Abstract
The use of assisted reproductive technologies (ART) has become increasingly common worldwide and is now responsible for 2–3% of children born in developed countries. Multiple reports have suggested that ART-conceived children are more likely to develop rare epigenetic disorders such as Beckwith-Wiedemann Syndrome or Angelman Syndrome, both of which involve dysregulation of imprinted genes. Anecdotal reports suggest that animals produced with ART that manifest apparent epigenetic defects typically do not transmit these epimutations to subsequent generations when allowed to breed naturally, but this hypothesis has not been directly studied. We analyzed allele-specific DNA methylation and expression at three imprinted genes, H19, Snrpn, and Peg3, in somatic cells from adult mice generated with the use of intracytoplasmic sperm injection (ICSI), a type of ART. Epimutations were detected in most of the ICSI-derived mice, but not in somatic cells of their offspring produced by natural mating. We examined germ cells from the ICSI mice that exhibited epimutations in their somatic cells and confirmed normal epigenetic reprogramming of the three imprinted genes analyzed. Collectively, these results confirm that ART procedures can lead to the formation of primary epimutations, but while such epimutations are likely to be maintained indefinitely in somatic cells of the ART-derived individuals, they are normally corrected in the germ line by epigenetic reprogramming and thus, not propagated to subsequent generations.
Keywords: gametogenesis, transgenerational inheritance
Early embryos and germ cells are unique in that they must undergo extensive epigenetic reprogramming to reestablish developmental potency during each generation. This reprogramming entails erasure of most inherited epigenetic modifications followed by the acquisition and subsequent maintenance of new epigenetic profiles (1). Genomic imprinting is an epigenetic phenomenon found in eutherian and metatherian mammals (2) that results in parent-of-origin-specific, monoallelic expression of a subset of genes (3). This functional asymmetry of imprinted genes is conferred through differential DNA methylation patterns established during gametogenesis in each parent that distinguish the maternal and paternal alleles in the ensuing offspring (4, 5). These regions are known as differentially methylated regions (DMRs), and the differential methylation profiles at these genetic elements play an important role in regulating allele-specific expression of imprinted genes (3).
Because the epigenome is distinct in each cell type and is readily reversible by developmental reprogramming, it is particularly susceptible to disruption by environmental influences (6). Such epigenetic defects are known as “epimutations” and can be of two types—primary epimutations or secondary epimutations (7). Primary epimutations result from a direct disruption of an epigenetic parameter such as DNA methylation that is then propagated through DNA replication to subsequent cells. Secondary epimutations result from an initial genetic mutation that leads to a functional defect in one or more gene products that normally establish and/or maintain epigenetic parameters such as a DNA methyltransferase (7).
Studies in animals have shown that certain aspects of assisted reproductive technology (ART) may induce imprinting errors that are maintained throughout fetal development (8) and/or postnatally (9) in ART offspring; however, the potential of propagating such epimutations to subsequent generations has not been thoroughly investigated. Secondary epimutations would be expected to be transmitted according to Mendelian inheritance patterns because of genetic transmission of the mutation that led to the observed epigenetic abnormalities, however, primary epimutations would potentially be corrected by germ-line–specific epigenetic reprogramming and, therefore, not transmitted to subsequent generations.
To determine whether epimutations induced by ART are transmitted to subsequent generations, we measured allele-specific DNA methylation and allele-specific transcript expression at three imprinted loci in both somatic and germ cells of mice generated by intracytoplasmic sperm injection (ICSI), a type of ART, and in offspring of these same mice produced by natural mating. We identified epimutations at multiple imprinted genes in the somatic tissues of mice produced by ICSI. However, we observed normal epigenetic programming at the same imprinted loci in somatic cells of offspring of these ICSI mice produced by natural mating. We then examined germ cells from the original ICSI mice and observed germ-line–specific correction of the epimutations.
Results
Epimutations Are Detected in Somatic Cells of Most ICSI Mice.
