Abstract
Protein-protein interactions mediated by ubiquitin-like (Ubl) modifications occur as mono-Ubl or poly-Ubl chains. Proteins that regulate poly-SUMO (small ubiquitin-like modifier) chain conjugates play important roles in cellular response to DNA damage, such as those caused by cancer radiation therapy. Additionally, high atomic number metals, such as gold, preferentially absorb much more X-ray energy than soft tissues, and thus augment the effect of ionizing radiation when delivered to cells. In this study, we demonstrate that conjugation of a weak SUMO-2/3 ligand to gold nanoparticles facilitated selective multivalent interactions with poly-SUMO-2/3 chains leading to efficient inhibition of poly-SUMO-chain-mediated protein-protein interactions. The ligand-gold particle conjugate significantly sensitized cancer cells to radiation but was not toxic to normal cells. This study demonstrates a viable approach for selective targeting of poly-Ubl chains through multivalent interactions created by nanoparticles that can be chosen based on their properties, such as abilities to augment radiation effects.
Keywords: polyubiquitin chain, radiation sensitization, SUMO-binding motif (SBM), SUMO-interacting motif (SIM)
Protein-protein interactions mediated by mono-Ubl (ubiquitin-like proteins) or poly-Ubl modifications are among the most important signaling and regulatory mechanisms that control nearly every aspect of cellular functions (1). It has been well established that the small ubiquitin-like modifiers (SUMO), SUMO-1, SUMO-2, and SUMO-3, play essential roles in cellular response to DNA damage (2). SUMO-2 and -3 are nearly identical, but share less than 50% sequence homology to SUMO-1 (3, 4). SUMO-2 and 3 are distinct from SUMO-1 in that they carry internal SUMO modification sites at their N-termini to form poly-SUMO chains (5).
In most cases, a Ubl serves as a platform for interactions with other proteins. For instance, ubiquitylation dependent processes are mediated by ubiquitin-binding motifs in receptor proteins, such as the proteosome (6). SUMO-dependent functions are mediated by a SUMO-interaction motif (SIM; also known as a SUMO-binding motif, SBM) (7, 8) that serves as receptor of SUMO modified proteins. Recent studies have shown that a SUMO-targeted ubiquitin ligase (STUBL) is important in DNA damage response, and the ligase specifically recognizes poly-SUMO-2/3 chains to ubiquitinate poly-SUMO modified proteins for degradation (9–14). These studies suggest that inhibiting poly-SUMO-2/3 interactions with down-stream effectors, such as STUBL, can inhibit DNA damage response. However, an effective approach to disrupting poly-Ubl chain-mediated cellular functions is lacking. In addition to proteosome inhibitors, a small molecule was identified that binds to and alters the conformation of polyubiquitin chains such that their interactions with the proteosome are inhibited (15). However, the negative charge of this small molecule prevents its translocation into cells.
Both ionizing radiation and most chemotherapeutic drugs kill cancer cells by inducing genotoxic stress. Thus, agents that can amplify radiation effects or inhibit DNA damage response will sensitize cancers to radiation therapy. High atomic number materials, such as gold, are known to be much more absorbent to ionizing radiation than soft tissues. Recent studies have shown that gold nanoparticles (AuNPs) do not cause cellular toxicity, but significantly enhance radiation effect (16, 17). AuNPs can translocate into cells and are not toxic to cells because of the inert property of gold (17). In this study, we explored the application of AuNPs as a platform to develop reagents that specifically target poly-SUMO chains and amplify radiation response. We show that a low affinity inhibitor for mono-SUMO can effectively inhibit poly-SUMO-mediated protein-protein interaction when the ligand is conjugated to AuNP. The selectivity for poly-SUMO chains is likely due to the AuNP providing a platform for multivalent interactions that result in much higher overall affinity between the AuNP-ligand conjugate and poly-SUMO chains. This study suggests an approach to modulating poly-Ubl-mediated protein-protein interactions in addition to amplifying the effects of ionizing radiation.
Results and Discussion
Identification of Peptide Analogues that Mimic the SIM
To identify a small molecular ligand of SUMO, virtual ligand screening was carried out based on the SIM in order to inhibit SUMO-mediated down-stream effects. The structure of SUMO-3 in complex with a SIM peptide was constructed by transferring the SIM sequence from the protein PIASx that is in the structure of its complex with SUMO-1 to the analogous site in the structure of SUMO-3 (8). The SIM sequence (KVDVIDLTIE) from PIASx binds all SUMO isoforms with dissociation constants between 4–8 μM (7). The analogous interactions of the SIM with different SUMO paralogues were confirmed by an independent NMR study (18). The SIM peptide forms a β-strand that extends the existing β-sheet in SUMO. Our previous studies have shown that the core consensus sequence of the SIM consists of 5 amino acid residues that bind to a shallow groove of SUMO (8). Hydrophobic interactions between the SIM and I33 and F31 of SUMO-3 appear to be critical. In addition, the SIM peptide forms backbone hydrogen bonds with SUMO-3, which extends the β-sheet of SUMO.
