Abstract
The rapid advance in genome sequencing presents substantial challenges for protein functional assignment, with half or more of new protein sequences inferred from these genomes having uncertain assignments. The assignment of enzyme function in functionally diverse superfamilies represents a particular challenge, which we address through a combination of computational predictions, enzymology, and structural biology. Here we describe the results of a focused investigation of a group of enzymes in the enolase superfamily that are involved in epimerizing dipeptides. The first members of this group to be functionally characterized were Ala-Glu epimerases in Eschericiha coli and Bacillus subtilis, based on the operon context and enzymological studies; these enzymes are presumed to be involved in peptidoglycan recycling. We have subsequently studied more than 65 related enzymes by computational methods, including homology modeling and metabolite docking, which suggested that many would have divergent specificities;, i.e., they are likely to have different (unknown) biological roles. In addition to the Ala-Phe epimerase specificity reported previously, we describe the prediction and experimental verification of: (i) a new group of presumed Ala-Glu epimerases; (ii) several enzymes with specificity for hydrophobic dipeptides, including one from Cytophaga hutchinsonii that epimerizes D-Ala-D-Ala; and (iii) a small group of enzymes that epimerize cationic dipeptides. Crystal structures for certain of these enzymes further elucidate the structural basis of the specificities. The results highlight the potential of computational methods to guide experimental characterization of enzymes in an automated, large-scale fashion.
Keywords: computational biology, enzymology, protein function
The number of sequences in the protein databases continues to expand, with almost 18 million entries in the nonredundant TrEMBL database (October 2011; www.ebi.ac.uk/trembl/). Despite this abundance of data, an increasingly large proportion of these sequences have uncertain, unknown, or incorrectly annotated functions (1). Without reliable functional assignments, the promise contained in Nature’s repertoire of enzymes and metabolic pathways for advances in medicine, chemistry, and industry cannot be fully realized. Because the number of protein sequences is large, a computation-guided strategy for discovering the functions of proteins discovered in genome projects will be required to address this challenge. Predicting a protein’s function from sequence remains challenging, in part because the “boundaries” between functions in sequence space can be difficult to define; that is, closely related sequences (e.g., 60% sequence identity) can have different functions, e.g., ref. (2), while highly divergent sequences with no detectable sequence similarity can have identical functions. e.g., refs. (3, 4).
Protein function can be characterized at many different levels; here we focus exclusively on the substrate specificity of enzymes, as determined by in vitro biochemistry. Assigning biochemical function is challenging in functionally diverse enzyme superfamilies. A recent survey by Babbitt and coworkers estimates that there are approximately 275 superfamilies encompassing two or more distinct biochemical functions, representing approximately one-third of the known enzyme universe (5). The functionally diverse enolase superfamily has provided a particularly challenging “model system” for developing methods for predicting and characterizing enzyme specificity (6). To date, more than 20 distinct substrates have been identified for members of the enolase superfamily [see the Structure-Function Linkage Database (SFLD; http://sfld.rbvi.ucsf.edu)]. The active sites of the members of this superfamily are located at the interface between a (β/α)7β barrel domain that contains functional groups involved in catalysis and an N-terminal (α + β) capping domain that contains side chains that determine substrate specificity. These enzymes catalyze unimolecular reactions initiated by abstraction of the α-proton of a carboxylate substrate by an active site base catalyst to form a Mg2+ stabilized enediolate intermediate that is directed to products by the participation of an acid catalyst (7). Therefore, prediction of the substrate specificity is sufficient to enable functional assignment.
A subset of proteins in the superfamily shares a pair of Lys acid/base catalysts at the ends of the second and sixth β-strands as well as an Asp-x-Asp (DxD) motif at the end of the eighth β-strand of the (β/α)7β barrel domain. The L-Ala-D/L-Glu epimerase (AEE) from Bacillus subtilis is the structure-function paradigm for these enzymes: the conserved Lys catalysts are the acid/base catalysts for the 1,1 proton transfer (epimerization) reaction (SI Appendix, Fig. S1), the α-ammonium group of the dipeptide substrate is hydrogen bonded to the DxD motif, and the γ-carboxylate group of the Glu residue is hydrogen bonded to Arg24 in the capping domain (8). Because the sequences of the recognition loops in the capping domains are not conserved in homologs, our expectation was that these are dipeptide epimerases that utilize different dipeptide substrates.
As of June 2011, more than 700 protein sequences in the enolase superfamily were predicted to have a dipeptide epimerase function based on the predicted presence of the DxD motif and Lys acid/base catalysts (e.g., examples in SI Appendix, Fig. S2). In 2006, when the computational results were generated, only 66 putative dipeptide epimerases (and 18 additional “environmental” sequences) were present in the sequence databases, so we created homology models of all of them. We set out to determine how many of these were likely to be specific for L-Ala-D/L-Glu and how many other specificities might be present. Even within this relatively small group of enzymes, it would not be possible to experimentally characterize every member, in part due to challenges and expense associated with expressing the proteins. To provide specific predictions of substrate specificity, we used virtual library screening against the solvent-sequestered active site, which provides a defined cavity for the substrate. We previously reported computational predictions and in vitro screening to assign the L-Ala-D/L-Phe epimerase activity to a protein from Thermotoga maritima (gi:156442781; TM006) (9). In addition, the N-succinyl-Arg racemase function was assigned to an enzyme from Bacillus cereus that clustered phylogenetically with the dipeptide epimerases (10). In both cases, the substrate specificities were predicted using homology models and then experimentally verified; structures were then determined of substrate-liganded complexes, confirming the validity of the homology models and predicted “poses” for the enzyme/substrate complexes.