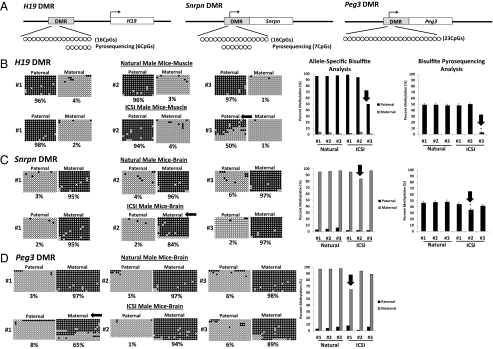
We assessed allele-specific DNA methylation and expression at multiple imprinted loci in somatic tissues of male and female mice produced by ICSI. DNA methylation at the H19 DMR was examined in a region within the DMR that contained 16 CpG sites on the paternal (B6) and 17 CpG sites on the maternal (Cast) allele (Fig. 1A). The imprinted H19 gene is paternally repressed and normally acquires paternal methylation at the DMR during spermatogenesis (10). Therefore, in somatic cells, the paternal allele is normally methylated, and the maternal allele is normally unmethylated. We observed hypermethylated paternal alleles (95–97% of all CpGs methylated) and hypomethylated maternal alleles (1–4% of all CpGs methylated) in muscle tissue of naturally conceived males and females (Fig. 1B and Fig. S1A). All three of the female mice and two of the male mice produced by ICSI displayed similar differential methylation. However, the third male mouse produced by ICSI (no. 3) showed a substantial reduction in methylation on the paternal allele in muscle tissue (Fig. 1B). We also used a custom pyrosequencing assay to examine six CpG sites of the region previously analyzed by allele-specific bisulfite sequencing (Fig. 1A). The same ICSI male mouse (no. 3) that showed relative hypomethylation on the paternal H19 allele in muscle tissue based on allele-specific bisulfite sequencing also displayed significantly reduced methylation (P < 0.005) when muscle tissue was examined by pyrosequencing (Fig. 1B). In addition, this same ICSI male (no. 3) showed significantly reduced methylation at the H19 DMR in liver and brain tissue as well, indicating the epimutation observed in the H19 gene was not tissue specific in this mouse (Fig. S2A).
Fig. 1.
Allele-specific DNA methylation of the H19, Snrpn, and Peg3 DMRs in somatic tissues of naturally conceived and ICSI-derived male mice. (A) Schematic representation of regions analyzed by allele-specific bisulfite sequencing or pyrosequencing. The B6 allele of paternally methylated H19 and maternally methylated Snrpn and Peg3 genes is shown. The bent arrow represents the transcription start site and circles represent individual CpG sites that were examined. Allele-specific bisulfite sequencing and pyrosequencing analyses of the methylation profiles for the H19 DMR in muscle tissue (B) and the Snrpn (C), and Peg3 (D) DMRs in brain tissue from naturally conceived and ICSI-derived mice. Allele-specific bisulfite sequencing data are shown on a per strand basis (circles) and as a bar graph; bisulfite pyrosequencing results are shown as a bar graph. For the per strand bisulfite sequencing results, each line corresponds to a single strand of DNA, and each circle represents a single CpG dinucleotide (black, methylated; white, unmethylated). Numbers to the left of each set of circles represent different individual mice and the percent of methylated CpG sites is indicated below each set of DNA strands. Pyrosequencing data are expressed as mean ± SD of methylated CpGs. *P < 0.005 compared with data from naturally conceived mice. Black arrows point to aberrant DNA methylation patterns that constitute epimutations in ICSI-derived mice. DMR, differentially methylated region.
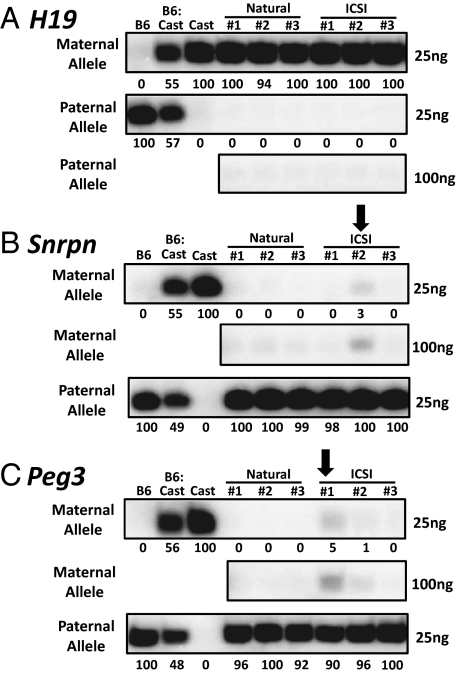
We used the single nucleotide primer extension (SNuPE) assay to quantify expression of paternal and maternal H19 alleles in naturally conceived and ICSI-derived mice (Fig. 2 and Figs. S3 and S4). All of the mice displayed normal maternal-specific expression in muscle tissue (Fig. 2A and Fig. S4A). Thus, the ICSI male (no. 3) that showed aberrant hypomethylation of paternal alleles at the H19 DMR (Fig. 1B) retained transcriptional repression of these alleles in muscle cells (Fig. 2A). Monoallelic expression of the H19 gene is also regulated by DNA methylation in the proximal promoter region (ref. 11; Fig. S5A). We analyzed DNA methylation in this region and found that despite the aberrant hypomethylation at the H19 DMR in ICSI male no. 3, hypermethylation was retained in the promoter region (Fig. S5B). These results suggest that hypermethylation in the promoter of the paternal allele of H19 can maintain allele-specific repression even when there is abnormal hypomethylation at the DMR.