Constrained docking was employed to find potential SIM mimetics. The initial virtual screening was performed by considering the shape and hydrophobic interactions of the SIM-binding groove of SUMO-3. However, this approach did not yield any hits. A second screening was carried out that included backbone hydrogen bonding constraints with K32 and K34 of SUMO-3, because hydrogen bonds provide high specificity in protein-protein and protein-ligand interactions. Known examples of the significance of hydrogen bonds include those that stabilize the double-strands of DNA and secondary structures of proteins. In addition to the two hydrogen bonds, a hydrophobic region was defined that included the contact areas of side chains of I33 and F31. The GLIDE program was used to search the 250,000 compound library provided by the Developmental Cancer Therapeutics program of the National Cancer Institute. The docked structures were chosen using the E-model scoring function of Cvdw, which is the sum of the van der Waals (Evdw) and electrostatic interaction energy terms (Eelec). A group of peptide analogues was identified among the top hits (Fig. 1A), and their GLIDE scores are shown in Table S1. The characteristics of this group of peptide analogues are consistent with our previous knowledge of SUMO-SIM interactions. It was shown previously that β-branched amino acid side chains, such as Val and Ile, as well as aromatic residues, have an intrinsic preference for the β-strand conformation (19). As seen in Fig. 1, the side chains of the identified analogues favor the β-strand conformation. They contain the proper backbone for hydrogen bond formation with SUMO that mimics the SIM. Furthermore, the hydrophobic side chains and the phenyl group can form hydrophobic interactions with the groove on SUMO.
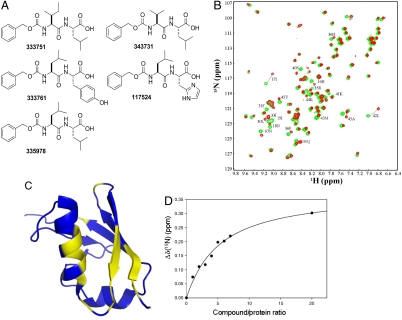
Fig. 1.
Top hits identified from the GLIDE program search of the NCI compound library. (A) The NSC compound identifier number is shown for each compound. (B) Superimposed HSQC spectra of SUMO-3 free (green) and in complex with 333751 (red) at 1∶10 protein:ligand ratio. (C) Structure of SUMO-3 in which residues that show significant CSP in the HSQC spectrum are indicated in yellow. (D) The 1H chemical shift of Leu42 of 15N-labeled SUMO-3 was monitored as a function of added compound 333751. The Kd of the complex was determined by a regression curve fitting of the chemical shift change as a function of the ligand∶protein ratio.
Interactions of the identified analogues with SUMO-1 and SUMO-3 were verified by NMR chemical shift perturbation analysis. These compounds generated negligible chemical shift perturbations (CSP) on SUMO-1, but much larger and specific CSP on SUMO-3 (Fig. 1B and Fig. S1). Compound 333751 was chosen for further development, because its binding affinity was among the strongest, as estimated by CSP, although all compounds have similarly weak affinities (Figs. 1B and 2B and Fig. S1). Based upon resonance assignments of SUMO-2 and -3, the compounds were determined as binding into the same groove as the SIM peptide (Fig. 1C). CSP was used to estimate the binding affinity of 333751 to SUMO-3, as described previously (20). The Kd of 333751 for binding SUMO-3 was 1.2 ± 0.4 mM (Fig. 1D). Thus, the affinity of this compound to SUMO-2 is more than 200-fold lower than that between the PIASx SIM and SUMO-2.
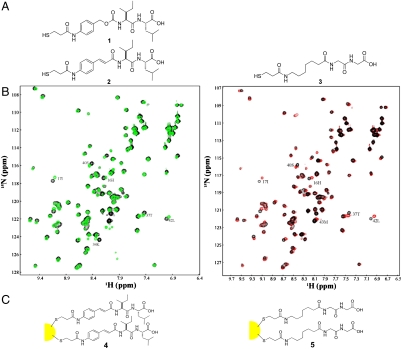
Fig. 2.
Synthesis of the AuNP-ligand conjugate. (A) Synthesis of thiol analogs of NSC333751 and a control for linking to AuNP. (B) Comparison of binding of the thiol analogue 2 (red spectrum) and NSC333751 (green spectrum) to SUMO-3, both at a ligand:protein ratio of approximately 3∶1. Both spectra are superimposed with that of free SUMO-3 (black). (C) Schematic diagram of the synthesis of the ligand-AuNP conjugates 4 and 5.
Synthesis of the AuNP-Ligand Conjugate.
Multivalency low affinity binding can achieve an overall high affinity. Therefore, we tested whether such specific but low affinity compounds can form high affinity interactions with poly-SUMO chains by creating multivalent interactions. We chose AuNPs as a platform for multivalent interactions, because they are commercially available and inert. Compound 333751 was modified for conjugation to an AuNP by adding a thiol tail. Initial attempts to synthesize thiol derivative 1 (Fig. 2A) proved problematic. Cleavage from the resin also resulted in cleavage of the aminobenzyl carbamate moiety, generating only trace amounts of derivative 1. Because of this instability, the aminobenzyl carbamate was replaced by a more hydrolysis resistant isostere, 4-aminocinnamic acid as contained in 2. The E-double bond geometry should mimic the S-trans configuration of the carbamate in 1. Derivative 2 was constructed on 2-chloro-chlorotrityl resin using standard Fmoc- synthesis conditions. N-Fmoc-cinnamic acid (21) and 3-(t-butoxycarbonylthio)propanioc acid (22) were then coupled to install the requisite thiol tail. Finally, resin cleavage and thiol deprotection led to thiol derivative 2 in good yield. A control, 3, that contains the same backbone but lacks side chains was also designed and synthesized using a similar strategy (Fig. 2A). Binding of derivative 2 to SUMO-1 and SUMO-3 was examined and compared to that of 333751. Fig. 2B shows a side-by-side comparison of the HSQC spectra of SUMO-3 binding to 333751 and the derivative 2 at a SUMO∶ligand ratio of 1∶3. Both compounds induced CSP on the same surface of SUMO-3 and to a very similar extent, indicating that their modes of interaction with SUMO-3 are nearly identical.
