shown in Fig. 2 were randomly shuffled among species on the phylogenetic tree and the value of 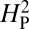 for that arrangement was calculated. This procedure was repeated 10,000 times to generate the distribution of
for that arrangement was calculated. This procedure was repeated 10,000 times to generate the distribution of 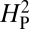 values shown in the histogram. As expected, random shuffling of bouton numbers among species, irrespective of phylogeny, results in mostly very low values for
values shown in the histogram. As expected, random shuffling of bouton numbers among species, irrespective of phylogeny, results in mostly very low values for 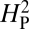 . However, 61% of the
. However, 61% of the 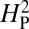 values from the random shuffling fall above the observed
values from the random shuffling fall above the observed 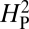 value calculated from our dataset. Therefore, the results are consistent with the absence of a strong phylogenetic contribution to NMJ phenotype and suggest that the observed variation in NMJ morphology cannot be explained by genetic drift alone.
value calculated from our dataset. Therefore, the results are consistent with the absence of a strong phylogenetic contribution to NMJ phenotype and suggest that the observed variation in NMJ morphology cannot be explained by genetic drift alone.
Synaptic Function Is Conserved Among Species.
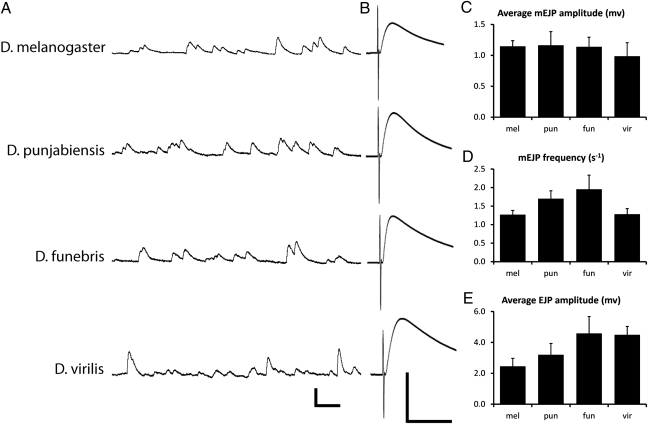
If selection is playing an important role to determine the differences in NMJ morphology among different Drosophila species, what are the targets of selection? The most obvious possibility is that differences in morphology would be associated with differences in synaptic function and that selection would act on synaptic function to meet the adaptive requirements of each particular species. We investigated this possibility by performing intracellular recording from muscle 4 in several different Drosophila species to assay various parameters of synaptic function including both spontaneous and evoked transmitter release. Spontaneous fusion of single synaptic vesicles with the presynaptic terminal elicits quantal depolarization events in the muscle called miniature excitatory junctional potentials (mEJPs). The amplitude of mEJPs is a measure of the size of synaptic vesicles and the amount of neurotransmitter packaged into each vesicle. The frequency of spontaneous fusion events provides an indication of the baseline properties of the synaptic release machinery. Electrical stimulation of the motor nerve generates an action potential that triggers the coordinated and simultaneous release of many synaptic vesicles that produce an excitatory junctional potential (EJP) in the muscle, whose amplitude is a measure of the total number of synaptic vesicles released in response to an action potential. We measured each of these parameters in D. melanogaster, D. punjabiensis, D. funebris, and D. virilis (Fig. 6). We chose these species because they span the range of NMJ phenotypes as well as the breadth of the phylogenetic tree for the collection of Drosophila species we have examined. We performed the recordings in a low Ca2+ (0.4 mM) Ringer's solution to avoid saturating muscle depolarization, thereby enabling better resolution of any subtle differences in neurotransmitter release.
Fig. 6.
Synaptic function of NMJ4 does not correlate with morphology. Intracellular recordings of miniature evoked excitatory junction potential (mEJP) amplitude and frequency and evoked excitatory junction potentials (EJPs) are from muscle 4 in 0.4 mM external calcium. (A) Representative mEJP recordings, quantified in C and D. Scale: 2 mv; 200 ms. (B) Representative EJPs evoked by electrical stimulation of the appropriate segmental nerve bundle. Scale: 5 mv; 200 ms. (E) Average EJP amplitude, a measure of the number of vesicles released in response to an evoked action potential, is quantified for each species. Error bars indicate SEM (minimum n = 12). mel, melanogaster; pun, punjabiensis; fun, funebris; vir, virilis.