Fig. 2.
Allele-specific expression of imprinted genes in somatic tissues of naturally conceived and ICSI-derived male mice. (A) The SNuPE assay for allele-specific expression was performed on RNA isolated from muscle tissue of natural and ICSI male mice for the H19 gene (A) and on RNA from brain tissue for the Snrpn (B) and Peg3 (C) genes. The expression of each parental allele was assessed separately and the parental origin is indicated to the left of each gel. Numbers below each lane indicate the percentage of expression compared with internal controls. The amount of purified RT-PCR product loaded onto the gel is indicated to the right of each gel. The 100ng image could not be quantified because the internal controls were saturated. Black arrows point to aberrant allele-specific gene expression patterns that constitute epimutations in ICSI-derived mice.
DNA methylation at the Snrpn DMR was also analyzed in the same naturally conceived and ICSI-derived mice. Allele-specific bisulfite sequencing was performed to examine a region that contained 16 CpG sites in the DMR on the paternal (B6) allele and 15 CpG sites in the DMR on the maternal (Cast) allele (Fig. 1A). The maternal allele of the Snrpn gene is typically hypermethylated, whereas the paternal allele is hypomethylated in somatic cells (12). As expected, naturally conceived male and female mice exhibited methylated maternal alleles (94–97% methylation) and unmethylated paternal alleles (1–6% methylation) in brain cells (Fig. 1C and Fig. S1B). All of the ICSI female mice and two of the ICSI male mice also displayed hypermethylation on the maternal allele (93–97% methylation) and hypomethylation on the paternal allele (1–2% methylation) (Fig. 1C and Fig. S1B). However, one ICSI-derived male mouse (no. 2) showed slightly reduced methylation (86%) on the maternal allele analyzed by allele-specific bisulfite sequencing (Fig. 1C). That this seemingly minor difference in overall methylation actually represented a valid epimutation was suggested by the following observations: (i) the aberrant, hypomethylated maternal allele was completely unmethylated (Fig. 1C), (ii) this same allele showed aberrant expression (Fig. 2B), and (iii) a significant reduction in methylation was also detected by bisulfite pyrosequencing (Fig. 1C).
We next analyzed DNA methylation at a second maternally repressed, imprinted gene, Peg3, in the same naturally conceived and ICSI-derived mice. The region examined contained 23 CpG sites on the paternal (B6) allele and 24 CpG sites on the maternal (Cast) allele (Fig. 1A). All naturally conceived males and females and all three of the ICSI-derived female mice and two of the ICSI-derived male mice exhibited the expected allele-specific methylation profile (Fig. 1D and Fig. S1C). However, ICSI male no. 1 displayed reduced methylation on the maternal allele in brain cells (Fig. 1D) and aberrant expression from this same allele (Fig. 2C). In addition, two ICSI females (nos. 1 and 3) that did not exhibit reduced methylation at the Peg3 DMR nevertheless showed abnormal expression from the maternal allele (Figs. S1C and S4C). Thus, it appears that expression of Peg3 can be induced from the normally repressed maternal allele with or without abnormal DNA methylation at the DMR.
Epimutations Are Not Transmitted to Naturally Conceived Progeny of ICSI Mice.
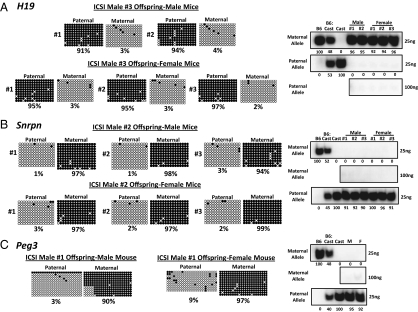
To assess transmission of primary epimutations induced in ICSI mice, we examined male and female offspring from the ICSI-derived mice that exhibited primary epimutations in somatic tissues. We analyzed offspring of ICSI male no. 3 for epimutations at the H19 DMR in muscle tissue (Fig. 3A), offspring of ICSI male no. 2 for epimutations at the Snrpn DMR in brain tissue (Fig. 3B), and offspring of ICSI male no. 1 for epimutations at the Peg3 gene in brain tissue (Fig. 3C). In each case, the somatic epimutations detected in each original ICSI-derived mouse (manifested as completely or nearly completely unmethylated strands of a normally methylated allele) were not detected in offspring of that mouse produced by natural reproduction.
Fig. 3.