Derivatives 2 and 3 conjugated to AuNPs were constructed from C4-alkanethiol AuNPs (Fig. 2C) (23). Derivative 2 or 3 was equilibrated with C4-AuNP in dichloromethane for 4 d, according to published procedures (24). The precipitated ligand-AuNP conjugate 4 or 5 was collected via filtration. NMR analysis (Fig. S2) indicated that derivitation was complete and all butanethiol groups on the AuNP were displaced. The number of ligands on each AuNP were determined using a similar procedure as previously described (25). Briefly, the size of the AuNPs was confirmed by transmission electron microscopy (Fig. S2) to be 2.0–2.5 nm. The molecular weight and molar amount of AuNPs were calculated by assuming ideal spherical particles. The number of ligands per AuNP was determined using ferrocene as an internal standard for calibration of ligand NMR signal intensity (Fig. S2) as described by Hostetler et al. (25). We estimated that each conjugated nanoparticle contains approximately 100 ligands. The sharp NMR resonances also indicated that the conjugated ligands maintained significant conformational flexibility (Fig. S2).
Inhibition of Poly-SUMO Chain-Mediated Protein-Protein Interactions
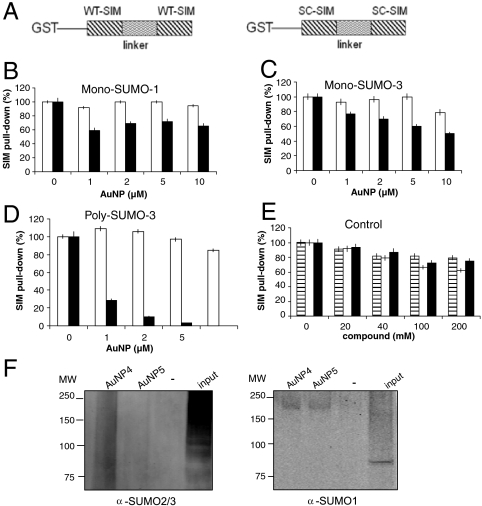
While the individual compounds shown in Fig. 1 do not have sufficient affinity for SUMO-2 or -3 to compete effectively with most SIM sequences (having Kd values ranging from 10 to 100 μM), the AuNP conjugate allows for multivalent interactions between the gold nanoparticle with multiple SUMO molecules in a poly-SUMO chain. To investigate the efficacy of AuNP 4 at inhibiting poly-SUMO-chain-mediated protein-protein interactions, a poly-SUMO binding peptide was designed as follows. The PIASx SIM was selected because this sequence has approximately 10-fold higher affinity for all SUMO isoforms than other characterized SIM sequences (7). This SIM sequence was used to replace two major SIM sequences in the protein rfp1, which is a ubiquitin ligase that specifically recognizes poly-SUMO modified proteins and ubiquitylates these proteins (11, 14). In this manner, a strong poly-SUMO-chain binding peptide was designed (Fig. 3A). In addition, a control was designed that contained two copies of the scrambled (SC) sequence of the SIM separated by the same linker (Fig. 3A). The GST-fusion SIM proteins were expressed and purified to greater than 90% homogeneity (Fig. S3A).
Fig. 3.
Inhibitory effect of the ligand-conjugated AuNP 4 on poly-SUMO chain-mediated protein-protein interactions. (A) Design of the SUMO-interacting motif (SIM) and control peptides. To target poly-SUMO-chain-mediated protein-protein interactions more effectively, a construct containing two SIMs separated by the sequence between the SIMs from the protein rfp1 was designed. In addition, a control was designed that contained two copies of the scrambled (SC) sequence of the SIM separated by the same linker. (B–C) AuNPs conjugated with (▪) or without (□) ligand were used to compete with the interactions between SIM and SUMO-1 (B), or SIM and SUMO-3 (C). Quantifications of all the SIM pull-down assays were scaled by the measurement of pull-down by the GST-tagged wild-type SIM peptide (100%) and GST-tagged control peptide (same amino acid content but with a scrambled sequence) (0%). (D) AuNP 4 significantly inhibited the SIM-poly-SUMO-chain interaction. AuNPs with (▪) or without (□) conjugated ligand 2 were used to compete with the interactions between SIM and poly-SUMO-3 chains. (E) The competition of protein-protein interactions between SIM and SUMO1 (dashed bars), SIM and SUMO3 (white bars) and SIM and poly-SUMO chains (black bars) by free derivative 2. (F) Western blot analysis of proteins pulled down by streptavidin agarose beads using an anti-SUMO-2/3 (Left) or anti-SUMO-1 (Right) antibody, showing that biotin-conjugated AuNP 4 preferentially binds to poly-SUMO2/3 conjugates in HeLa cell lysates. The input fraction represents 6% of inputs.
