Abstract
Quantitative trait loci (QTL) mapping is a powerful tool for investigating the genetic basis of natural variation. QTL can be mapped using a number of different population designs, but recombinant inbred lines (RILs) are among the most effective. Unfortunately, homozygous RIL populations are time consuming to construct, typically requiring at least six generations of selfing starting from a heterozygous F1. Haploid plants produced from an F1 combine the two parental genomes and have only one allele at every locus. Converting these sterile haploids into fertile diploids (termed “doubled haploids,” DHs) produces immortal homozygous lines in only two steps. Here we describe a unique technique for rapidly creating recombinant doubled haploid populations in Arabidopsis thaliana: centromere-mediated genome elimination. We generated a population of 238 doubled haploid lines that combine two parental genomes and genotyped them by reduced representation Illumina sequencing. The recombination rate and parental allele frequencies in our population are similar to those found in existing RIL sets. We phenotyped this population for traits related to flowering time and for petiole length and successfully mapped QTL controlling each trait. Our work demonstrates that doubled haploid populations offer a rapid, easy alternative to RILs for Arabidopsis genetic analysis.
Exploring the genetic basis of phenotypic natural variation is a cornerstone for biological research. Knowledge gained from these studies can identify genes controlling a phenotype, provide insight into evolution and adaptation, and facilitate the improvement of crops through selective breeding. Many traits of interest vary quantitatively and display complex inheritance. Quantitative trait loci (QTL) mapping is an important approach commonly used to dissect the genetic basis of complex traits. It is especially valuable in species such as the model plant Arabidopsis thaliana, for which a wide range of accessions have been collected from diverse geographical regions.
To map QTL, phenotype and genotype information for a large population are needed. In the past, genotyping has been a significant limitation for QTL mapping. Current high-throughput DNA sequencing technology allows rapid, inexpensive identification of informative genome-wide markers. Now, the major limitation for identifying loci contributing to phenotypic variation is the construction of mapping populations. There are a number of population structures that have been used to map QTL, and each has its own benefits. F2 and backcross (BC) populations have the advantage of speed, as they both enable mapping within a few generations of the initial parental cross. Unfortunately, because of the high levels of heterozygosity in these designs, genotypic and phenotypic information is not stable and must be captured in each subsequent generation. In contrast, recombinant inbred line (RIL) populations are developed through multiple generations of selfing until each line is a nearly homozygous mosaic of the parental genomes. RIL populations in A. thaliana are commonly selfed until the F8 generation. The substantial amount of time spent developing these populations is rewarded by the immortality of each line. Every generation is essentially genetically identical, allowing the lines to be genotyped just once. Additionally, isogenic lines are advantageous because they allow replication, which facilitates dissection of the genetic and environmental components contributing to phenotypic variation. Estimating the interaction between genotype and environment (G × E) is also possible when using isogenic lines because replication can occur across multiple environments. Replication also allows investigators to study subtle trait differences that are clarified when many individuals are analyzed for each line. The extensive amount of time required to make new isogenic populations is currently a major impediment to using A. thaliana for studying the genetic basis of natural variation.
In this paper, we demonstrate a unique method for developing an A. thaliana mapping population that integrates speed (the advantage of F2 and BC populations) with the immortality of RILs. These benefits are achieved by quickly generating individuals that contain two identical copies of each chromosome, known as doubled haploids (DHs). Similar to RILs, doubled haploids are homozygous throughout their genome, a property that facilitates the detection of QTL. Doubled haploids have been used for QTL mapping in a variety of species, especially the grasses (1–4), as they represent the fastest method to achieve homozygosity from any F1 or heterozygote. DH mapping populations have been made in agronomically important crops such as rice, maize, rapeseed, wheat, and barley. However, haploid induction in these and other plants usually requires tissue culture of microspores, ovules, or rescued haploid embryos following distant hybridization (5). Such cell-culture–based methods are highly influenced by genotype and/or environmental factors. Hence their practical usefulness is limited by the technical difficulty of producing a large doubled haploid population in a desired genetic background, even within a species. The A. thaliana recombinant doubled haploid population described here was created via centromere-mediated genome elimination (6). This approach is genotype independent and involves simple crossing of a F1 to a haploid inducer parent to obtain haploid seeds. Before the discovery of this method, it was difficult to generate doubled haploids in A. thaliana despite the common use of such lines in other species (6). To induce haploidy, this method relies on a transgenic line, cenh3-1 GFP-tailswap, which contains a modified version of the centromere-specific histone protein CENH3. After fertilization, competition occurs between chromosomes containing the wild-type and mutant CENH3 proteins, leading to the loss of mutant-derived chromosomes and the production of haploid embryos. The resulting haploid plants spontaneously produce a low number of seed, most of which are fertile doubled haploids. Here, we describe the development and genetic characterization of a recombinant doubled haploid population in A. thaliana and its use in mapping QTL for flowering time and petiole length.
Results
Development of an A. thaliana Doubled Haploid Population.