Despite the variation in NMJ morphology and time since divergence among these species, we did not observe substantial differences in any of the electrophysiological parameters we measured (Fig. 6). Although there are some minor differences in mEJP frequency and EJP amplitude, such as a higher frequency of mEJPs in D. funebris and smaller EJP amplitude in D. melanogaster, there is no apparent correlation between the number and arrangement of boutons or muscle size in the various species and the measured parameters of synaptic function. One caveat is that the ionic concentrations of the hemolymph of species other than D. melanogaster are not known. Therefore, it is possible that our electrophysiological analyses on dissected preparations bathed in a standard Ringer's solution fail to detect some differences in synaptic function that may occur in vivo if there are significant differences in the normal concentrations of ions in the hemolymph of these species. However, any such differences are not expected to be large. Thus, to a first approximation, the functional output of the NMJs of different species is comparable despite their very different morphologies. This result is consistent with previous studies of mutants affecting NMJ growth in D. melanogaster, where EJP amplitudes do not correlate with NMJ size or complexity. Consequently, if selection is acting on synaptic function, it must be operating on parameters more subtle than those we have measured here, such as synaptic fatigue, following frequency, etc., which could be of major significance under natural conditions.
Discussion
The Drosophila larval NMJ has been used extensively as an experimental system for studies of synaptic function, development, and plasticity, using the wide array of genetic, molecular, histological, and electrophysiological tools available in this organism. Here, we add another dimension to previous studies by taking an evolutionary perspective to examine phenotypic diversity of NMJ morphology among different species of Drosophila. The surprisingly high degree of morphological variation that we have discovered has the potential to lead to important insights concerning the molecular mechanisms that regulate synaptic development and growth, the relationship between synaptic structure and synaptic function, and the evolution of nervous systems and behavior.
Given the complete conservation of the larval body plan, musculature, and pattern of innervation among all species of Drosophila and even among other very distantly related families of Diptera, the great diversity of NMJ morphologies we have found was wholly unexpected. Although we have attempted to represent this diversity by quantifying parameters such as number of boutons and number of branch points, these measures alone do not fully capture the extent of the diverse phenotypes we observe, which include differences in bouton size, morphology, and distribution as well as differences in the overall topography and geometry of the NMJs. The NMJ phenotypes span the range from the most undergrown to the most overgrown mutants characterized in D. melanogaster and include several distinctive phenotypes that have not yet been found in mutant screens. In this case, however, these phenotypes have not been generated as a result of deliberate mutagenesis and screening but rather represent the typical appearance of NMJs for each species.
For most species we were limited to the single stocks that were available; therefore there is some question of whether the lines are truly representative of that particular species. To resolve this question with certainty, it will be necessary to collect and characterize multiple additional representatives of each of these species from nature. However, for several reasons, we believe that a more exhaustive analysis is unlikely to alter our results in any substantive way. First, for at least six of the species examined, more than one isolate was available and in each case, we observed no significant difference in phenotypes among the isolates. Second, for D. melanogaster we have characterized >20 natural isolates from diverse climates and geographic locations and again found only limited variation in synaptic morphology. Third, although it is possible to imagine that for a few of the species the established isolates happen to carry a random mutation that confers an unusual NMJ phenotype, it seems highly unlikely that this would be true for the majority of species examined. Fourth, even large-scale screens in D. melanogaster have only rarely resulted in the identification of mutants with extreme or unusual NMJ phenotypes so the probability that such variants would by chance be incorporated into the isolates established for most Drosophila species should be extremely small. Consequently, we believe that the phenotypes we describe are likely to be representative for each species.