The SIM and control peptides were used in pull-down assays to investigate the effects of the conjugation of inhibitor to AuNPs. Poly-SUMO-3 chains were synthesized by enzymatic reactions of GST-tagged SUMO-3 with His-tagged SUMO-3 followed by affinity purification with glutathione beads and then Ni-NTA beads (Fig. S3B). The amount of SUMO pulled down by the SIM peptide was detected by enzyme-linked immunosorbent assay (ELISA) in an EIA microtiter plate. Ligand-conjugated AuNP 4 did not efficiently inhibit binding between monomers of SUMO-3 or SUMO-1 and the tandem-SIM-containing peptide (Fig. 3B and 3C). This was not surprising since the ligand has much lower affinity (Kd in the mM range, as described above) than the PIASX SIM sequence used for pull-down. On the other hand, the same concentration of AuNP 4 was highly effective at inhibiting the interaction between a poly-SUMO-3 chain and the tandem-SIM peptide (Fig. 3D), and had an IC50 less than 1 μM. It is difficult to accurately estimate the binding affinity between AuNP 4 and poly-SUMO chains. Similar to naturally occurring poly-SUMO or polyubiquitin chains, the in vitro synthesized poly-SUMO chains were a mixture of different lengths. As such, the binding affinity likely varied with the length of SUMO chains. Based on the concentrations of SUMO chains and the experimental design, we estimated that the IC50 value is larger than the average Kd value of the complex between AuNP and SUMO chains. AuNP that was not conjugated with derivative 2 did not exhibit the inhibitory effect (Fig. 3D). In addition, when not conjugated to the AuNP, derivative 2 was not significantly inhibitory to interactions between single SUMO-1 or SUMO-3 and the SIM peptide, nor did it inhibit poly-SUMO chain and SIM interactions (Fig. 3E).
To further investigate the specificity of AuNP 4 for poly-SUMO-2/3 chains, both AuNP 4 and 5 were conjugated with a small portion of biotin, using thiol-biotin (described in Methods). NMR data demonstrated the successful conjugation of ligands 2 and 3 to AuNP, and the presence of a small portion of biotin conjugated to AuNP was detected by FITC-conjugated streptavidin. Biotin-conjugated AuNP 4 and 5 were used to pull down SUMOylated proteins from HeLa cell extracts with streptavidin agarose beads, followed by immunoblotting with an anti-SUMO-1 or anti-SUMO-2/3 antibody. Biotin-conjugated AuNP 4, but not biotin-conjugated AuNP 5, pulled down poly-SUMO-2/3 conjugates (Fig. 3F). AuNP 4 and 5 do not appear to pull down significantly different amounts of SUMO1 conjugates. The faint signal seen with anti-SUMO1 antibodies in both the AuNP4 and AuNP5 pull-down lanes may be nonspecific. This result further demonstrates the specificity of AuNP 4 for poly-SUMO-2/3-modified proteins.
Effect on Inhibition of Poly-SUMO Chain-Mediated Protein-Protein Interactions in Cells
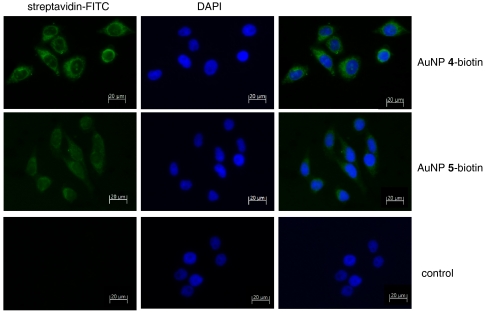
Localization of AuNP 4 and 5 was detected by FITC-conjugated streptavidin (Fig. 4). Fluorescence microscopy indicated that both AuNP 5 and AuNP 4 localized to both the cytoplasm and nucleus. However, AuNP 4 consistently formed brighter foci in the nuclei (Fig. 4). The foci do not completely colocalize with the PML nuclear bodies that are known to contain poly-SUMO2 and 3 conjugates. The nature of the foci that are not colocalized with the PML nuclear bodies requires further investigation. These observations demonstrate that both AuNP conjugates can permeate cells and localize to the nucleus.
Fig. 4.