Here, we describe a large-scale computational prediction of specificity for representative members of the dipeptide epimerase group (all members that were available in 2006 when initial predictions were made), in vitro testing of these predictions for 17 members of the group, and determination of a 18 crystal structures for six members of the group, including 11 structures of substrate-liganded complexes. The homology models and docking results led to predictions of a rich diversity of substrate specificity for several previously uncharacterized groups of dipeptide epimerases. Subsequent biochemical and structural studies confirmed the key predictions, including (i) a group of AEE’s, phylogenetically distinct from the two previously characterized groups; (ii) several groups of epimerases with specificity for hydrophobic dipeptides, including a group dominated by sequences from plants; and (iii) a small group of epimerases with specificity for positively charged dipeptides. The sequences with divergent (i.e., non-AEE) specificity are very likely to have distinct in vivo functions, which remain unknown, although one particular member with specificity for D-Ala-D-Ala is likely to be involved in processing peptidoglycan, which is also the assumed function of the AEE’s.
Results
Sequence Analysis and Clustering.
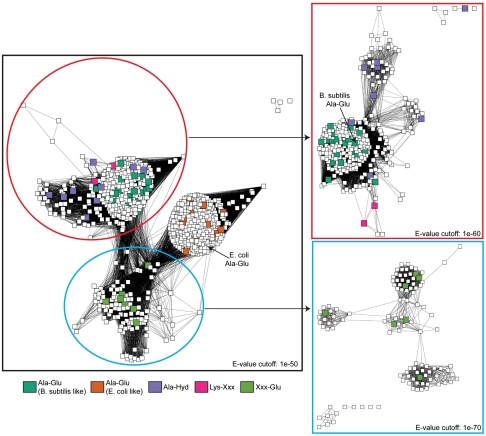
Relationships among the sequences are depicted in the sequence similarity networks shown in Figs. 1 and 2, where each edge (line) in the Cytoscape-generated representation indicates a pair of proteins (nodes) that have a Blast e-value more stringent than a certain cutoff value. Advantages of sequence similarity networks include the ability to visualize relationships among large numbers of sequences, such as the 735 represented here, with modest computational expense (11). As shown elsewhere, this type of clustering analysis tracks closely with phylogenetic trees generated using more rigorous methods, and we have confirmed this finding by comparing the networks to trees for representative members of this group [e.g., Fig. 4 in ref. (9)]. Representing the networks using different e-value cutoffs makes it possible to visualize sequence and functional relationships at different levels of granularity.
Fig. 1.
Sequence similarity network for the putative dipeptide epimerases in the enolase superfamily. Each node represents a single sequence; edges represents connections with BLAST e-value better than the cutoff value shown in each panel. Enzymes whose sequences were available in 2006 are colored according to their computationally predicted specificity, using homology models and ligand docking (larger nodes). The previously characterized Ala-Glu epimerases from E. coli and B. subtilis are labeled.
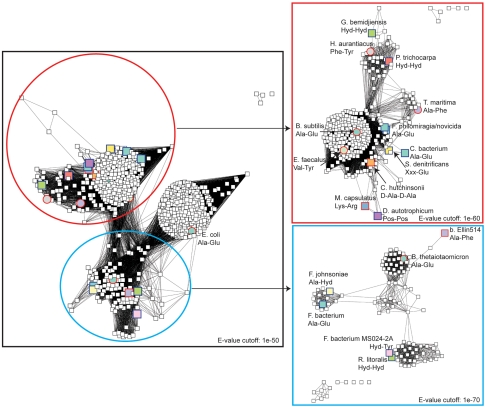
Fig. 2.
Sequence similarity network with experimentally characterized enzyme specificities labeled. Larger nodes represent sequences that have been experimentally characterized in this or earlier studies. Circles represent sequences for which crystal structures have been determined. Node border colors represent the type of experiment used to establish substrate specificity (red, kinetics; blue, mass spectrometry). In cases where there is no clear specificity for a single amino acid, “Pos” refers to specificity for positively charged amino acids (Lys, Arg, and His), “Hyd” refers to specificity for hydrophobic amino acids, and “Xxx” refers to no significant specificity observed.
Fig. 1 indicates the two previously characterized AEE’s from B. subtilis and Escherichia coli (12), which represent distinct clusters and use different constellations of amino acids in the binding sites to achieve a common specificity for L-Ala-L-Glu. As shown in SI Appendix, Fig. S3, although the putative dipeptide epimerases are strongly dominated by sequences from bacteria, a relatively small number of the sequences are found in archaea and eukaryotes, specifically plants and protozoa. With few exceptions, the archaeal and eukaryotic sequences do not cluster tightly with the characterized AEE’s, suggesting potentially divergent function, since only prokaryotes have peptidoglycan that contains L-Ala-D-Glu. (We note, however, that some eukaryotic sequences are from organisms such as Dictyostelium, which “eat” bacteria; we predict that these sequences do in fact have specificity for L-Ala-D/L-Glu (SI Appendix, Fig. S4), but thus far have been unable to obtain these proteins to confirm this prediction.)