Somatic epimutations found in ICSI-derived mice are not transmitted to offspring. DNA methylation profiles of the H19 (A), Snrpn (B), and Peg3 (C) DMRs were examined in somatic cells from male and female offspring of ICSI mice that displayed somatic epimutations. No aberrant, allele-specific DNA methylation patterns were detected in any of the offspring. Allele-specific gene expression patterns were also examined for the H19 (A), Snrpn (B), and Peg3 (C) genes in somatic cells of naturally produced offspring of ICSI mice. Normal maternal-specific expression of the H19 gene in muscle cells, and paternal-specific expression of the Snrpn and Peg3 genes in brain cells, was observed in offspring of ICSI mice.
Epimutations Are Corrected in Male Germ Cells of ICSI Mice.
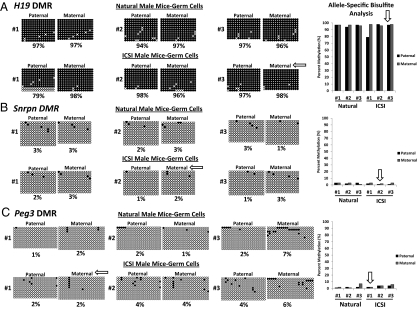
To determine whether primary epimutations are corrected by germ-line–specific epigenetic reprogramming, we analyzed allele-specific methylation and expression profiles of the same imprinted genes in purified germ cell populations from the same naturally conceived and ICSI-derived male mice. As expected, the advanced germ cells of naturally conceived and ICSI-derived male mice exhibited hypermethylation on both alleles at the H19 DMR (Fig. 4A). Interestingly, there was one ICSI-derived male mouse (no. 1) that displayed a modest reduction in methylation at the H19 DMR on some paternal alleles in male germ cells; however, we did not detect any abnormal methylation in muscle tissue, and we did not observe any abnormal expression in the germ line of this mouse (Fig. 1A and Fig. S6A). The Snrpn and Peg3 DMRs are maternally methylated so both alleles are unmethylated in male germ cells. We found that maternal and paternal alleles were both hypomethylated at the Snprn and Peg3 DMRs in germ cells of all natural and ICSI male mice (Fig. 4 B and C). Thus, all three of the ICSI-derived male mice that exhibited epimutations in somatic tissues at different imprinted genes displayed normal methylation profiles at these same loci in advanced male germ cells demonstrating normal germ-line–specific epigenetic reprogramming at both paternally and maternally imprinted loci (Fig. 4 and Fig. S6 A and B).
Fig. 4.
Allele-specific DNA methylation at the H19, Snrpn, and Peg3 DMRs in purified germ cells of naturally conceived and ICSI-derived male mice. DNA methylation profiles of the H19 (A), Snrpn (B) and Peg3 (C) DMRs in germ cells from naturally conceived and ICSI-derived male mice are shown. Analyses of DNA methylation are shown for germ cells from the same mice for which somatic cell data are presented in Fig. 1. Biallelic hypermethylation of the H19 DMR and hypomethylation of the Snrpn and Peg3 DMRs were observed in advanced male germ cells from all naturally conceived mice and ICSI mice. White arrows point to normally reprogrammed patterns in germ cells from mice that displayed epimutations in somatic cells.
Discussion
Although the use of ART has allowed millions of otherwise infertile couples to conceive children of their own, there remains some concern about the safety of these procedures because of reports of an increased propensity for imprinting disorders in children born after the use of these complex techniques (13, 14). Of particular concern is the possibility that manipulations used during ART procedures may disrupt the epigenetic programming required in each cell type to facilitate its proper function. Studies in animals have demonstrated that the use of ART methods can induce epigenetic abnormalities or epimutations in offspring, particularly at imprinted genes (15, 16). However, the possibility of transmitting these imprinting errors through the ensuing germ line to subsequent generations has not been thoroughly investigated.