Subcellular localization of biotinylated AuNPs. HeLa cells were treated with biotinylated AuNP 4 or 5 for 24 h and the AuNPs were visualized with FITC-conjugated streptavidin (green). Cell nuclei were visualized with DAPI (blue). Cells without AuNP treatment were the control. Bar, 20 μm
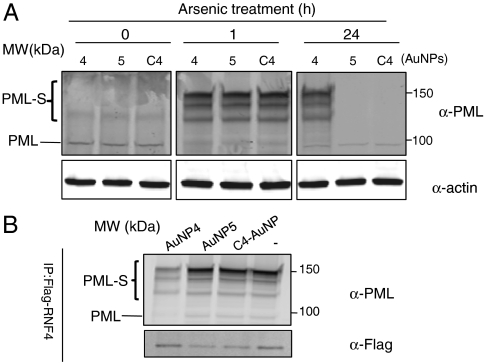
To investigate whether AuNP 4 inhibits SUMO-mediated protein-protein interactions in cells, we evaluated whether it inhibited arsenic-induced degradation of PML protein. In acute promyelocytic leukemia, PML forms a fusion protein with the retinoic acid receptor alpha (RAR). Arsenic trioxide (As2O3) can effectively treat this disease by inducing poly-SUMO-chain-dependent ubiquitylation and proteasomal degradation of the PML-RAR fusion protein (2, 26, 27, 30). The tandem repeats of SIM in RNF4, as discussed above, are responsible for recognition of the poly-SUMOylated PML to target it for ubiquitylation and degradation by the proteasome. Thus, this is a well-established mechanism that specifically depends on poly-SUMOylation-mediated protein-protein interactions. Arsenic induced an initial increase in PML body formation on both cells treated with AuNP 4 or the control C4-AuNP, as observed previously (2). The number of PML nuclear bodies in control (C4-AuNP)-treated cells decreased significantly over time, but not in the AuNP 4-treated cells. The difference was most pronounced after 24 h of treatment (Fig. S4). To further validate this finding, Western blot analysis was conducted, including both C4-AuNP and AuNP 5 as controls. Because AuNP 5 was a much more expensive control, most experiments were conducted using C4-AuNP as a control, which required that both controls be compared to validate C4-AuNP as a control. Treatment of all AuNP-treated cells with As2O3 led to a significant increase in PML SUMOylation (higher molecular weight bands in Fig. 5A) after 1 h, indicating that AuNP, by itself, does not exert a nonspecific effect. In contrast, after 24 h arsenic treatment, AuNP 4-treated cells had significant higher levels of modified PML than AuNP 5- and C4-AuNP-treated cells (Fig. 5A). This suggests that AuNP 4 specifically inhibits SUMOylation-dependent PML degradation by inhibiting poly-SUMO-mediated protein-protein interactions between PML and RNF4 in cells. To further demonstrate that AuNP 4 inhibits the interaction of SUMOylated PML with RNF4, coimmunoprecipitation (IP) experiments were conducted using HeLa cells that stably expressed SUMO-2. After overnight treatment with C4-AuNP, or AuNP 4 or 5, cells were exposed to As2O3 for 1 h, followed by co-IP. RNF4 pulled down much less SUMOylated PML in AuNP 4-treated cells than in C4-AuNP- and AuNP 5-treated cells (Fig. 5B). Again, both controls produced similar results.
Fig. 5.
(A) Comparison of the inhibitory effect of AuNP 4 with the two AuNP controls on arsenic trioxide induced PML degradation. Western blot analysis of PML protein modification and degradation in HeLa-SUMO2 cells treated with C4-AuNP or AuNP 4 or 5 (4 μM) for 1 h, and then were exposed to 1 μM arsenic trioxide for 1 or 24 h. (B) AuNP 4 inhibited protein-protein interactions between SUMOylated PML and RNF4. HeLa-SUMO2 cells were transfected with a plasmid expressing Flag-tagged RNF4, and after 24 h, were treated with C4-AuNP, AuNP 4 or 5 (4 μM) for 24 h followed by exposure to 1 μM arsenic trioxide for 1 h. The Flag-tagged RNF4 protein was immunoprecipitated using an anti-Flag antibody and its association with PML protein was analyzed by Western blot. In both panels A and B, unmodified PML bands and SUMO modified PML bands (labeled with PML-S) are indicated.
These findings further demonstrate the specificity of AuNP 4 at inhibiting poly-SUMO2/3-mediated protein-protein interactions by targeting the SIM-binding surface of SUMO-2/3. The ligand specifically targets the SIM-binding site of SUMO instead of other hydrophobic patches that are involved in binding the E1 and E2 enzymes, as demonstrated by NMR chemical shift perturbation (Figs. 1 and 2B). The potency of AuNP 4 is likely due to binding of multiple SUMO proteins in a chain. Poly-Ubl chains are usually long and contain more Ubl modules than necessary for binding receptor proteins. Although rfp1 (Schizosaccharomyces pombe homologue of RNF4) contains two SIM sites and presumably binds two SUMOs in a chain (2, 14), poly-SUMO chains formed in vitro and in vivo are generally much longer (26). Similarly, binding of the proteosome requires a chain of 4 ubiquitins (28), but polyubiquitin chains formed in vitro and in vivo are generally much longer (29). The Ubl chains are usually flexible, and the extensive conformational flexibility of the conjugated ligands and poly-SUMO chains would allow the binding of multiple SUMO modules in a chain with AuNP 4.