Computational Predictions of Specificity.
We created homology models of all members of the group for which sequences were available in 2006, 84 sequences in total (16 of which were environmental sequences, which were not studied further), using the structure of the B. subtilis AEE in complex with L-Ala-L-Glu as a template (SI Appendix, Table S1). Several of these models suggested important substitutions in specificity-determining residues. To predict specific dipeptide substrates, we then used virtual screening methods, more commonly used in structure-based drug design, to dock all 400 possible L/L-dipeptides to each homology model, thus predicting which dipeptides are likely to bind as well as their “poses” in the binding site. The results were analyzed by generating a consensus profile that indicated the dominant amino acid preferences in the N- and C-terminal positions, averaged over the putative specificity groups (SI Appendix, SI Methods).
As described previously (9), L-Ala-L-Glu ranked among the top hits for the B. subtilis AEE, as well as the E. coli AEE homology model, despite the different sequence determinants of specificity among these two enzymes and the relatively low sequence identity between them (approximately 30%). As shown in Fig. 1, the top docking hits for the sequences closely related to the two characterized AEE’s were dominated by dipeptides with small amino acids in the N-terminal position and either Asp or Glu in the epimerized position (e.g., Ser-Asp). The key specificity-determining residues were also conserved within these two groups, so we predicted that all of these enzymes are AEE’s.
One other uncharacterized group of enzymes was also dominated by hit lists with negatively charged amino acids in the C-terminal position, but some showed relaxed specificity at the N-terminal position, e.g., Phe, Asn, or other larger amino acids among the top-ranked hits. We labeled this group as Xxx-Glu. Note that it is now clear that this group splits into several clusters at a Blast e-value of 1e-70 (Fig. 1), but only a subset of these sequences was available at the time the predictions were made, and they were treated as one cluster based on the initial phylogenetic tree at that time.
Two other clearly distinct specificities were also predicted on the basis of the modeling. First, sequences from Gloeobacter violaceus and Methylococcus capsulatus (as well as four closely related environmental sequences) were predicted to be specific for positively charged dipeptides, with a strong preference for positive amino acids in the N-terminal position. We labeled these as Lys-Xxx (pink nodes in Fig. 1). Finally, a group of sequences, including the previously reported T. maritima sequence (9), was predicted to prefer hydrophobic dipeptides. We labeled these as predicted Ala-hydrophobic epimerases, because many of these showed a predicted preference for Ala in the N-terminal position.
Determination of in Vitro Biochemical Function.
Protein expression and purification were attempted for dozens of representative sequences, heavily weighted towards the groups predicted to have divergent specificity; i.e., not closely related to the characterized AEE’s. A total of 17 enzymes was obtained for in vitro characterization, with several in each “new” specificity group (SI Appendix, Table S2). Of these, only five were among those in the original homology model-based predictions, from M. capsulatus (from the Lys-Xxx group), B. thetaiotaomicron (from the Xxx-Glu group), C. hutchinsonii (from the Ala-Hyd group), E. faecalis (from the B. subtilis-like group), and T. maritima (Ala-Hyd, described previously). Protein expression and purification for many others failed, but the other 12 enzymes that were successfully obtained also populate the regions of sequence space predicted to have divergent specificity.
These were first subjected to a mass spectroscopy-based assay that detects epimerization by incorporation of deuterium and can be used in a multiplexed fashion with mixtures of dipeptides. The first library to be screened consisted of all L-Ala-L-Xxx dipeptides other than L-Ala-Gly or L-Ala-L-Cys; the results are summarized in SI Appendix, Table S3. A few of these enzymes showed specificity for L-Ala-L-Glu in this assay, such as the dipeptide epimerase from Bacteroides thetaiotamicron (a dominant member of the human gut microbiome), from the group of enzymes predicted based on the modeling to be Xxx-Glu epimerases, and Campylobacterales bacterium which clusters with the B. subtilis AEE group (Fig. 2). However, the majority of the other enzymes showed divergent (i.e., not Glu) specificities at the C-terminal position, which were mostly consistent with the computational predictions for the groups to which they belong. For example, the M. capsulatus and Desulfobacterium autotrophicum dipeptide epimerases showed clear specificity for positively charged dipeptides; the enzymes from clusters predicted to be “Ala-Hyd” did, in fact, show epimerization with hydrophobic amino acids at the C-terminal position.
In all cases, the enzymes subsequently were assayed with additional libraries to define the N-terminal specificity; the complete results are reported in SI Appendix, Tables S4–S20. For five of the dipeptide epimerases, additional experiments were carried out to determine kinetic constants for various dipeptide substrates. Selected results, primarily for the best substrates for each enzyme, are summarized in Table 1; detailed results for all substrates tested are reported in SI Appendix, Tables S21–S25. The results are summarized on Fig. 2. In cases where kinetic constants were obtained, we label the enzyme with the single best substrate, as judged by kcat/KM. In cases where only mass spec screening was performed, the assigned specificity is necessarily more qualitative. In many cases, no clear preference for a single amino acid was found at either position; in such cases we use the abbreviations “Hyd” to represent little selectivity among various hydrophobic amino acids; “Pos” to represent Arg, Lys, and His; and “Xxx” in cases where most amino acids are tolerated. In cases where Glu is preferred in the C-terminal position and Ala is tolerated in the N-terminal position, we label the enzyme as Ala-Glu, because we assume that that is the relevant substrate in vivo. In all cases, the concise labels clearly cannot capture all of the specificity information provided by the screening.