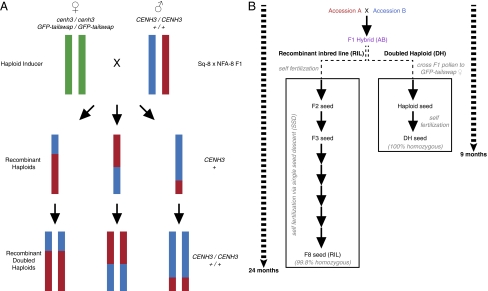
To determine whether doubled haploid populations generated via genome elimination are comparable to standard recombinant inbred lines for QTL mapping, we focused on flowering time, a well-described trait in A. thaliana (7, 8). The parental accessions of the population, NFA-8 (CS22598) and Sq-8 (CS22601), exhibit variation in flowering time, although both accessions flower in less than 23 days without vernalization. To create recombinant doubled haploid lines, an F1 derived by crossing the two parental accessions was used as the male parent in a cross to the transgenic haploid inducer line, cenh3-1 GFP-tailswap, in the Columbia (Col-0) background (6) (Fig. 1A). Haploid F2 plants, which combine the genomes of NFA-8 and Sq-8 yet contain no genetic contribution from the haploid inducer (Col-0), were selected phenotypically using three criteria: the vegetative phenotype of haploids before flowering, sterility after flowering, and the small floral size distinctive of A. thaliana haploids (6). Although haploid A. thaliana plants are sterile, they can produce rare diploid progeny through random chromosome segregation in haploid meiosis (and occasionally through spontaneous somatic doubling) (6). We collected the spontaneous diploid seed produced by each haploid plant and propagated them as doubled haploid lines. Because doubled haploids are fertile and homozygous for every locus in the genome, each plant in the population is fixed for unique recombination events between the two original parents. To compare RIL and DH construction strategies, see Fig. 1B. Simulations were performed to identify a population size that would provide reasonable estimates of QTL position, LOD score, and Bayesian interval size (SI Materials and Methods and Fig. S1). For this experiment, 260 doubled haploid lines derived from an Sq-8 × NFA-8 F1 were phenotyped and genotyped to map QTL controlling flowering time differences between parental accessions.
Fig. 1.
(A) Construction of a recombinant doubled haploid population. Wild-type F1 plants (Sq-8 × NFA-8) were crossed to cenh3-1 GFP-tailswap plants to produce recombinant haploids. Chromosomal doubling generated fertile doubled haploid lines with homozygous chromosome regions derived from either NFA-8 or Sq-8. (B) Schematic showing a timeline comparison for the development of DH and RIL populations. The generation time (seed to seed) will vary for each combination of parental accessions used. For this schematic, a single generation was represented with a black arrow and assumed to be 3 mo.
Rapid, Inexpensive Genotyping by Reduced Representation Sequencing.
High-throughput DNA sequencing has revolutionized the ability to rapidly characterize genetic variation, particularly when a high-quality reference genome is available (9). Reduced representation sequencing (10) offers an alternative to whole genome sequencing, which further increases the efficiency of high-throughput genotyping. By sequencing a reproducible subfraction of the genome of each individual, it becomes economical to use short read sequencing, not only for SNP discovery but also to genotype large mapping populations. For each doubled haploid line in our population, reduced representation restriction enzyme sequence comparative analysis (RESCAN) libraries were constructed for Illumina sequencing using barcoded adapter oligonucleotides (SI Materials and Methods) (11). Using the restriction enzyme NlaIII, which cleaves at CATG sequences, we were able to reproducibly sequence ∼10% of the genome from each DH line. The 260 putative doubled haploid lines and the two parental accessions were sequenced using 40 base pair single end reads on three lanes of the Illumina GAII with 96 individuals pooled per lane. After filtering, 10,173 codominant, biallelic sites were defined and used to genotype the population (SI Materials and Methods and Table S1). The mean distance between selected markers was 11,700 base pairs (SD = 27,054) (Table S2). On average, each line had read coverage at 3,373 of the 10,173 sites (SD = 1,258) (Table S2). In summary, reduced representation high-throughput sequencing provides an efficient and cost-effective approach for densely genotyping every individual in a mapping population.
A Large Majority of Doubled Haploids Contained only Genetic Material from Their Heterozygous F1 Parent.