In the current study, we analyzed epigenetic profiles of three imprinted genes in somatic and germ cells from naturally conceived mice and mice produced by ICSI as well as in the offspring of ICSI-derived mice that exhibited epimutations in somatic cells. We found that the ICSI procedure enhanced the occurrence of epimutations manifest as either disruption of allele-specific DNA methylation patterns at DMRs and/or aberrant allele-specific expression of imprinted genes in somatic cells of most mice produced by this method, whereas no such epimutations were observed in age-matched, naturally conceived mice. The DNA methylation epimutations we observed on the basis of allele-specific bisulfite genomic sequencing all involved loss of methylation from the normally hypermethylated allele of each imprinted gene investigated. Additional analysis by bisulfite pyrosequencing confirmed each epimutation detected by allele-specific bisulfite genomic sequencing in the H19 and Snrpn genes. Moreover, the pyrosequencing analysis indicated that the same epimutation found on the paternal H19 allele in muscle tissue of ICSI male no. 3 was also present in brain and liver tissue. However, the epimutation observed on the maternal allele of the Snrpn gene in brain tissue of ICSI male no. 2 was not detected in liver or muscle tissue. Thus, we observed at least one epimutation in each of the three ICSI males we investigated, although these epimutations occurred in different imprinted genes in each mouse. ICSI male no. 3 exhibited epimutations at H19 and Snrpn DMRs in muscle tissue demonstrating that the ICSI procedure can, on occasion, induce epigenetic defects at multiple loci within the same individual. In addition, we observed variation in the extent of tissue specificity of each epimutation detected. This variation may be a reflection of tissue-specific differences in maintenance of epimutations rather than tissue-specific differences in their initial induction.
Our data indicate that the occurrence of epimutations in mice generated by ICSI is stochastic and may be detected ubiquitously or tissue-specifically in individual mice. Our results showed losses of methylation on the normally hypermethylated allele of imprinted genes but not gain of methylation on the normally hypomethylated allele in ART-generated offspring. These results are consistent with a study by Market-Velker et al. (17) that analyzed offspring from mice subjected to ovarian stimulation, but contrast with the pattern of aberrant hypermethylation observed by Turan et al. (18) in human tissues from offspring generated by ART.
Epimutations will typically only lead to a detectable phenotype if they produce a disruption in the expression of one or more genes. Thus, we also evaluated allele-specific expression. We observed abnormal expression from the maternal allele of the Snrpn and Peg3 genes in the somatic cells of the same ICSI-derived male mice that exhibited hypomethylation of the corresponding maternal alleles. Abnormal expression from the maternal allele of the Peg3 gene was also detected in two of the three ICSI-derived female mice, although we did not observe a corresponding decrease in maternal methylation at the Peg3 DMR. Nevertheless, we interpret this aberrant expression to be indicative of the presence of an epimutation, just as a mutant phenotype is considered to be indicative of a genetic mutation. We also detected a significant reduction of paternal-specific methylation at the H19 DMR in muscle cells of one ICSI-derived male mouse that was not correlated with abnormal expression from the repressed allele in these same cells. This observation is consistent with previous reports that loss of DNA methylation at a DMR is not always associated with ectopic expression of the normally repressed allele (19, 20). Importantly, none of the ICSI-derived mice produced in this study exhibited any obvious morphological or functional abnormalities, and all were able to reproduce naturally. These results suggest that many epimutations induced by ART may impose no significant deleterious effects.
The absence of transgenerational transmission of the epimutations we observed in ICSI-derived males suggests that these were primary epimutations that were subject to correction by epigenetic reprogramming in the germ line before production of progeny by natural reproduction. In the case of epimutations detected at the Snrpn and Peg3 DMRs, this correction presumably involved the standard erasure of inherited methylation on either allele of these imprinted genes followed by maintenance of this biallelic hypomethylation. However, correction of epimutations detected on the paternal allele of the H19 gene must have involved erasure of inherited paternal methylation followed by resetting and maintenance of complete methylation on both alleles in spermatogenic cells.
In conclusion, we found that most mice produced by ICSI in our study sustained primary epimutations, detected as abnormalities in either allele-specific DNA methylation or allele-specific expression of one or more imprinted genes, whereas control mice generated by natural reproduction did not. All offspring of ICSI-derived males exhibited normal epigenetic profiles in their somatic tissues, suggesting correction of the observed epimutations by germ-line–specific epigenetic reprogramming, and our analysis of germ cells from these mice supported this notion. This observation is consistent with the findings of Popp et al. (21), who demonstrated that male primordial germ cells are globally demethylated, thus limiting the potential for transgenerational epigenetic inheritance. However, because of the relatively low numbers of animals examined in this study, we cannot conclude that all primary epimutations will be consistently corrected. Indeed, certain types of epimutations may escape germ-line correction. Thus, in utero exposure to certain endocrine disruptors appears to induce epimutations that are transmitted to subsequent generations at high frequencies (22). However, these epimutations appear to be exceptions to the effect exemplified by the results provided in our study, indicating that primary epimutations are typically corrected by germ-line–specific epigenetic reprogramming.
Methods
Animals.