Table 1.
Enzyme kinetics with selected dipeptide substrates
| Enzyme | Dipeptide substrate | kcat [s-1] | KM [mM] | kcat/KM [M-1 s-1] |
| B. thetaiotamicron | L-Ala-L-Glu | 147 ± 10 | 2.0 ± 1 | 7.4 × 104 |
| L-Ala-D-Glu | 59 ± 7 | 6.0 ± 2 | 9.8 × 103 | |
| L-Val-L-Glu | 96 ± 7 | 1.7 ± 0.2 | 5.6 × 104 | |
| C. hutchinsonii | D-Ala-D-Ala | 43 ± 1.3 | 1.9 ± 0.2 | 2.3 × 104 |
| D-Ala-L-Ala | 58 ± 4 | 1.1 ± 0.3 | 5.3 × 104 | |
| D-Ala-L-Val | 19 ± 0.8 | 0.50 ± 0.1 | 3.7 × 104 | |
| L-Ala-L-Ala | 37 ± 3 | 5.0 ± 0.7 | 7.5 × 103 | |
| E. faecalis | L-Ile-L-Tyr | 9.2 ± 0.7 | 15 ± 0.07 | 1.2 × 104 |
| L-Val-L-Tyr | 8.7 ± 2 | 0.70 ± 0.4 | 1.4 × 104 | |
| L-Arg-L-Tyr | 15 ± 0.07 | 1.4 ± 0.2 | 1.0 × 104 | |
| H. aurantiacus | L-Phe-L-Tyr | 4.7 ± 1 | 4.8 ± 1 | 980 |
| M. capsulatus | L-Lys-L-Arg | 8.4 ± 1 | 0.44 ± 0.1 | 1.9 × 104 |
| L-Lys-L-Lys | 0.029 ± 0.001 | 0.15 ± 0.02 | 200 | |
| L-Arg-L-Arg | 0.72 ± 0.4 | 0.19 ± 0.03 | 3.6 × 103 |
In addition, as summarized in Table 2, a total of 18 crystal structures were obtained for six of the dipeptide epimerases, including 11 structures with Mg2+ and five with dipeptide substrates (SI Appendix, Tables S26–S31). These structures reveal the basis for the observed specificity; depictions of the binding sites appear in Figs. 3 and 4, and SI Appendix, Figs. S5–S11. Comparisons of these structures with the previously predicted models, with optimal substrates bound, are shown in SI Appendix, Fig. S12. Although the predicted enzyme-substrate complexes show some differences from the subsequently determined structures, most of the key interactions between the substrate and the active site were correctly predicted.
Table 2.
New dipeptide epimerase crystallographic structures
| Species | Ligands | PDB entry | Resolution (Å) | Rfree |
| E. faecalis | apo | 3JVA | 1.7 | 0.224 |
| L-Ile-L-Tyr | 3JW7 | 1.8 | 0.254 | |
| L-Leu-L-Tyr | 3JZU | 2.0 | 0.281 | |
| L-Ser-L-Tyr | 3K1G | 2.0 | 0.282 | |
| L-Arg-L-Tyr | 3KUM | 1.9 | 0.270 | |
| B. thetaiotamicron | L-Ala-D-Glu | 3IJI | 1.6 | 0.218 |
| L-Pro-D-Glu | 3IJL | 1.5 | 0.205 | |
| L-Ala-D-Glu | 3IJQ | 2.0 | 0.287 | |
| C. hutchinsonii | D-Ala-L-Ala | 3Q4D | 3.0 | 0.254 |
| D-Ala-L-Val | 3Q45 | 3.0 | 0.251 | |
| M. capsulatus | apo | 3RO6 | 2.2 | 0.229 |
| L-Arg-D-Lys | 3RIT | 2.7 | 0.222 | |
| F. philomiragia | apo | 3R10 | 2.0 | 0.191 |
| fumarate | 3R11 | 2.0 | 0.181 | |
| L-Ala-D/L-Glu | 3R1Z | 1.9 | 0.185 | |
| tartrate | 3R0U | 1.9 | 0.195 | |
| tartrate (no Mg2+) | 3R0K | 2.0 | 0.197 | |
| H. aurantiacus | apo (no Mg2+) | 3IK4 | 2.1 | 0.275 |
All structures contain Mg2+ unless otherwise indicated.
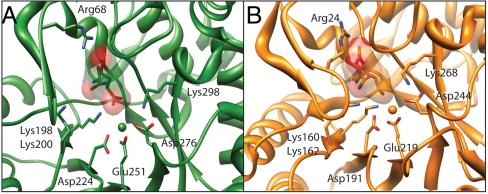
Fig. 3.
Comparison of the active sites of the dipeptide epimerases from (A) B. thetaiotamicron (3IJQ) and (B) B. subtilis (1TKK). The dipeptide substrates and key residues in the catalytic site are shown in stick representation, and the Mg2+ is shown as a sphere.