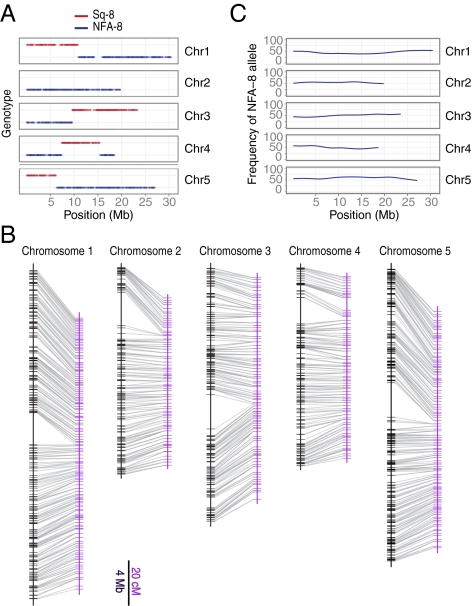
In addition to producing haploids, cenh3-1 GFP-tailswap × wild-type crosses yield diploid and aneuploid progeny with chromosomes from both parents (6). Further, a low frequency of haploid progeny arising from genome elimination crosses in maize have been shown to contain limited traces of the inducer parent genome (12).Therefore, we sought to determine whether our putative doubled haploids contained any genetic material from their cenh3-1 GFP-tailswap parent, which is in the Col-0 accession. Data from Illumina sequencing was used to ask whether potential doubled haploids contained genetic information from Col-0 in addition to alleles from their wild-type parent (an Sq-8 and NFA-8 F1). To perform this analysis, it was important that the parents of the F1 did not share a genetic background with the haploid inducer. We identified a further 3,873 SNPs that discriminate between Col-0 and the two parental accessions (SI Materials and Methods). Using these SNPs, we found that 238 of the 260 sequenced lines were true recombinant doubled haploids with only NFA-8 and Sq-8 genetic material. An example of a typical doubled haploid genotype is shown in Fig. 2A. The remaining 22 lines (8.46%) were contaminated with genetic material from Col-0. Nineteen of these lines contained Col-0 alleles throughout their genomes and likely resulted from outcrossing of recombinant haploid plants with either diploid hybrid siblings or wild-type Col-0 in a mixed growth chamber. Because haploid plants are sterile, their propensity for outcrossing is increased relative to diploid plants. In the future, diploid plants identified by their fertility (or better, by a dominant Col-0 marker) should be culled to alleviate this problem.
Fig. 2.
Genetic characterization of doubled haploid population. (A) Genotype of a typical doubled haploid. Each point represents a single SNP with either an NFA-8 allele (blue) or Sq-8 allele (red). Chromosomes 1, 3, and 5 are recombinant products of a single crossover event in the parental meiosis (Sq-8 × NFA-8 F1 hybrid). Chromosome 4 is a double recombinant and chromosome 2 is a nonrecombinant NFA-8 parental chromosome. (B) Comparisons of marker locations between the physical (black) and genetic (purple) maps. Only the 1,043 markers used to build the genetic map are shown. (C) Sliding window (250 kb) average of genome-wide NFA-8 allele frequency for each chromosome (1-kb step size).
The remaining three lines contained only short fragments of the Col-0 genome confined to one of the five chromosomes in a region spanning the centromere. The exact nature of these lines is unknown, but they may be the result of incomplete genome elimination. We have shown that such contamination can be identified and avoided by densely genotyping putative A. thaliana doubled haploid lines with centromere-proximal markers.
In summary, our analysis demonstrates that it is easy to identify haploid progeny from A. thaliana genome elimination crosses that contain only genetic material from their wild-type parent. Therefore, doubled haploid production in Arabidopsis offers a rapid alternative to developing traditional recombinant inbred lines by recurrent inbreeding.
Genetic Characterization of a Doubled Haploid Population.
The genetic properties of our recombinant doubled haploid population were examined to ask whether the construction technique influenced the manner in which parental genomes were combined. Because the number of SNPs discovered exceeds the recombination resolution in our population, only markers flanking recombination breakpoints were included in the genetic map. In total, 1,043 markers scored in 238 individuals were used to construct a map with a total genetic distance of 498.19 cM (Fig. 2B). Doubled haploids constructed via genome elimination are expected to have recombination rates comparable to standard F2 gametes. Each doubled haploid has a unique paternally derived recombinant chromosome set, representing approximately half of the crossover events per generation seen in an F2 diploid. The mean number of recombination events per doubled haploid line is 4.89 (SD = 2.05), which is approximately half of the average of 8.9 crossovers previously estimated in Arabidopsis F2 tetrads (13) and confirms that paternal recombination occurs as expected during the doubled haploid construction process. Once crossing design is considered, recombination rates are similar and the overall genetic distance is comparable to previously published A. thaliana RIL map distances, which range from 358 to 475 cM (14–17). Additionally, the order of markers on the physical and genetic maps are identical (Fig. 2B), indicating that there are no detectable rearrangements in the parents.
The genetic behavior of a mapping population can also be evaluated by measuring preferential segregation for parental alleles at each marker. As the markers scored in any given doubled haploid were different, we imputed missing genotypes before calculating allele frequencies (SI Materials and Methods). The mean allele frequency from the 250-kb sliding window analysis shown in Fig. 2C is 51.9% (SD = 6.5%). Looking at segregation distortion more closely, we find that at P < 0.05, ∼34.5% of markers deviate from the expected segregation of 1:1, 75% of which reside on either chromosome 1 or chromosome 5. The maximally distorted marker exhibits 1.8:1 segregation. In existing RIL populations, the largest magnitudes of distortion range from 1.8:1 to 2.7:1 (14–16, 18, 19). The distorted regions in existing populations can also span entire chromosome arms, but resolving the length of these regions is constrained by the limited marker density as many populations were genotyped with fewer than 100 markers (14, 16, 18). High-density genotyping of our doubled haploid populations provided accurate estimates of allele frequency variation and improved the resolution of genomic regions exhibiting minor distortion (Fig. 2C). In summary, the degree of allele frequency variation in our population is similar to the magnitude of segregation distortion measured in other A. thaliana RIL populations. Most importantly, haploid formation does not induce a high frequency of severe segregation distortion in our population.