Effect on Cell Proliferation and Radiation Sensitization of AuNP 4
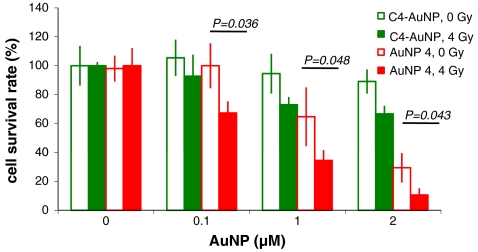
The effect of AuNP 4 on cytotoxicity and radiation response was tested (Fig. 6). The control C4-AuNP did not have significant toxicity to either the breast cancer cell line MCF-7 or the normal mammary epithelial cell line MCF-10A, consistent with previous findings that gold nanoparticles have low cytotoxicity to cells (17). The minimal reduction in cell viability of C4-AuNP-treated cells after radiation treatment was due to the short time after radiation at which cells were observed; 48 h post radiation, an approximately 20% reduction in cell viability was observed on average. AuNP 4 reproducibly stimulated the growth of MCF-10 cells slightly (Fig. S5A), but inhibited the growth of MCF-7 cells (Fig. S5B). In addition, it sensitized MCF-7 cells to radiation much more than it sensitized MCF-10 cells (Fig. S5). To further confirm the radiation sensitization effect, clonogenic assays were conducted to evaluate long-term cell survival (Fig. 6). AuNP 4 inhibited growth of the cancer cells (MCF-7) and enhanced their sensitivity to radiation. These findings are consistent with previous reports that RNF4 and its homologues are important in DNA repair and genome stability (11, 31). The effects of AuNP 4 on proliferation of MCF-7 and MCF-10A cells also suggest that SUMOylation plays a more important role in the growth of the cancer cells than normal cells, which is consistent with previous findings of the key role of SUMOylation in tumorigenesis (32, 33). AuNP 4 also sensitized MCF-7 cells to genotoxic stress generated by chemotherapeutic drug doxorubicin (Fig. S6), and sensitized the prostate cancer cell line Pc-3 to radiation and doxorubicin treatment (Figs. S6 and S7).
Fig. 6.
Comparison of cytotoxic and radiation sensitization effects of C4-AuNP and AuNP 4 on MCF-7 cell as assessed by long-term cell survival assay. MCF-7 cells were treated with either C4-AuNP or AuNP 4 at the indicated concentrations for 1 h before irradiation at 4 Gy. After irradiation, cells were incubated for 12 d until colonies formed. Unirradiated MCF-7 cells were also treated with either C4-AuNP or AuNP 4 and incubated for 12 d to determine cell survival rates. Experiments were performed in triplicate, and error bars represent standard deviations.
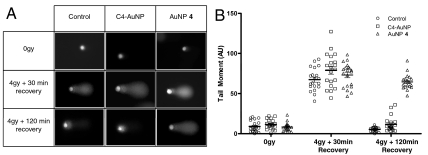
To further confirm the inhibitory effect of the SIM peptide on DNA repair, we used the comet assay to measure the amount of DNA damage in cells treated with AuNP 4 or the control C4-AuNP overnight, followed by 4 Gy γ-radiation and recovery for 30 or 120 min. In the comet assay, damaged DNA fragments migrate out of the cell nucleus as a streak similar to the tail of a comet, and the quantified tail moments are directly proportional to the amount of DNA damage. Immediately after irradiation, all cells produced comet tails of similar lengths and intensities (Fig. 7). However, after 120 min, the tail moments were significantly larger in cells treated with AuNP 4 than those treated with C4-AuNP or in untreated cells. These data were consistent with the radiation sensitivity data described above, indicating that repair of the damaged DNA was delayed by AuNP 4.
Fig. 7.
Detection of DNA damage using comet assays. Comet assays were used to measure the amount of unrepaired DNA damage in untreated cells or cells treated with AuNP 4 or C4-AuNP after γ-irradiation (4 Gy) and recovery for 30 or 120 min. (A) Representative images of the three sets of cells. (B) Tail moments (Tail DNA% × Length of Tail) as quantified for each cell using Comet Assay IV (Perceptive Instruments). Each spot represents a single cell; 20 comet images were measured for each treatment.
Conclusions
This study demonstrates a strategy to selectively inhibit protein-protein interactions mediated by posttranslational modifications with poly-Ubl, although the data does not exclude the possibility that AuNP 4 bind other molecules in addition to polySUMOylated proteins. With the rapid development of nanotechnology, a wide variety of nanoparticles has become available. As shown here, combining the properties of nano materials with nanoparticles as platforms for multivalent interactions creates a significant potential for future research and therapeutic applications.
Methods
(A detailed version is provided as Supporting Information)
Virtual Ligand Screening and NMR Studies
Docking was performed with GLIDE 3.5 (34, 35). A grid box of default size (18 × 18 × 18 Å3) was centered on three core amino acids in the SIM peptide (D3, V4, and I5) (7, 8). All NMR samples contained 10 mM phosphate buffer (pH 7.0) in 90% H2O/10%D2O.
Synthesis of Derivatives 2 and 3 and AuNP 4 and 5
Derivative 2 was constructed on 2-chlorotrityl resin loaded with Fmoc-Leu-OH at 0.8 mmol/g. Fmoc-Ile-OH, Fmoc-cinnaminic acid (21) and 3-(t-butoxycarbonylthio)propanioc acid (22) were sequentially coupled on using standard Fmoc synthesis conditions using HCTU (3 eq) and diisopropylethylamine (8 eq). The negative control, 3, was constructed in a similar manner as 2 with 2-chlorotrityl resin loaded with Fmoc-Gly-OH, Fmoc -7-aminoheptanoic acid and 3-(t-butoxycarbonylthio)propanioc acid.
The AuNP-ligand conjugate was synthesized according to a procedure described previously (36). To conjugate a small amount of biotin onto AuNP 4 or 5, C4-AuNP (6 mg) was combined with derivative 2 (10 mg) or 3 (8 mg) and thiol-biotin (2 mg; Nanoscience Instruments, CMT015).