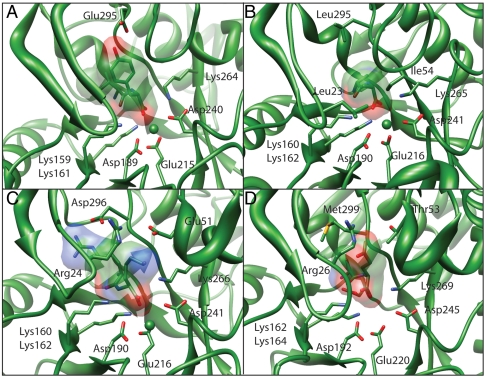
Fig. 4.
Binding sites of the dipeptide epimerases from (A) E. faecalis (3JW7), (B) C. hutchinsoni (3Q4D), (C) M. capsulatus (3RIT) and (D) F. philomiragia (3R1Z).
A Unique Class of Ala-Glu Epimerases.
The computational modeling suggested that, in addition to the sequences clustering with the E. coli and B. subtilis AEE’s, a third distinct group of dipeptide epimerases would have specificity for negatively charged amino acids at the C-terminal position, but somewhat relaxed specificity at the N-terminal position (Xxx-Glu in Fig. 1). Among the sequences included in the modeling study, the dipeptide epimerase from B. thetaiotamicron (BT1313) was characterized by mass spectroscopy-based screening followed by more detailed enzyme kinetics with selected substrates. These results for BT1313 are consistent with its assignment as an AEE, with similar kinetic constants to the previously characterized AEE’s from B. subtilis and E. coli. However, L-Val-L-Glu showed similar kinetics to L-Ala-L-Glu, and both Ile and Leu were also tolerated in the N-terminal position with kcat/KM > 103 M-1 s-1, confirming the prediction that amino acids larger than Ala would be tolerated in the N-terminal position. The promiscuous substrate specificity observed in vitro would be physiologically unimportant if dipeptides containing the variants in the N-terminal position are not present in the organism; metabolomic experiments could provide support for this expectation.
Crystals could be obtained for BT1313 in the presence of Mg2+ and either L-Ala-D-Glu (3IJI) or L-Pro-D-Glu (3IJL). These structures revealed that the dipeptide substrate/product was bound in a nonproductive complex, with the carboxylate group in the second coordination sphere of the Mg2+. A second monoclinic crystal form was then obtained in the presence of Mg2+ and L-Ala-D-Glu (3IJQ), with two polypeptides in the asymmetric unit. Although the carboxylate group of the dipeptide is located in the second coordination sphere of the Mg2+ in polypeptide A, in polypeptide B the carboxylate group is a bidentate ligand of the Mg2+, and the Nξ atoms of Lys200 and Lys298 are in appropriate locations relative to the α-carbon of the Glu residue to function as the acid/base catalysts.
As expected from the sequence identity (31%), the structures of BT1313 and the template for its homology modeling, the AEE from B. subtilis, are well superimposed (rmsd = 1.1 Å for 206 Cα pairs), although BT1313 is a monomer and the AEE from B. subtilis is an octamer. Focusing on the active sites, however, these structures reveal differing structural bases for the conserved specificity for L-Ala-D/L-Glu. In the AEE from B. subtilis, Arg24 in the 20’s loop of the capping domain provides key specificity-determining interactions with the α-carboxylate group of the Glu moiety of the substrate; the analogous residue in BT1313 is Arg68. As illustrated in Fig. 3, these two Arg are not spatially conserved. The crystal structure of the AEE from E. coli has not been determined with a dipeptide substrate (12), but there are two Arg in the 20’s loop that may coordinate the glutamate side chain, neither of which aligns to Arg24 of the B. subtilis AEE, and one of which may align to Arg68 from B. thetaiotaomicron (SI Appendix, Fig. S2). Thus, the AEE substrate specificity can be assigned to three distinct groups, highlighting the ability of divergent evolution within the enolase superfamily to deliver the same function. Previous examples of the malleability of structure to enable conserved substrate specificity within the enolase superfamily are provided by N-succinylamino acid racemases (10, 13), cis,cis-muconate lactonizing enzymes (13), and galactarate dehydratases (producing enantiomeric products) (14).
Finally, the dipeptide epimerase from Francisella philomiragia (Fphi1647), a pathogenic bacterium, is an example of an AEE that does not cluster tightly with any of the three major clades of AEE’s; a second example is the dipeptide epimerase from Campylobacterales bacterium, which was not structurally characterized. In the sequence network in Fig. 2, these sequences can be found on the periphery of the B. subtilis AEE group, and, judging by overall sequence identity, they are as similar to some of the non-AEE dipeptide epimerases as they are to the B. subtilis AEE. For example, Fphi1647 shares 37% sequence identity to the L-Leu-L-Tyr epimerase from E. faecalis discussed below, and 35% sequence identity to the AEE from B. subtilis. However, the dipeptide epimerase activity screening of Fphi1647 showed strict specificity for only Glu (preferred) or Asp in the epimerized position (SI Appendix, Table S3). The structure of Fphi1647 in complex with L-Ala-L/D-Glu (3R1Z) shows that, despite the relatively low overall sequence identity, the interactions between the dipeptide and protein are virtually identical to those seen for the B. subtilis AEE. Specifically, Arg26 of the 20 s loop is spatially and sequentially conserved with Arg24 of the B. subtilis 20 s loop (Fig. 4).
Cationic Dipeptide Epimerases.