High-density SNP information can also be used to address the frequency of gene conversion (GC) events. For each doubled haploid, SNPs in disagreement with the surrounding majority genotype were identified (SI Materials and Methods). Sanger resequencing was performed on 34 of the 1,242 discordant loci identified to determine whether the SNPs are due to Illumina sequencing error or are products of GC (SI Materials and Methods). Of the resequenced loci, only two, or 5.9%, are likely GC events. If this frequency is extrapolated, there may be ∼73 GC events detected in our reduced representation sequencing data. Gene conversion frequency was recently assessed in two A. thaliana tetrads using whole genome sequencing (20).The authors concluded that up to six GC events could occur per meiosis (20). Although only a fraction of the genome was sequenced for each doubled haploid, the frequency of GC events per individual is of comparable magnitude to GC measured using whole genome sequencing. Thus, the construction process used to develop our doubled haploids does not seem to distort the frequency of GC.
In conclusion, our recombinant doubled haploid population shares typical genetic characteristics such as total map distance, marker order, and allele segregation distortion with RIL populations constructed by the traditional inbreeding method.
Phenotypic Analysis of Flowering Time and Petiole Length Traits.
The phenotypic behavior of the doubled haploids was evaluated using six traits, five of which relate to flowering time. Because the five flowering time traits are correlated (SI Materials and Methods), we measured petiole length of the longest leaf to examine QTL controlling an unrelated trait. The petiole is a plant organ that connects the stem to the leaf blade. Its length varies in response to environmental conditions (21, 22). The transformed phenotypic distributions for all traits (Materials and Methods) are plotted in Fig. S2. As expected, the genetic correlation between flowering time traits was high, ranging from 0.69 to 0.86 (Fig. S3). Additionally, petiole length showed very little genetic correlation with the flowering traits, supporting the independence of these phenotypes (Fig. S3). For each trait, broad sense heritability was calculated (Table 1). Although the estimates of heritability for the flowering time traits are lower than other published values (23), the discrepancy is probably due to low genetic variation in flowering time between the parents. Indeed the parents exhibit very similar phenotypic values: the fitted values for days to flowering between the parental accessions only differ by 1 d. Despite the low level of phenotypic variation in the parents, the doubled haploid progeny display notable transgressive segregation for each trait, which is typical of recombinant inbred populations (18) and a desirable characteristic for mapping QTL.
Table 1.
Phenotypic analysis of flowering time and petiole length
| Trait | Heritability | Percent of phenotypic variance explained | No. of QTL |
| Diameter at bolting | 0.504 | 56.85 | 7 |
| Days to bolting | 0.457 | 58.80 | 9 |
| Leaves at bolting | 0.530 | 51.19 | 6 |
| Days to flowering | 0.442 | 54.52 | 8 |
| Leaves at flowering | 0.512 | 48.31 | 5 |
| Petiole length | 0.188 | 38.70 | 5 |
Identification of Flowering Time QTL.
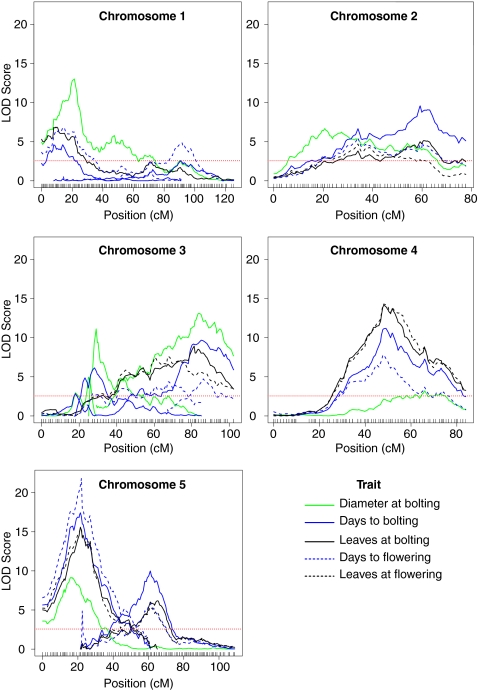
We next asked whether we could detect QTL for each measured trait in our doubled haploid population. For all traits, QTL mapping was performed using both raw fitted phenotypic values as well as transformed fitted values (Materials and Methods). Three methods for detecting QTL were used, with similar results: standard interval mapping, composite interval mapping, and multiple QTL mapping (24, 25). The results discussed below are derived from the multiple QTL model as this method was the most sensitive for detecting small effect QTL and genetic interactions in our data. All significant interactions between QTL were minor and contributed minimally to the overall phenotypic variance explained by each model (Table S3). The location of significant QTL, maximum LOD score, and percent of phenotypic variation explained by each QTL detected are summarized in Table S4. For the flowering time traits, between five and nine QTL were identified. Intervals containing major effect QTL coincide across the traits (Fig. 3). The final number of QTL specified in each model explain an average of 53.93% of the total phenotypic variation (SD = 4.23%). The contribution of any individual QTL ranged from 2.08 to 23.6% of the phenotypic variation. Considering only QTL with an effect on variation greater than 5%, the average Bayesian credible interval length (SI Materials and Methods) is 12.1 cM (SD = 10.94 cM), corresponding to a physical distance of ∼3.8 megabases and encompassing 780 genes on average (SD = 703). QTL were also mapped for petiole length, demonstrating that significant QTL are detectable in this population even for low heritability traits (Table S5 and Fig. S4). Therefore, using our A. thaliana doubled haploid population, QTL mapping was able to identify both small and large effect QTL for multiple traits with varying levels of heritability.