Poly-SUMO Chain Formation and Expression of GST-WT-SIM and GST-SC-SIM
Poly-SUMO chains were synthesized by an in vitro SUMOylation reaction and purified by glutathione affinity chromatography followed by NTA chromatography. GST-tagged wild-type (GST-WT-SIM) or scrambled SIM sequence (GST-SC-SIM) was subcloned into the pGex4T-S expression vector (Amersham), overexpressed as an N-terminal GST-fusion protein in Escherichia coli strain BL21 (DE3) (Invitrogen), and purified using affinity chromatography.
Biotin-AuNP Pull-down and Subcellular Location
HeLa cell extracts were precleared with streptavidin agarose beads before incubation with biotinylated AuNPs, which were pulled down by streptavidin agarose beads. To examine AuNP location, HeLa cells were treated with Biotin-AuNP 4 and -AuNP 5, and the detected with streptavidin-FITC using a fluorescence microscope (AX10, Zeiss).
Comet Assay
MCF-7 cells were then treated with either 0.2% DMSO, 2 μM C4-AuNP, or 2 μM AuNP 4 for 24 h before irradiation. Cells received either no irradiation or 4 Gy irradiation followed by recovery at 37 °C for 30 min or 2 h. Cells were collected and prepared using the OxiSelect™ Comet Assay Kit (Cell Biolabs, Inc.). Comets were imaged by fluorescence microscopy (Olympus Inverted IX81). The tail moment (Tail DNA% × Length of Tail) was quantified for each cell using Comet Assay IV (Perceptive Instruments).
Supplementary Material
Acknowledgments.
This work was supported by NIH grants R01GM074748 and R01GM086171 to Y. Chen, P30 CA33572 from the National Cancer Institute, F32CA134180 to Y.-J. Li, and in part by an AMMI grant.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1109131109/-/DCSupplemental.
References
- 1.Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol. 2006;22:159–180. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- 2.Tatham MH, et al. RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat Cell Biol. 2008;10:538–546. doi: 10.1038/ncb1716. [DOI] [PubMed] [Google Scholar]
- 3.Saitoh H, Hinchey J. Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J Biol Chem. 2000;275:6252–6258. doi: 10.1074/jbc.275.9.6252. [DOI] [PubMed] [Google Scholar]
- 4.Ayaydin F, Dasso M. Distinct in vivo dynamics of vertebrate SUMO paralogues. Mol Biol Cell. 2004;15:5208–5218. doi: 10.1091/mbc.E04-07-0589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tatham MH, et al. Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J Biol Chem. 2001;21:35368–35374. doi: 10.1074/jbc.M104214200. [DOI] [PubMed] [Google Scholar]
- 6.Hicke L, Schubert HL, Hill CP. Ubiquitin-binding domains. Nat Rev. 2005;6:610–621. doi: 10.1038/nrm1701. [DOI] [PubMed] [Google Scholar]
- 7.Song J, Durrin LK, Wilkinson TA, Krontiris TG, Chen Y. Identification of a SUMO-binding motif that recognizes SUMO-modified proteins. Proc Natl Acad Sci USA. 2004;101:14373–14378. doi: 10.1073/pnas.0403498101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Song J, Zhang Z, Hu W, Chen Y. Small ubiquitin-like modifier (SUMO) recognition of a SUMO binding motif: A reversal of the bound orientation. J Biol Chem. 2005;280:40122–40129. doi: 10.1074/jbc.M507059200. [DOI] [PubMed] [Google Scholar]
- 9.Burgess RC, Rahman S, Lisby M, Rothstein R, Zhao X. The Slx5-Slx8 complex affects sumoylation of DNA repair proteins and negatively regulates recombination. Mol Cell Biol. 2007;27:6153–6162. doi: 10.1128/MCB.00787-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ii T, Mullen JR, Slagle CE, Brill SJ. Stimulation of in vitro sumoylation by Slx5-Slx8: evidence for a functional interaction with the SUMO pathway. DNA Repair. 2007;6:1679–1691. doi: 10.1016/j.dnarep.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prudden J, et al. SUMO-targeted ubiquitin ligases in genome stability. Embo J. 2007;26:4089–4101. doi: 10.1038/sj.emboj.7601838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nagai S, et al. Functional targeting of DNA damage to a nuclear pore-associated SUMO-dependent ubiquitin ligase. Science. 2008;322:597–602. doi: 10.1126/science.1162790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cook CE, Hochstrasser M, Kerscher O. The SUMO-targeted ubiquitin ligase subunit Slx5 resides in nuclear foci and at sites of DNA breaks. Cell Cycle. 2009;8:1080–1089. doi: 10.4161/cc.8.7.8123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun H, Leverson JD, Hunter T. Conserved function of RNF4 family proteins in eukaryotes: targeting a ubiquitin ligase to SUMOylated proteins. Embo J. 2007;26:4102–4112. doi: 10.1038/sj.emboj.7601839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Verma R, et al. Ubistatins inhibit proteasome-dependent degradation by binding the ubiquitin chain. Science. 2004;306:117–120. doi: 10.1126/science.1100946. [DOI] [PubMed] [Google Scholar]