We have used a phylogenetic quantitative model (14) to ask whether random accumulation of neutral mutations is sufficient to explain the observed phenotypic diversity in NMJ morphology. The underlying premise of the model is that if adaptively neutral mutations are the primary force driving the evolution of the quantitative trait under analysis, the similarity in phenotypes between two different species should reflect their phylogenetic distance: Species that are more closely related should be more comparable in phenotype than more distantly related species. That is, the phenotypes should be structured in a way that reflects phylogeny. This model has been used previously to examine other quantitative traits that vary between species such as rates of meiotic recombination in different species of mice (14). In that case, the phenotypes are phylogenetically structured, suggesting that neutral mutations are largely responsible for the evolutionary divergence of this phenotype. We applied this analysis to 11 species of Drosophila whose genomes have been completely sequenced and whose phylogenetic relationships are very well defined. We found that the key parameter, phenotypic heritability, calculated from our dataset did not differ significantly from the values obtained by computer simulations where the phenotypic values were randomly shuffled along the phylogenetic tree. These results indicate that there is no apparent phylogenetic structure for the NMJ morphological phenotypes we examined and suggest that neutral mutations are unlikely to account for all of the observed phylogenetic diversity in NMJ morphology. Instead, it seems likely that the phenotypic appearance of NMJs in different Drosophila species is under selection. However, the source of selective pressure remains unclear. For example, we do not know whether selection might be acting directly on NMJ morphology or whether changes in this phenotype merely reflect a pleiotropic by-product of selection acting elsewhere. Previous mutational analysis has revealed that NMJ development is dependent upon many regulatory pathways and signal transduction mechanisms including BMP (9, 15) and Wg signaling (16), protein turnover via proteasomal- and autophagy-dependent mechanisms (17), and neuronal activity (18), all of which affect many aspects of fly development and physiology aside from the NMJ. One of these other developmental processes could be the actual target of selection with changes in NMJ morphology occurring as a secondary consequence. If this situation were the case, we would expect to see some other phenotype that shows an equally extensive range of phylogenetic variation but we are aware of no other phenotype for which this is true. This result is particularly notable in comparisons of NMJ morphology in sibling species such as D. simulans and D. sechellia, which are not yet completely sexually isolated. These species are almost indistinguishable on the basis of external morphology, but their NMJs are highly divergent with NMJs in D. sechellia containing twice as many boutons arranged in a much more elaborate branching pattern compared with D. melanogaster. Although we cannot rule out selection with pleiotropic effects, our results can be explained more readily if selection is acting on NMJ morphology more directly rather than on some other developmental process.
Given the substantial differences in natural history among the various Drosophila species, it would seem likely that selection is acting upon synaptic function, modifying it as required to provide each species with a selective advantage in its respective environment. Form would then follow function. However, contrary to this expectation, we have not observed any obvious differences among the various species in synaptic function as measured by the amplitude or frequency of mEJPs or by the amplitude of evoked potentials that correlate with the size or shape of the NMJ. The lack of any simple relationship between NMJ morphology and synaptic function has been previously encountered in studies of mutants affecting NMJ growth in D. melanogaster, where EJP amplitudes do not correlate in any predictable way with NMJ size or complexity. In addition, homeostatic regulatory systems can potentially compensate for some differences in synaptic size and complexity by modulating other aspects of synaptic function to maintain stable synaptic output (19).
We have not observed any obvious differences in simple behaviors such as crawling ability. One possibility is that the precise size and layout of the NMJ have no physiological significance and a functional NMJ of any type is all that matters. However, this interpretation seems unlikely on the basis of several different observations: The NMJ phenotype is relatively uniform within a species; NMJ morphology varies for different muscles within a body segment but is stereotypic for any given muscle; and our phylogenetic analysis suggests that random genetic drift is an insufficient explanation for the observed phenotypic variation. Thus, we suspect that a more likely explanation is that the key differences in synaptic function and behavior are more subtle than those we have examined here. Under natural conditions where fitness would depend strongly on appropriate responses to changes in temperature and humidity, ability to forage for food, and successfully avoiding predation, even small differences in parameters such as speed of transmission or synaptic fatigue could have profound effects on fitness but be easily missed in our initial basic examination of synaptic function and behavior. Consequently, more extensive and systematic behavioral studies, particularly under conditions that approximate the natural environment, will be very desirable to seek behavioral correlates for the different NMJ phenotypes.
In screening the literature for possible ecological or behavioral parameters that correlate with NMJ morphology, we have identified one intriguing connection that may be relevant. Among the species we examined, three are feeding specialists: D. sechellia (20), D. erecta (21), and D. mojavensis (22). In fact, both D. sechellia and D. erecta feed exclusively on plants that are toxic to their sibling species (20, 21). All three of these species have large and complex NMJs by comparison with their close relatives, which are generalists. This trait cannot fully account for the diversity of NMJ phenotypes because some generalists also have large, complex NMJs. Nonetheless, these results suggest that it may be worthwhile to identify other parameters that may correlate with NMJ morphology.