We used adult (age 70–150 d) B6(CAST7) mice (19) to analyze allele-specific DNA methylation and expression of imprinted genes. B6(CAST7) mice possess a chromosome 7 from the Mus musculus castaneus (Cast) strain on a C57BL/6 (B6) background. B6(CAST7) females were crossed with DBA males and the ensuing F1 progeny carried several previously characterized strain-specific polymorphisms that were used to distinguish the parental alleles of imprinted genes on chromosome 7 (17, 23, 24). Because of the significant labor involved in generating mice by ICSI and analyzing allele-specific DNA methylation and expression patterns, our study was limited to examining six mice (three male, three female) conceived through natural reproduction and six mice (three male, three female) created through the use of ICSI. We crossed the B6(CAST7)/DBA ICSI mice with naturally conceived C57BL/6 mice to produce progeny. All of the progeny inherited the Cast (maternal) allele from the ICSI-derived mice and the B6 allele from the naturally conceived mate. Protocols for the handling and treatment of all animals were reviewed and approved by the Institutional Animal Care and Use Committee at the University of Texas at San Antonio or the University of Hawaii, as appropriate.
ICSI and Embryo Transfer.
ICSI was performed according to Kimura and Yanagimachi (25) with minor modifications (26). Mature oocytes at metaphase II were recovered from 10- to 12-wk-old C57BL/6(CAST7) females that had been subjected to endocrine stimulation by consecutive injections of equine CG (5 IU) and human CG (hCG)(5 IU) administered 48 h apart. Approximately 15–16 h after the injection of hCG, cumulus–oocyte complexes were collected from the oviducts and treated with 0.1% hyaluronidase to remove cumulus cells. Cauda epididymal sperm of DBA males were suspended in Hepes-CZB medium for 20 min, and a drop of sperm suspension was then mixed with 12% (wt/vol) polyvinylpyrrolidone) in Hepes-CZB. The head of a single sperm was injected into each oocyte. Resulting embryos were cultured for 3 d to obtain morula/blastocyst stage embryos that were transferred into the oviducts of CD-1 surrogate mothers (day 0.5) that had been mated with vasectomized CD-1 males the previous night. Newborn pups were obtained on day 19.5 after embryo transfer.
Purification of Spermatogenic Cells.
Germ cells were isolated from each individual male mouse by using a 50-mL “mini” Sta-Put chamber as described (27). Gonadal tissue was subjected to sequential digestions of 15 min each at 37 °C with 0.5 mg/mL collagenase and 0.5 mg/mL trypsin. Cells were treated briefly with 2 mg/mL DNase, resuspended in 0.5% BSA, and loaded onto a 50-mL gradient of 2–4% (wt/vol) BSA in KREBS buffer (27) and allowed to settle at unit gravity for 2 h. Fractions were collected and examined under phase optics to identify the desired cell type, and these fractions were then pooled. A combined population of pachytene spermatocytes plus round spermatids at purity of >95% was isolated from testes of adult mice at ≥60 d postpartum (dpp).
DNA and RNA Isolation.
DNA and RNA were isolated simultaneously from somatic tissue or germ cells by using a TRIzol-based protocol. Somatic tissue (0.1 g) was homogenized in 1-mL TRIzol (Invitrogen). Purified populations of germ cells were suspended in 200μL of TRIzol and pipetted to lyse cells before adding an additional 600 μL of TRIzol. After organic extraction, the aqueous phase was used for RNA isolation and the interphase/organic phase was used for DNA isolation, both as described by the manufacturer (Invitrogen) with slight modifications (SI Methods).
DNA Methylation Analysis.
Allele-specific bisulfite sequencing was performed on genomic DNA isolated from somatic tissue and germ cells by using the EZ Gold DNA methylation kit according to manufacturer's instructions (Zymo Research). Bisulfite-converted DNA was amplified by using nested PCR for H19, Snrpn, and Peg3 DMRs as described (refs. 17, 23, and 24; SI Methods and Table S1). Amplified products were directly cloned into the pCR2.1-TOPO vector (Invitrogen), and then transformed into chemically competent Escherichia coli cells (Invitrogen) and plated on LB plates with ampicillin (0.1 mg/mL). Recombinant plasmids were isolated (28) and sequenced with the M13 reverse primer at the University of Texas Health Science Center at San Antonio DNA sequencing facility. Raw bisulfite sequences were analyzed by using QUMA software (29) to eliminate replicate sequences with identical location and number of unconverted non-CpG–associated cytosines. Maternal and paternal alleles were distinguished by using multiple polymorphisms that are distinct between B6 and Cast sequences as described (17, 23, 24). At least 10 distinct bisulfite sequences from each parental allele were obtained from each sample for each imprinted gene analyzed. We also used custom pyrosequencing assays for the H19 and Snrpn DMRs on six and seven CpG sites, respectively, of the region previously analyzed by allele-specific bisulfite sequencing (SI Methods).
RT-PCR.