Fig. 3.
Flowering time QTL. Results of the multiple QTL model for each measured flowering time trait. Significance threshold of 2.54 is the mean threshold for all flowering traits (based on 1,000 permutations). For each chromosome, traces of the same color indicate multiple QTL were identified.
Using the intervals defined by multiple QTL mapping, we identified candidate genes for some of the major effect QTL. For flowering time, candidates were identified for three QTL that were consistently detected in at least four of the flowering time traits (Table S4). Each of these candidates, FLOWERING LOCUS C (FLC), TWIN SISTER OF FT (TSF), and CYCLING DOF FACTOR 2 (CDF2), have known roles in flowering time pathways (7, 8, 26–29). FLC, which resides under the QTL with the largest phenotypic effect (SI Materials and Methods, Fig. S5, and Table S6), was also a candidate gene in a previous F2 QTL mapping experiment using one of the parental accessions, NFA-8, in a cross to two other natural accessions (Van-0 and Bor-4) (30). We were also able to identify a number of candidates for the largest effect QTL controlling petiole length located on chromosome 2. The most likely candidate for this QTL is PHYTOCHROME INTERACTING FACTOR 4 (PIF4). PIF4 is involved in hypocotyl and petiole elongation in response to high temperatures and to changes in light quality (31–34). Although a large number of genes reside within each QTL interval, we can identify candidate genes for the largest effect QTL in our doubled haploid population.
Discussion
We have demonstrated that doubled haploid populations in A. thaliana offer an appealing alternative to traditional inbred populations for QTL mapping. The value of using centromere-mediated genome elimination for generating recombinant mapping populations rests in its ability to achieve complete homozygosity much more rapidly than conventional inbreeding. The genotypic immortality of inbred lines improves QTL detection by enabling replication, thereby increasing the power to detect QTL (35, 36). Although inbred populations are preferred for QTL mapping, the number of generations required to achieve near homozygosity and genetic contamination by outcrossing or seed admixture of other genotypes are an impediment for many researchers. Doubled haploids offer a solution to this problem. Genome elimination can produce homozygous lines in only two generations starting from an F1 (Fig. 1B). One disadvantage of using an inbred population such as RILs or DH lines for QTL analysis is that the dominance deviation of an allele cannot be estimated due to the complete homozygosity of the population. Despite this disadvantage, the significant time saving offered by doubled haploids has remarkable benefits for those interested in the natural variation of complex traits.
The technical simplicity of genome elimination contributes to the ease of creating doubled haploid populations in A. thaliana. The only requirement for haploid induction using genome elimination is the ability to perform a genetic cross, making the method accessible to any laboratory. Another asset of haploid production is the ability to induce homozygosity at any generation—inducing haploidization at later generations will increase mapping resolution because each generation adds another round of meiotic recombination. Thus, although doubled haploids can be developed very quickly, there is a compromise between speed and the resolution of QTL mapping. In the population described here, the size of the intervals around each QTL reflects the single generation of recombination. On the basis of our simulations, increasing the size of the population has a greater impact on the QTL interval size than additional generations of recombination (Fig. S1). Consistent with these simulations, a survey of recombination rates in five A. thaliana RIL populations at the F8 generation, showed that the mean number of recombination events ranged from 6.23 to 8.89 per individual (37). Relative to our doubled haploid population (4.89 recombination events per individual) these RILs have undergone an additional six generations of selfing and the average number of recombination events has increased approximately twofold. Theoretical calculations (38–40) and further empirical evidence in maize (41) support the twofold increase of recombination in A. thaliana RILs relative to DHs. Due to the rapid decrease in heterozygosity during RIL construction most recombination events are masked. At completion, a single RIL consists of two identical gametes containing the equivalent of two rounds of meiotic recombination (39). Therefore, to obtain a genetic map with higher resolution, the best choice is either to increase the population size or to begin by using an advanced intercross scheme (42) and induce haploidy at a later generation.