One of the most striking predictions made using computational modeling was the existence of a small group of proteins with specificity for positively charged dipeptides. Among the modeled sequences, the dipeptide epimerase from M. capsulatus (MCA1834) was successfully characterized by in vitro biochemistry and crystallography; the closely related enzyme from D. autotrophicum was also characterized by mass spectroscopy-based screening, with similar results. The in vitro kinetics for MCA1834 confirmed the predicted preference for positively charged dipeptides. Dicationic dipeptides were actually preferred, with L-Lys-L-Arg being the best substrate, with kcat/KM = 1.9 × 104 M-1 s-1, which is similar to other members of the enolase superfamily. This strong preference for dicationic dipeptides was not predicted.
Crystals structures for MCA1834 were solved either in complex with Mg2+ ion alone (3RO6) or complexed both with L-Arg-D-Lys and Mg2+ (3RIT). Interestingly, the binding site of MCA1834 contains an Arg at the same position as Arg24 in the B. subtilis AEE, where it is used to stabilize the δ-carboxylate group of the glutamate portion of the Ala-Glu substrate. In the structure of MCA1834, the same residue is involved in a salt bridge with Glu51 (Fig. 4), perhaps to help position that side chain and bring about a closed conformation of the active site upon substrate binding. The conservation of this Arg between two enzymes with completely different specificities highlights a challenge of functional annotation; in this case, conservation of an amino acid critical for specificity in the AEE does not imply conservation of enzyme specificity (and presumably in vivo function). Conversely, nonconservation of the same amino acid does not necessarily imply a new specificity, as illustrated by the group of AEE’s discussed above that achieve specificity in a different manner than the B. subtilis AEE. These examples highlight the utility of computationally predicted and experimentally determined structures for helping to characterize specificity.
The side chain of Glu51 is likely the key specificity determinant for a positively charged residue in the C-terminal side of the dipeptide substrate, while the positively charged side chain at the N-terminal position of the substrate is stabilized via interactions with the side chains of Asp296 and Asp325. The hydroxyl group of Tyr19 is hydrogen-bonded to one of the carboxylate oxygens of Asp296 and secludes the binding pocket for the N-terminal portion of the substrate from the C-terminal side.
Several Groups of Hydrophobic Dipeptide Epimerases.
The computational modeling suggested that a group of enzymes, distantly (approximately 30% sequence identity) related to the B. subtilis AEE, would be specific for hydrophobic dipeptides. One of these dipeptide epimerases, from T. maritima, was reported previously (9); to show specificity for L-Ala-L-Phe and similar dipeptides. Four of the predicted hydrophobic dipeptide epimerases belong to a group that includes a number of plant enzymes (SI Appendix, Fig. S3). Three enzymes in this cluster were characterized in vitro, all of which showed specificity for hydrophobic dipeptides generally with fairly broad specificity in one or both positions. including the dipeptide epimerase from the black cottonwood tree, which showed specificity primarily for hydrophobic dipeptides (Populus trichocarpa, SI Appendix, Table S15; Herpetosiphon aurantiacus, SI Appendix, Tables S7 and S24; Geobacter bemidjiensis, SI Appendix, Table S10).
The E. faecalis dipeptide epimerase (EF1511) clustered closely with the B. subtilis AEE group in both the initial definition of the specificity groups and the sequence networks in Fig. 1, and so was erroneously predicted to have an AEE specificity. Instead, specificity in the C-terminal (epimerized) position is similar to that of the previously reported T. maritima dipeptide epimerase, with Phe, His, and especially Tyr preferred; the two enzymes are not closely related, however. N-Terminal specificity is broad, with L-Ile-L-Tyr, L-Val-L-Tyr, and L-Arg-L-Tyr being the best substrates. Several structures of EF1511 in complex with dipeptide substrates were determined, revealing the basis for specificity (Fig. 4 and SI Appendix, Fig. S5). The pocket for the C-terminal Tyr residue is, as expected, primarily hydrophobic, but the OH group on the Tyr side chain is hydrogen-bonded to Glu295. Interestingly, EF1511 showed no turnover with dipeptides containing Lys at the C-terminal position, which would also be capable of forming seemingly favorable electrostatic interactions with Glu295, based on the docking results. We speculate that the specificity for Tyr may result, in part, from its ability to fill the large and hydrophobic pocket, which Lys would not do.
The dipeptide epimerase from C. hutchinsonii (CHU2140) is unique in preferring Ala at both positions, although larger hydrophobic residues are somewhat tolerated. The best substrates are D-Ala-D/L-Ala (Table 1), strongly suggesting a biological role related to peptidoglycan, which contains D-Ala-D-Ala. Crystal structures of CHU2140 were solved in complex with Mg2+ ion and either D-Ala-L-Ala (3Q4D) or D-Ala-L-Val (3Q45) dipeptide substrates. The substrate-binding pocket is formed by hydrophobic residues including Phe19, Ile21, Phe51, Ile54, and Phe294. A small hydrophobic pocket containing Phe301 facilitates binding of the methyl side chain of D-Ala (Fig. 4). We hypothesize that this hydrophobic pocket is responsible for the stereochemical preference of D-Ala in the N-terminal position of the substrate molecule. Substituting L-Ala into the same position would introduce steric clashes with Thr321, resulting in less than optimal binding and therefore an increased KM value. The latter was validated experimentally with a range of dipeptide substrates with an N-terminal L-Ala (Table 1 and SI Appendix, Tables S20 and S22).