PCR was carried out by the addition of 2 μL of cDNA to the PCR mix [1× PCR buffer, 250 μM dNTPs, 0.5 μM forward and reverse primers (Tables S1 and S2), 1.25 U per reaction Hotmaster Taq DNA Polymerase (Eppendorf)]. The reaction conditions for all genes were as follows: 94 °C for 20 s, 55 °C for 20 s, 70 °C for 30 s (30 cycles), and 70 °C for 10 min. PCR products were run on a 1% agarose gel followed by gel-purification using the QIAquick nucleotide removal kit (Qiagen).
SNuPE Assay.
SNuPE reactions were carried out on RT-PCR products as described (30). Either 25 or 100 ng of amplified product were added to the SNuPE reaction [1× PCR buffer, 2 μCi of [32P] dNTP, 1 μM SNuPE primer (Table S2), and 1 U Hotmaster Taq DNA polymerase] and subjected to 95 °C for 1 min, 55 °C for 2 min, and 72 °C for 1 min. Extension products were electrophoresed through 15% polyacrylamide-urea gel to detect incorporation of radioactive nucleotides by phosphorimaging, and the relative quantity of each band was determined by using Quantity One Analysis software (Bio-Rad).
Statistical Analysis.
To evaluate the significance of the bisulfite pyrosequencing data, the Student t test was used and P values were considered to be significant at P < 0.005.
Supplementary Material
Acknowledgments
We thank Jacey Hornecker and Christopher Krapp for technical assistance. This work was supported by National Institutes of Health Grants HD42772 (to J.R.M. and R.Y.) and HD068157 (to M.B.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1201990109/-/DCSupplemental.
References
- 1.Morgan HD, Santos F, Green K, Dean W, Reik W. Epigenetic reprogramming in mammals. Hum Mol Genet. 2005;14(Spec No 1):R47–R58. doi: 10.1093/hmg/ddi114. [DOI] [PubMed] [Google Scholar]
- 2.Renfree MB, Hore TA, Shaw G, Graves JA, Pask AJ. Evolution of genomic imprinting: Insights from marsupials and monotremes. Annu Rev Genomics Hum Genet. 2009;10:241–262. doi: 10.1146/annurev-genom-082908-150026. [DOI] [PubMed] [Google Scholar]
- 3.Reik W, Walter J. Genomic imprinting: Parental influence on the genome. Nat Rev Genet. 2001;2:21–32. doi: 10.1038/35047554. [DOI] [PubMed] [Google Scholar]
- 4.Brandeis M, et al. The ontogeny of allele-specific methylation associated with imprinted genes in the mouse. EMBO J. 1993;12:3669–3677. doi: 10.1002/j.1460-2075.1993.tb06041.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li E, Beard C, Jaenisch R. Role for DNA methylation in genomic imprinting. Nature. 1993;366:362–365. doi: 10.1038/366362a0. [DOI] [PubMed] [Google Scholar]
- 6.McCarrey JR. The epigenome as a target for heritable environmental disruptions of cellular function. Mol Cell Endocr. September 25, 2011 doi: 10.1016/j.mce.2011.09.014. [DOI] [PubMed] [Google Scholar]
- 7.Horsthemke B. Epimutations in human disease. Curr Top Microbiol Immunol. 2006;310:45–59. doi: 10.1007/3-540-31181-5_4. [DOI] [PubMed] [Google Scholar]
- 8.Fortier AL, Lopes FL, Darricarrère N, Martel J, Trasler JM. Superovulation alters the expression of imprinted genes in the midgestation mouse placenta. Hum Mol Genet. 2008;17:1653–1665. doi: 10.1093/hmg/ddn055. [DOI] [PubMed] [Google Scholar]
- 9.Hori N, et al. Aberrant CpG methylation of the imprinting control region KvDMR1 detected in assisted reproductive technology-produced calves and pathogenesis of large offspring syndrome. Anim Reprod Sci. 2010;122:303–312. doi: 10.1016/j.anireprosci.2010.09.008. [DOI] [PubMed] [Google Scholar]
- 10.Tremblay KD, Saam JR, Ingram RS, Tilghman SM, Bartolomei MS. A paternal-specific methylation imprint marks the alleles of the mouse H19 gene. Nat Genet. 1995;9:407–413. doi: 10.1038/ng0495-407. [DOI] [PubMed] [Google Scholar]
- 11.John RM, Lefebvre L. Developmental regulation of somatic imprints. Differentiation. 2011;81:270–280. doi: 10.1016/j.diff.2011.01.007. [DOI] [PubMed] [Google Scholar]
- 12.Barr JA, Jones J, Glenister PH, Cattanach BM. Ubiquitous expression and imprinting of Snrpn in the mouse. Mamm Genome. 1995;6:405–407. doi: 10.1007/BF00355641. [DOI] [PubMed] [Google Scholar]