NMJ morphology appears to evolve rapidly relative to the evolution of other morphological traits. This rapid evolution is again best illustrated by the differences between sibling species, such as D. melanogaster and D. sechellia, which have very distinct NMJ morphologies. Although this rapid evolution is consistent with strong selection, further studies are necessary to understand the basis for it. One appealing speculation is that synaptic structure is not only a target on which selection acts but also itself an evolutionary driving force. For example, one could imagine that a genetic variant altering the structure of NMJs (and presumably some central synapses as well) confers some behavioral changes that affect selection of mates or preferred microenvironment. These behavioral changes could restrict gene flow with other members of the population by creating an artificial mating barrier leading to the accumulation of additional genetic variants that further modify synapses and behavior. The net result would be a positive feedback loop in which changes in synaptic morphology would lead increasingly to reproductive isolation and incipient speciation even in the absence of geographic barriers. Whether any mechanism of this type actually operates in natural populations awaits future studies.
Ultimately, it will be important to identify the genes and molecular pathways responsible for the remarkable variation in NMJ phenotypes among Drosophila species. One possibility is that the same genes that have been identified via mutational analyses in the laboratory mediate these phenotypes in nature. Alternatively, the naturally occurring phenotypes may lead to the discovery of new mechanisms that regulate synaptic growth and development. Either of these outcomes will be of substantial interest.
The extraordinary phenotypic variation in NMJ morphology that we have discovered among species of Drosophila is a remarkable and unexpected feature of the biology of these organisms that challenges our understanding and poses many more questions than we have been able to answer. At the same time, the existence of this phenomenon affords unique opportunities for further investigation that can provide novel insights into mechanisms that regulate synaptic growth, the interrelationships between synaptic morphology, neural function and behavior, and the evolution of nervous systems and behavior in natural populations. We believe that concerted efforts by investigators from the various disciplines touched upon by this work will be required to address the challenges and to extract all of the information that the discovery of extensive evolutionary divergence in NMJ morphology has the potential to provide.
Materials and Methods
Fly Stocks.
Canton-S was used as the laboratory wild-type stock of D. melanogaster. Other stocks of D. melanogaster were established from wild isolates trapped in Madison, Wisconsin. Stocks of various Drosophila species were obtained from Sean Carroll (University of Wisconsin, Madison) or from the San Diego Stock Center (University of California, San Diego).
Imaging.
All larvae were obtained from crosses of 10 females and 10 males incubated at 25 °C. Wandering third-instar larvae were dissected in ice-cold Ca2+-free saline and fixed for 15 min in 4% formaldehyde in PBS. Larvae were incubated in FITC-conjugated anti-HRP (Jackson ImmunoResearch) at 1:100 overnight at 4 °C. NMJs were imaged using a Zeiss 510 confocal microscope. Body wall images were obtained with bright-field optics on a compound microscope.
Quantification of boutons and branch points was performed at NMJ4 in segments A2–A4 as described in ref. 23. Branch points were determined for each bouton with branch points defined as the number of projections from that bouton minus one (excluding terminal boutons). Branch points for each bouton were then added to determine the total number of branch points per NMJ. Muscle 4 surface area was measured using ImageJ software.
Computational Modeling.
The phylogenetic tree was obtained from ref. 12. The computational model used was adapted from ref. 14, which uses Lynch's mixed-model approach (24) to partition the phenotypic value, z, for a quantitative trait into three components: zi = u + ai + ei, where u is the underlying phenotypic component shared by all members of a phylogeny; ai is the heritable additive effect for a particular species; and ei is the residual error, which includes measurement error, phylogenetic uncertainty, fluctuating selection, etc. Phylogenetic heritability, 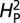 , is then defined as
, is then defined as 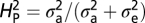 , where
, where 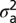 is the variance associated with the heritable additive effect and
is the variance associated with the heritable additive effect and 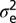 is the variance associated with the residual error. In our analysis
is the variance associated with the residual error. In our analysis 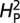 estimates the proportion of variation in synaptic morphology that can be attributed to the phylogenetic relationship. Quantitative measures of synaptic morphology (primarily bouton number) were randomly shuffled along the tips of the phylogenetic tree and
estimates the proportion of variation in synaptic morphology that can be attributed to the phylogenetic relationship. Quantitative measures of synaptic morphology (primarily bouton number) were randomly shuffled along the tips of the phylogenetic tree and 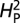 was recalculated. A total of 10,000 permutations were performed to calculate P values and assess significance of the estimated
was recalculated. A total of 10,000 permutations were performed to calculate P values and assess significance of the estimated 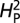 . When using branch number as the quantitative parameter, only 1,000 permuations were calculated owing to computational limitations. All computational analyses were performed in the R environment, using base package functions and function calls within the APE contributed package (25, 26).