As construction of a new RIL population by conventional inbreeding is very time consuming and labor intensive, investigators often map QTL for their trait of interest using a small number of preexisting populations derived from commonly used parents (35). With centromere-mediated genome elimination, small research groups can rapidly create recombinant doubled haploid populations from parental accessions that are best suited to answer their biological question. Doubled haploids can even be used to quickly generate populations for validation of results from genome-wide association studies. These advantages promote the use of genome elimination as an excellent approach for building recombinant mapping populations.
Traditional methods for producing haploid plants, such as microspore culture or wide crosses, have been used to create doubled haploids in many plant species (although these methods have been ineffective for Arabidopsis) (6, 43). However, these approaches are technically challenging, often inefficient, and not portable across species. Additionally, the success of traditional methods is usually genotype specific, preventing their use with a wide variety of biologically interesting germplasm. It is possible that genome elimination based on CENH3 engineering can be applied to QTL mapping in other plant species, due to the conserved function of CENH3 in all eukaryotes. As the method has produced haploids from every A. thaliana accession tested so far, genome elimination may be able to overcome the genotype specificity of traditional haploid production methods.
The extensive genetic and genomic resources generated for A. thaliana continue to make this species an outstanding model for understanding general principles in plant biology. The ability to construct doubled haploid populations via centromere-mediated genome elimination synergizes with a high-quality genome sequence and vast natural collections with corresponding genotypic information to strengthen A. thaliana as a premier species for exploring the genetic basis of natural variation.
Materials and Methods
Population Development.
The initial F1 was generated by crossing two A. thaliana accessions, NFA-8 (CS22598) and Sq-8 (CS22601). Sq-8 was used as the female parent. To generate recombinant haploids, the F1 was used as the male parent in a cross to the transgenic haploid inducer line with the cenh3-1 GFP-tailswap genotype (6). Haploid progeny from this cross were selected phenotypically as previously described (6). Haploid Arabidopsis plants are mostly sterile because unbalanced reductional segregation during meiosis generates aneuploid gametes (6). In rare instances, meiotic nonreduction during anaphase I can produce viable haploid gametes (6). Upon self-fertilization these gametes give rise to fertile diploid seed (6). Spontaneous somatic doubling in haploid plants can also lead to production of fertile diploid seed, but this is rarer than meiotic nonreduction (6). Plants grown from these fertile diploid seeds constitute the first doubled haploid generation. This generation was genotyped and phenotyped to allow QTL mapping.
Phenotyping.
A total of 238 putative doubled haploid lines and the two parental accessions were grown in a randomized block design with five replicates under 16-h light: 8-h dark conditions. The average fluence rate for all shelves was 124.5 μE m−2 sec−1 (SD = 9.8 μE m−2 sec−1). Seeds were stratified at 4 °C for 4 d before sowing on soil. Each plant was phenotyped for five traits associated with flowering time: (i) rosette diameter at bolting (defined as when the inflorescence had risen 0.5 cm above the rosette), (ii) vegetative leaf number at bolting, (iii) days from sowing to bolting, (iv) vegetative leaf number at first open flower, and (v) days from sowing to first open flower. To measure petiole length, we took images of all plants 11 d after sowing. Petiole length of the longest leaf was measured using ImageJ (44).
Phenotypic Analysis.
The phenotypic data collected for each flowering time trait was analyzed using the R package lme4. For each trait, a linear mixed-effect model was specified containing two random factors, genotype and block. From each model, the fitted values for each genotype were extracted and used for subsequent QTL mapping. In addition to fitting a model to the original data, for each trait a model was also fit to data transformed using the Box Cox power transformation. The transformed fitted phenotypic values were used for the QTL mapping described in this paper. The fitted values from the original data are plotted in Fig. S6.
Estimation of Heritability.
Variance components for each trait were estimated in the mixed-effect models described above. Broad sense heritability was calculated for each trait by dividing the variance due to genotype by the total phenotypic variance.
QTL Mapping.
QTL mapping was performed in R/qtl (45) using the fitted phenotypic values (both transformed and untransformed) for 238 doubled haploid lines and 1,043 markers. Markers were selected on the basis of informativeness; each marker flanked a recombination breakpoint in at least one doubled haploid line. The doubled haploid crossing scheme was accounted for by setting the cross type to “dh” in R/qtl. When building the genetic map, est.map() considered the crossing design and adjusted the genetic distances accordingly. Standard 1-D and 2-D interval mapping was performed in addition to multiple QTL model analyses (stepwiseqtl()) and composite interval mapping. The results for each method (location, LOD score, and percent variation explained) were highly similar for the major effect QTLs across the two datasets. However, multiple QTL models were able to detect additional small effect QTL in the transformed dataset. LOD thresholds for each method were estimated using 1,000 permutations.