- 13.Cox GF, et al. Intracytoplasmic sperm injection may increase the risk of imprinting defects. Am J Hum Genet. 2002;71:162–164. doi: 10.1086/341096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeBaun MR, Niemitz EL, Feinberg AP. Association of in vitro fertilization with Beckwith-Wiedemann syndrome and epigenetic alterations of LIT1 and H19. Am J Hum Genet. 2003;72:156–160. doi: 10.1086/346031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doherty AS, Mann MR, Tremblay KD, Bartolomei MS, Schultz RM. Differential effects of culture on imprinted H19 expression in the preimplantation mouse embryo. Biol Reprod. 2000;62:1526–1535. doi: 10.1095/biolreprod62.6.1526. [DOI] [PubMed] [Google Scholar]
- 16.Sato A, Otsu E, Negishi H, Utsunomiya T, Arima T. Aberrant DNA methylation of imprinted loci in superovulated oocytes. Hum Reprod. 2007;22:26–35. doi: 10.1093/humrep/del316. [DOI] [PubMed] [Google Scholar]
- 17.Market-Velker BA, Zhang L, Magri LS, Bonvissuto AC, Mann MR. Dual effects of superovulation: Loss of maternal and paternal imprinted methylation in a dose-dependent manner. Hum Mol Genet. 2010;19:36–51. doi: 10.1093/hmg/ddp465. [DOI] [PubMed] [Google Scholar]
- 18.Turan N, et al. Inter- and intra-individual variation in allele-specific DNA methylation and gene expression in children conceived using assisted reproductive technology. PLoS Genet. 2010;6:e1001033. doi: 10.1371/journal.pgen.1001033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mann MR, et al. Disruption of imprinted gene methylation and expression in cloned preimplantation stage mouse embryos. Biol Reprod. 2003;69:902–914. doi: 10.1095/biolreprod.103.017293. [DOI] [PubMed] [Google Scholar]
- 20.Fauque P, et al. Assisted Reproductive Technology affects developmental kinetics, H19 Imprinting Control Region methylation and H19 gene expression in individual mouse embryos. BMC Dev Biol. 2007;7:116. doi: 10.1186/1471-213X-7-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Popp C, et al. Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature. 2010;463:1101–1105. doi: 10.1038/nature08829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anway MD, Skinner MK. Epigenetic transgenerational actions of endocrine disruptors. Endocrinology. 2006;147(6, Suppl):S43–S49. doi: 10.1210/en.2005-1058. [DOI] [PubMed] [Google Scholar]
- 23.Tremblay KD, Duran KL, Bartolomei MS. A 5′ 2-kilobase-pair region of the imprinted mouse H19 gene exhibits exclusive paternal methylation throughout development. Mol Cell Biol. 1997;17:4322–4329. doi: 10.1128/mcb.17.8.4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lucifero D, Mertineit C, Clarke HJ, Bestor TH, Trasler JM. Methylation dynamics of imprinted genes in mouse germ cells. Genomics. 2002;79:530–538. doi: 10.1006/geno.2002.6732. [DOI] [PubMed] [Google Scholar]
- 25.Kimura Y, Yanagimachi R. Intracytoplasmic sperm injection in the mouse. Biol Reprod. 1995;52:709–720. doi: 10.1095/biolreprod52.4.709. [DOI] [PubMed] [Google Scholar]
- 26.Yamazaki Y, Wakayama T, Yanagimachi R. Contribution of cumulus cells and serum to the maturation of oocyte cytoplasm as revealed by intracytoplasmic sperm injection (ICSI) Zygote. 2001;9:277–282. doi: 10.1017/s0967199401001307. [DOI] [PubMed] [Google Scholar]
- 27.McCarrey JR, et al. Differential transcription of Pgk genes during spermatogenesis in the mouse. Dev Biol. 1992;154:160–168. doi: 10.1016/0012-1606(92)90056-m. [DOI] [PubMed] [Google Scholar]
- 28.Birnboim HC, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumaki Y, Oda M, Okano M. QUMA: Quantification tool for methylation analysis. Nucleic Acids Res. 2008;36(Web Server issue):W170–175. doi: 10.1093/nar/gkn294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Singer-Sam J, LeBon JM, Dai A, Riggs AD. A sensitive, quantitative assay for measurement of allele-specific transcripts differing by a single nucleotide. PCR Methods Appl. 1992;1:160–163. doi: 10.1101/gr.1.3.160. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.