. When using branch number as the quantitative parameter, only 1,000 permuations were calculated owing to computational limitations. All computational analyses were performed in the R environment, using base package functions and function calls within the APE contributed package (25, 26).
Electrophysiology.
Electrophysiology was performed on muscle 4 (segments 3 and 4) of wandering third-instar larvae. Standard HL-3 saline with 0.4 mM external calcium was used for recording. Preparations were visualized on an inverted Nikon DIAPHOT200, using Normarski optics. Microelectrodes with R ∼ 25 MΩ were filled with 3 M KCl and nerve stimulation electrodes were filled with bath saline. Average muscle input resistances were as follows: D. melanogster, 14.4 MΩ; D. punjabiensis, 16.7 MΩ; D. funebris, 13.9 MΩ; and D. virilis, 12.5 MΩ. Evoked EJPs were recorded in current-clamp, using the Axoclamp 2B amplifier (Axon Instruments). Mean EJP amplitudes were determined from 100 consecutive evoked EJPs at 2 Hz stimulation. Traces were analyzed using AxonClamp software. mEJP amplitude and frequency were analyzed using Mini Analysis software 5.6.4 (Synaptosoft).
Acknowledgments
We thank Sean Carroll and his laboratory for providing stocks of many of the Drosophila species used in these analyses. We also thank Bret Payseur and Beth Dumont for their assistance with the computational model, all members of the B.G. laboratory for helpful discussion and critical comments on the manuscript, and the San Diego Stock Center for supplying stocks of several species examined here. This research was supported by the National Institutes of Health through a predoctoral training grant (T32 GM007507) to the Neuroscience Training Program and a research grant (RO1NS15390) (to B.G.).
Footnotes
The authors declare no conflict of interest.
See Author Summary on page 4037 (volume 109, number 11).
References
- 1.Ryan TJ, Grant SGN. The origin and evolution of synapses. Nat Rev Neurosci. 2009;10:701–712. doi: 10.1038/nrn2717. [DOI] [PubMed] [Google Scholar]
- 2.Collins CA, DiAntonio A. Synaptic development: Insights from Drosophila. Curr Opin Neurobiol. 2007;17:35–42. doi: 10.1016/j.conb.2007.01.001. [DOI] [PubMed] [Google Scholar]
- 3.Lachaise D, Tsacas L. In: The Genetics and Biology of Drosophila. Ashburner M, Carson HL, Thompson JN, editors. Vol 3D. New York: Norton; 1986. pp. 221–332. [Google Scholar]
- 4.Russo CAM, Takezaki N, Nei M. Molecular phylogeny and divergence times of drosophilid species. Mol Biol Evol. 1995;12:391–404. doi: 10.1093/oxfordjournals.molbev.a040214. [DOI] [PubMed] [Google Scholar]
- 5.Gompel N, Prud'homme B, Wittkopp PJ, Kassner VA, Carroll SB. Chance caught on the wing: Cis-regulatory evolution and the origin of pigment patterns in Drosophila. Nature. 2005;433:481–487. doi: 10.1038/nature03235. [DOI] [PubMed] [Google Scholar]
- 6.Jeong S, Rokas A, Carroll SB. Regulation of body pigmentation by the Abdominal-B Hox protein and its gain and loss in Drosophila evolution. Cell. 2006;125:1387–1399. doi: 10.1016/j.cell.2006.04.043. [DOI] [PubMed] [Google Scholar]
- 7.Jeong S, et al. The evolution of gene regulation underlies a morphological difference between two Drosophila sister species. Cell. 2008;132:783–793. doi: 10.1016/j.cell.2008.01.014. [DOI] [PubMed] [Google Scholar]
- 8.McGregor AP, et al. Morphological evolution through multiple cis-regulatory mutations at a single gene. Nature. 2007;448:587–590. doi: 10.1038/nature05988. [DOI] [PubMed] [Google Scholar]
- 9.Marqués G, et al. The Drosophila BMP type II receptor Wishful Thinking regulates neuromuscular synapse morphology and function. Neuron. 2002;33:529–543. doi: 10.1016/s0896-6273(02)00595-0. [DOI] [PubMed] [Google Scholar]