Finally, several sequences that cluster with the new, third group of Ala-Glu epimerases (exemplified by BT1313, from B. thetaiotamicron) also show specificity for hydrophobic dipeptides (Fig. 2 and SI Appendix, Table S3). None of these sequences were included in the initial modeling; we had assumed, incorrectly, that this entire group of dipeptide epimerases would have the AEE specificity. This group, like the large group of sequences that cluster with the B. subtilis AEE, contains multiple specificities, with no clear boundary in the sequence similarity network between the different specificities.
Discussion
Through a combination of bioinformatics, computational modeling, enzymology, and structural biology, we have identified several unique classes of dipeptide epimerases and characterized the structural and chemical determinants that underlie their specificity. The diversity of specificities, summarized in Fig. 2, was unexpected and highlights challenges for functional assignment. Most of these sequences are annotated in Genbank generically as “mandelate racemase/muconate lactonizing enzyme” (SI Appendix, Table S2), correctly establishing them as members of the enolase superfamily, but providing no information about their substrates, a critical first step in establishing their function. We were able to correctly identify these proteins as dipeptide epimerases based on the conservation of key residues in the binding sites, and homology models of the enzymes suggested a remarkable diversity of specificities. Some of the enzymes with divergent (i.e., not Ala-Glu) specificity are from plants or archaea, as well as a diverse group of bacterial species. The biological roles of these divergent specificities are unknown, but the in silico and in vitro studies presented here provide a starting point for in vivo studies.
The most important role of computational modeling in this work was to identify groups of enzymes likely to have novel substrate specificities, thereby guiding the experimental efforts by prioritizing specific enzymes for expression, purification, and screening. It is not feasible to experimentally characterize all or even most of the enzymes in this group, which is itself only a small fraction of the much larger functionally diverse enolase superfamily. Moreover, as is clear from Fig. 2, there are no clear boundaries between specificities in the sequence map. That is, as has been seen for other classes of enzymes in the enolase superfamily (4), highly divergent enzymes can have the same substrates, such as the dominant specificity for L-Ala-L-Glu, which can be achieved with at least three different constellations of active site residues. Conversely, relatively closely related enzymes can exhibit different specificities.
For these reasons, functional annotation can benefit significantly from structural information, in addition to bioinformatics analysis of protein sequences, as also shown elsewhere (15–17). By constructing homology models and docking dipeptide substrates, we were able to generate specific hypotheses regarding substrate specificities. In particular, we correctly predicted the existence of three specificity groups, although the predictions were clearly qualitative; e.g., “cationic dipeptide epimerase.” It is not yet possible, in our hands at least, to predict the very best substrate; e.g., L-Lys-L-Arg, which has a value for kcat/KM 100 times larger than that of L-Lys-L-Lys for the dipeptide epimerase from M. capsulatus. Nonetheless, because these computational methods can be scaled to hundreds or thousands of sequences, this approach holds the potential to help guide large-scale functional assignment, particularly to identify enzymes likely to have novel substrate specificities, as we have demonstrated here.
The current efforts resulted in the identification of catalytic activities for a significant number of members of the enolase superfamily. However, more important than the discovery of these activities is the demonstrated synergy between computational modeling and informatics approaches, which represents an important step towards the realization of general strategies for functional annotation. The assignment of in vitro catalytic activities to individual enzymes affords an entry point for defining entire metabolic pathways. For example, in favorable cases, where interpretable genome/operon context exists, the determination of a single catalytic activity provides direct insights into the activities of proteins encoded by physically proximal genes and may ultimately lead to predictions regarding new metabolic pathways. In turn, these predictions offer significant cues for the design and interpretation of experiments to define the true in vivo functions of the initially annotated enzyme, as well as its associated metabolic pathway. Thus, the computational approaches described here are likely to represent an important component of the overall strategies needed to systematically define the metabolic repertoire present in nature.
Materials and Methods
The computational and experimental methods have been described previously (9) in the context of assigning the function of one member of this group. Detailed methods are described in SI Appendix.
Briefly, the computational predictions were generated using a multiple sequence alignment generated by Muscle (18). Homology models were created for all putative dipeptide epimerases with Prime (Schrodinger LLC), using the substrate-bound structure of B. subtilis AEE (PDB ID 1TKK) as the template. All 400 possible L/L-dipeptides were docked to the models using Glide SP (v4.0108, Schrodinger LLC). Consensus results were generated by analyzing the top hits (top 5%, i.e., top 20 hits) of all members of a phylogenetic group, characterizing the most prominent amino acids in the N-terminal and C-terminal positions for the dipeptides.
Network analyses were performed as previously described (11), modified as given in SI Appendix. Cytoscape networks (19) were created from these BLAST results at several different e-value cutoffs. Tools used for visualization of protein networks were created by the UCSF Resource for Biocomputing, Visualization, and Informatics and are available from the Resource (http://www.rbvi.ucsf.edu). All of the sequences considered in this study are available in the SFLD along with additional metadata for each by searching with the associated gi numbers, given in SI Appendix, Table S1.
Supplementary Material
ACKNOWLEDGMENTS.
This research was supported by National Institutes of Health P01 GM071790 (to P.C.B., J.A.G., M.P.J., and S.C.A.) and U54 GM094662 (to S.C.A.). Molecular graphics images were produced using the University of California, San Francisco Chimera package from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by National Institutes of Health P41 RR-01081).