Supplementary Material
Acknowledgments
We thank Daniel Koenig for discussions on bioinformatics and technical support. This work was funded by National Science Foundation (NSF) IOS-1026094 and a Basil O'Connor Starter Scholar Award from the March of Dimes (to S.W.L.C.) and NSF IOS-0923752 and IOS-0820854 (to J.N.M.). I.M.H., J.M.-M., and L.C. were supported by NSF Plant Genome Grant DBI-0733857. S.W.L.C. is a Howard Hughes Medical Institute–Gordon and Betty Moore Foundation Investigator.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the National Center for Biotechnology Information Sequence Read Archive (SRA) (www.ncbi.nlm.nih.gov/sra/) (accession no. SRP010765.1).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1117277109/-/DCSupplemental.
References
- 1.Yang DL, Jing RL, Chang XP, Li W. Identification of quantitative trait loci and environmental interactions for accumulation and remobilization of water-soluble carbohydrates in wheat (Triticum aestivum L.) stems. Genetics. 2007;176:571–584. doi: 10.1534/genetics.106.068361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xu S, Jia Z. Genomewide analysis of epistatic effects for quantitative traits in barley. Genetics. 2007;175:1955–1963. doi: 10.1534/genetics.106.066571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Presterl T, et al. Quantitative trait loci for early plant vigour of maize grown in chilly environments. Theor Appl Genet. 2007;114:1059–1070. doi: 10.1007/s00122-006-0499-4. [DOI] [PubMed] [Google Scholar]
- 4.Fan CC, et al. The main effects, epistatic effects and environmental interactions of QTLs on the cooking and eating quality of rice in a doubled-haploid line population. Theor Appl Genet. 2005;110:1445–1452. doi: 10.1007/s00122-005-1975-y. [DOI] [PubMed] [Google Scholar]
- 5.Dunwell JM. Haploids in flowering plants: Origins and exploitation. Plant Biotechnol J. 2010;8:377–424. doi: 10.1111/j.1467-7652.2009.00498.x. [DOI] [PubMed] [Google Scholar]
- 6.Ravi M, Chan SW. Haploid plants produced by centromere-mediated genome elimination. Nature. 2010;464:615–618. doi: 10.1038/nature08842. [DOI] [PubMed] [Google Scholar]
- 7.Amasino R. Seasonal and developmental timing of flowering. Plant J. 2010;61:1001–1013. doi: 10.1111/j.1365-313X.2010.04148.x. [DOI] [PubMed] [Google Scholar]
- 8.Turck F, Fornara F, Coupland G. Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu Rev Plant Biol. 2008;59:573–594. doi: 10.1146/annurev.arplant.59.032607.092755. [DOI] [PubMed] [Google Scholar]
- 9.Davey JW, et al. Genome-wide genetic marker discovery and genotyping using next-generation sequencing. Nat Rev Genet. 2011;12:499–510. doi: 10.1038/nrg3012. [DOI] [PubMed] [Google Scholar]
- 10.Altshuler D, et al. An SNP map of the human genome generated by reduced representation shotgun sequencing. Nature. 2000;407:513–516. doi: 10.1038/35035083. [DOI] [PubMed] [Google Scholar]
- 11.Monson-Miller J, et al. Reference genome-independent assessment of mutation density using restriction enzyme-phased sequencing. BMC Genomics. 2012 doi: 10.1186/1471-2164-13-72. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Z, et al. Chromosome elimination and in vivo haploid production induced by Stock 6-derived inducer line in maize (Zea mays L.) Plant Cell Rep. 2008;27:1851–1860. doi: 10.1007/s00299-008-0601-2. [DOI] [PubMed] [Google Scholar]
- 13.Copenhaver GP, Browne WE, Preuss D. Assaying genome-wide recombination and centromere functions with Arabidopsis tetrads. Proc Natl Acad Sci USA. 1998;95:247–252. doi: 10.1073/pnas.95.1.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lister C, Dean C. Recombinant inbred lines for mapping RFLP and phenotypic markers in Arabidopsis thaliana. Plant J. 1993;4:745–750. doi: 10.1046/j.1365-313x.1996.10040733.x. [DOI] [PubMed] [Google Scholar]
- 15.Alonso-Blanco C, et al. Development of an AFLP based linkage map of Ler, Col and Cvi Arabidopsis thaliana ecotypes and construction of a Ler/Cvi recombinant inbred line population. Plant J. 1998;14:259–271. doi: 10.1046/j.1365-313x.1998.00115.x. [DOI] [PubMed] [Google Scholar]
- 16.Loudet O, Chaillou S, Camilleri C, Bouchez D, Daniel-Vedele F. Bay-0 × Shahdara recombinant inbred line population: A powerful tool for the genetic dissection of complex traits in Arabidopsis. Theor Appl Genet. 2002;104:1173–1184. doi: 10.1007/s00122-001-0825-9. [DOI] [PubMed] [Google Scholar]