- 16.Chithrani DB, et al. Gold nanoparticles as radiation sensitizers in cancer therapy. Radiat Res. 2010;173:719–728. doi: 10.1667/RR1984.1. [DOI] [PubMed] [Google Scholar]
- 17.Butterworth KT, et al. Evaluation of cytotoxicity and radiation enhancement using 1.9 nm gold particles: potential application for cancer therapy. Nanotechnology. 2010;21:295101. doi: 10.1088/0957-4484/21/29/295101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sekiyama N, et al. Structure of the small ubiquitin-like modifier (SUMO)-interacting motif of MBD1-containing chromatin-associated factor 1 bound to SUMO-3. J Biol Chem. 2008;283:35966–35975. doi: 10.1074/jbc.M802528200. [DOI] [PubMed] [Google Scholar]
- 19.Street AG, Mayo SL. Intrinsic beta-sheet propensities result from van der Waals interactions between side chains and the local backbone. Proc Natl Acad Sci USA. 1999;96(16):9074–9076. doi: 10.1073/pnas.96.16.9074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang J, et al. The intrinsic affinity between E2 and the Cys domain of E1 in ubiquitin-like modifications. Mol Cell. 2007;27(2):228–237. doi: 10.1016/j.molcel.2007.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miyawaki A, Miyauchi M, Takashima Y, Yamaguchi H, Harada A. Formation of supramolecular isomers; poly[2]rotaxane and supramolecular assembly. Chem Commun. 2008;4:456–458. doi: 10.1039/b713588b. [DOI] [PubMed] [Google Scholar]
- 22.Boyce RJ, Mulqueen GC, Pattenden G. Total synthesis of thiangazole, a novel naturally occurring HIV-1 inhibitor from Polyangium sp. Tetrahedron. 1995;51:7321–7330. [Google Scholar]
- 23.Brust M, Walker M, Bethell D, Schiffrin DJ, Whyman R. Synthesis of thiol-derivatised gold nanoparticles in a two-phase liquid-liquid system. J Chem Soc Chem Comm. 1994:801–802. [Google Scholar]
- 24.You C-C, De M, Han G, Rotello VM. Tunable inhibition and denaturation of a-chymotrypsin with amino acid-functionalized gold nanoparticles. J Am Chem Soc. 2005;127:12873–12881. doi: 10.1021/ja0512881. [DOI] [PubMed] [Google Scholar]
- 25.Hostetler MJ, Templeton AC, Murray RW. Dynamics of place-exchange reactions on monolayer-protected gold cluster molecules. Langmuir. 1999;15:3782–3789. [Google Scholar]
- 26.Weisshaar SR, et al. Arsenic trioxide stimulates SUMO-2/3 modification leading to RNF4-dependent proteolytic targeting of PML. FEBS Lett. 2008;582:3174–3178. doi: 10.1016/j.febslet.2008.08.008. [DOI] [PubMed] [Google Scholar]
- 27.Lallemand-Breitenbach V, et al. Arsenic degrades PML or PML-RARalpha through a SUMO-triggered RNF4/ubiquitin-mediated pathway. Nat Cell Biol. 2008;10:547–555. doi: 10.1038/ncb1717. [DOI] [PubMed] [Google Scholar]
- 28.Thrower JS, Hoffman L, Rechsteiner M, Pickart CM. Recognition of the polyubiquitin proteolytic signal. Embo J. 2000;19:94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Piotrowski J, et al. Inhibition of the 26 S proteasome by polyubiquitin chains synthesized to have defined lengths. J Biol Chem. 1997;272:23712–23721. doi: 10.1074/jbc.272.38.23712. [DOI] [PubMed] [Google Scholar]
- 30.Percherancier Y, et al. Role of SUMO in RNF4-mediated promyelocytic leukemia protein (PML) degradation: sumoylation of PML and phospho-switch control of its SUMO binding domain dissected in living cells. J Biol Chem. 2009;284:16595–16608. doi: 10.1074/jbc.M109.006387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kosoy A, Calonge TM, Outwin EA, O’Connell MJ. Fission yeast Rnf4 homologs are required for DNA repair. J Biol Chem. 2007;282:20388–20394. doi: 10.1074/jbc.M702652200. [DOI] [PubMed] [Google Scholar]
- 32.Mo YY, Moschos SJ. Targeting Ubc9 for cancer therapy. Expert Opin Ther Tar. 2005;9:1203–1216. doi: 10.1517/14728222.9.6.1203. [DOI] [PubMed] [Google Scholar]
- 33.Zheng Z, et al. SUMO-3 enhances androgen receptor transcriptional activity through a sumoylation-independent mechanism in prostate cancer cells. J Biol Chem. 2006;281:4002–4012. doi: 10.1074/jbc.M509260200. [DOI] [PubMed] [Google Scholar]
- 34.Halgren TA, et al. Glide: A new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. J Med Chem. 2004;47:1750–1759. doi: 10.1021/jm030644s. [DOI] [PubMed] [Google Scholar]
- 35.Friesner RA, et al. Glide: A new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J Med Chem. 2004;47:1739–1749. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- 36.You CC, De M, Han G, Rotello VM. Tunable inhibition and denaturation of alpha-chymotrypsin with amino acid-functionalized gold nanoparticles. J Am Chem Soc. 2005;127:12873–12881. doi: 10.1021/ja0512881. [DOI] [PubMed] [Google Scholar]
- 37.Franken NA, Rodermond HM, Stap J, Haveman J, van Bree C. Clonogenic assay of cells in vitro. Nat Protoc. 2006;1:2315–2319. doi: 10.1038/nprot.2006.339. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.