Footnotes
The authors declare a conflict of interest. M.P.J. is a consultant to Schrodinger LLC, which developed or licensed some of the software used in this study.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 3JVA, 3JW7, 3JZU, 3K1G, 3KUM, 3IJI, 3IJL, 3IJQ, 3Q4D, 3Q45, 3RO6, 3RIT, 3R10, 3R11, 3R1Z, 3R0U, 3R0K, and 3IK4).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1112081109/-/DCSupplemental.
References
- 1.Schnoes AM, Brown SD, Dodevski I, Babbitt PC. Annotation error in public databases: Misannotation of molecular function in enzyme superfamilies. PLoS Comput Biol. 2009;5:e1000605. doi: 10.1371/journal.pcbi.1000605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Seffernick JL, de Souza ML, Sadowsky MJ, Wackett LP. Melamine deaminase and atrazine chlorohydrolase: 98 percent identical but functionally different. J Bacteriol. 2001;183:2405–2410. doi: 10.1128/JB.183.8.2405-2410.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burroughs AM, Allen KN, Dunaway-Mariano D, Aravind L. Evolutionary genomics of the HAD superfamily: Understanding the structural adaptations and catalytic diversity in a superfamily of phosphoesterases and allied enzymes. J Mol Biol. 2006;361:1003–1034. doi: 10.1016/j.jmb.2006.06.049. [DOI] [PubMed] [Google Scholar]
- 4.Glasner ME, et al. Evolution of structure and function in the o-succinylbenzoate synthase/N-acylamino acid racemase family of the enolase superfamily. J Mol Biol. 2006;360:228–250. doi: 10.1016/j.jmb.2006.04.055. [DOI] [PubMed] [Google Scholar]
- 5.Almonacid DE, Babbitt PC. Toward mechanistic classification of enzyme functions. Curr Opin Chem Biol. 2011;15:435–442. doi: 10.1016/j.cbpa.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gerlt JA, Babbitt PC, Jacobson MP, Almo SC. Divergent evolution in the enolase superfamily: Strategies for assigning functions. J Biol Chem. doi: 10.1074/jbc.R111.240945. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Babbitt PC, et al. The enolase superfamily: A general strategy for enzyme-catalyzed abstraction of the alpha-protons of carboxylic acids. Biochemistry. 1996;35:16489–16501. doi: 10.1021/bi9616413. [DOI] [PubMed] [Google Scholar]
- 8.Klenchin VA, Schmidt DM, Gerlt JA, Rayment I. Evolution of enzymatic activities in the enolase superfamily: Structure of a substrate-liganded complex of the L-ala-D/L-glu epimerase from bacillus subtilis. Biochemistry. 2004;43:10370–10378. doi: 10.1021/bi049197o. [DOI] [PubMed] [Google Scholar]
- 9.Kalyanaraman C, et al. Discovery of a dipeptide epimerase enzymatic function guided by homology modeling and virtual screening. Structure. 2008;16:1668–1677. doi: 10.1016/j.str.2008.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Song L, et al. Prediction and assignment of function for a divergent N-succinyl amino acid racemase. Nat Chem Biol. 2007;3:486–491. doi: 10.1038/nchembio.2007.11. [DOI] [PubMed] [Google Scholar]
- 11.Atkinson HJ, Morris JH, Ferrin TE, Babbitt PC. Using sequence similarity networks for visualization of relationships across diverse protein superfamilies. PLoS One. 2009;4:e4345. doi: 10.1371/journal.pone.0004345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gulick AM, Schmidt DM, Gerlt JA, Rayment I. Evolution of enzymatic activities in the enolase superfamily: Crystal structures of the L-ala-D/L-glu epimerases from escherichia coli and bacillus subtilis. Biochemistry. 2001;40:15716–15724. doi: 10.1021/bi011641p. [DOI] [PubMed] [Google Scholar]
- 13.Sakai A, et al. Evolution of enzymatic activities in the enolase superfamily: N-succinylamino acid racemase and a new pathway for the irreversible conversion of D- to L-amino acids. Biochemistry. 2006;45:4455–4462. doi: 10.1021/bi060230b. [DOI] [PubMed] [Google Scholar]
- 14.Rakus JF, et al. Computation-facilitated assignment of the function in the enolase superfamily: A regiochemically distinct galactarate dehydratase from oceanobacillus iheyensis. Biochemistry. 2009;48:11546–11558. doi: 10.1021/bi901731c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hermann JC, et al. Structure-based activity prediction for an enzyme of unknown function. Nature. 2007;448:775–779. doi: 10.1038/nature05981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Favia AD, Nobeli I, Glaser F, Thornton JM. Molecular docking for substrate identification: The short-chain dehydrogenases/reductases. J Mol Biol. 2008;375:855–874. doi: 10.1016/j.jmb.2007.10.065. [DOI] [PubMed] [Google Scholar]
- 17.Juhl PB, Trodler P, Tyagi S, Pleiss J. Modelling substrate specificity and enantioselectivity for lipases and esterases by substrate-imprinted docking. BMC Struct Biol. 2009;9:39. doi: 10.1186/1472-6807-9-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edgar RC. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cline MS, et al. Integration of biological networks and gene expression data using cytoscape. Nat Protoc. 2007;2:2366–2382. doi: 10.1038/nprot.2007.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.