- 10.Wan HI, et al. Highwire regulates synaptic growth in Drosophila. Neuron. 2000;26:313–329. doi: 10.1016/s0896-6273(00)81166-6. [DOI] [PubMed] [Google Scholar]
- 11.Schuster CM, Davis GW, Fetter RD, Goodman CS. Genetic dissection of structural and functional components of synaptic plasticity. I. Fasciclin II controls synaptic stabilization and growth. Neuron. 1996;17:641–654. doi: 10.1016/s0896-6273(00)80197-x. [DOI] [PubMed] [Google Scholar]
- 12.Stark A, et al. Harvard FlyBase curators; Berkeley Drosophila Genome Project Discovery of functional elements in 12 Drosophila genomes using evolutionary signatures. Nature. 2007;450:219–232. doi: 10.1038/nature06340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu J, et al. Genetic analysis of a morphological shape difference in the male genitalia of Drosophila simulans and D. mauritiana. Genetics. 1996;142:1129–1145. doi: 10.1093/genetics/142.4.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dumont BL, Payseur BA. Evolution of the genomic rate of recombination in mammals. Evolution. 2008;62:276–294. doi: 10.1111/j.1558-5646.2007.00278.x. [DOI] [PubMed] [Google Scholar]
- 15.O'Connor-Giles KM, Ho LL, Ganetzky B. Nervous wreck interacts with thickveins and the endocytic machinery to attenuate retrograde BMP signaling during synaptic growth. Neuron. 2008;58:507–518. doi: 10.1016/j.neuron.2008.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Budnik V, Salinas PC. Wnt signaling during synaptic development and plasticity. Curr Opin Neurobiol. 2011;21:151–159. doi: 10.1016/j.conb.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shen W, Ganetzky B. Autophagy promotes synapse development in Drosophila. J Cell Biol. 2009;187:71–79. doi: 10.1083/jcb.200907109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Bivort BL, Guo HF, Zhong Y. Notch signaling is required for activity-dependent synaptic plasticity at the Drosophila neuromuscular junction. J Neurogenet. 2009;23:395–404. doi: 10.3109/01677060902878481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davis GW. Homeostatic control of neural activity: From phenomenology to molecular design. Annu Rev Neurosci. 2006;29:307–323. doi: 10.1146/annurev.neuro.28.061604.135751. [DOI] [PubMed] [Google Scholar]
- 20.R'Kha S, Capy P, David JR. Host-plant specialization in the Drosophila melanogaster species complex: A physiological, behavioral, and genetical analysis. Proc Natl Acad Sci USA. 1991;88:1835–1839. doi: 10.1073/pnas.88.5.1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lachaise D, Tsacas L. Les Drosophilidae de savanes preforestieres de la region tropicale de Lamto II. Le peuplement des fruits de Pandanus candelabrum (Pandanacees). [The Drosophila of savannahs of the tropical region of Lamto II. The settlement on the fruits of Pandanus candelabrum (Pandanacees).] Ann Univ Abidjan. 1974;7:153–192. French. [Google Scholar]
- 22.Fogleman JC, Starmer WT, Heed WB. Larval selectivity for yeast species by Drosophila mojavensis in natural substrates. Proc Natl Acad Sci USA. 1981;78:4435–4439. doi: 10.1073/pnas.78.7.4435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Coyle IP, et al. Nervous wreck, an SH3 adaptor protein that interacts with Wsp, regulates synaptic growth in Drosophila. Neuron. 2004;41:521–534. doi: 10.1016/s0896-6273(04)00016-9. [DOI] [PubMed] [Google Scholar]
- 24.Lynch M. Methods for the analysis of comparative data in evolutionary biology. Evolution. 1991;5:1065–1080. doi: 10.1111/j.1558-5646.1991.tb04375.x. [DOI] [PubMed] [Google Scholar]
- 25.Paradis E, Claude J, Strimmer K. APE: Analyses of phylogenetics and evolution in R language. Bioinformatics. 2004;20:289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- 26.R Development Core Team . T: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing; 2006. [Google Scholar]