- 17.West MA, et al. High-density haplotyping with microarray-based expression and single feature polymorphism markers in Arabidopsis. Genome Res. 2006;16:787–795. doi: 10.1101/gr.5011206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.el-Lithy ME, et al. New Arabidopsis recombinant inbred line populations genotyped using SNPWave and their use for mapping flowering-time quantitative trait loci. Genetics. 2006;172:1867–1876. doi: 10.1534/genetics.105.050617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Törjék O, et al. Segregation distortion in Arabidopsis C24/Col-0 and Col-0/C24 recombinant inbred line populations is due to reduced fertility caused by epistatic interaction of two loci. Theor Appl Genet. 2006;113:1551–1561. doi: 10.1007/s00122-006-0402-3. [DOI] [PubMed] [Google Scholar]
- 20.Lu P, et al. 2011. Analysis of Arabidopsis genome-wide variations before and after meiosis and meiotic recombination by re-sequencing Landsberg erecta and all four products of a single meiosis. Genome Res. [DOI]
- 21.van Zanten M, Voesenek LA, Peeters AJ, Millenaar FF. Hormone- and light-mediated regulation of heat-induced differential petiole growth in Arabidopsis. Plant Physiol. 2009;151:1446–1458. doi: 10.1104/pp.109.144386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kozuka T, et al. Involvement of auxin and brassinosteroid in the regulation of petiole elongation under the shade. Plant Physiol. 2010;153:1608–1618. doi: 10.1104/pp.110.156802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brachi B, et al. Linkage and association mapping of Arabidopsis thaliana flowering time in nature. PLoS Genet. 2010;6:e1000940. doi: 10.1371/journal.pgen.1000940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lander ES, Botstein D. Mapping mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics. 1989;121:185–199. doi: 10.1093/genetics/121.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zeng ZB. Precision mapping of quantitative trait loci. Genetics. 1994;136:1457–1468. doi: 10.1093/genetics/136.4.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Michaels SD, Amasino RM. FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell. 1999;11:949–956. doi: 10.1105/tpc.11.5.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee I, Michaels SD, Masshardt AS, Amasino RM. The late-flowering phenotype of FRIGIDA and mutations in LUMINIDEPENDENS is suppressed in the Landsberg erecta strain of Arabidopsis. Plant J. 1994;6:903–909. [Google Scholar]
- 28.Koornneef M, Blankestijndevries H, Hanhart C, Soppe W, Peeters T. The phenotype of some late-flowering mutants is enhanced by a locus on chromosome 5 that is not effective in the Landsberg erecta wild-type. Plant J. 1994;6:911–919. [Google Scholar]
- 29.Fornara F, et al. Arabidopsis DOF transcription factors act redundantly to reduce CONSTANS expression and are essential for a photoperiodic flowering response. Dev Cell. 2009;17:75–86. doi: 10.1016/j.devcel.2009.06.015. [DOI] [PubMed] [Google Scholar]
- 30.Salomé PA, et al. Genetic architecture of flowering-time variation in Arabidopsis thaliana. Genetics. 2011;188:421–433. doi: 10.1534/genetics.111.126607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koini MA, et al. High temperature-mediated adaptations in plant architecture require the bHLH transcription factor PIF4. Curr Biol. 2009;19:408–413. doi: 10.1016/j.cub.2009.01.046. [DOI] [PubMed] [Google Scholar]
- 32.Lucyshyn D, Wigge PA. Plant development: PIF4 integrates diverse environmental signals. Curr Biol. 2009;19:R265–R266. doi: 10.1016/j.cub.2009.01.051. [DOI] [PubMed] [Google Scholar]
- 33.Hornitschek P, Lorrain S, Zoete V, Michielin O, Fankhauser C. Inhibition of the shade avoidance response by formation of non-DNA binding bHLH heterodimers. EMBO J. 2009;28:3893–3902. doi: 10.1038/emboj.2009.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brock MT, Maloof JN, Weinig C. Genes underlying quantitative variation in ecologically important traits: PIF4 (phytochrome interacting factor 4) is associated with variation in internode length, flowering time, and fruit set in Arabidopsis thaliana. Mol Ecol. 2010;19:1187–1199. doi: 10.1111/j.1365-294X.2010.04538.x. [DOI] [PubMed] [Google Scholar]
- 35.Koornneef M, Alonso-Blanco C, Vreugdenhil D. Naturally occurring genetic variation in Arabidopsis thaliana. Annu Rev Plant Biol. 2004;55:141–172. doi: 10.1146/annurev.arplant.55.031903.141605. [DOI] [PubMed] [Google Scholar]
- 36.Knapp SJ, Bridges WC. Using molecular markers to estimate quantitative trait locus parameters: Power and genetic variances for unreplicated and replicated progeny. Genetics. 1990;126:769–777. doi: 10.1093/genetics/126.3.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Esch E, Szymaniak JM, Yates H, Pawlowski WP, Buckler ES. Using crossover breakpoints in recombinant inbred lines to identify quantitative trait loci controlling the global recombination frequency. Genetics. 2007;177:1851–1858. doi: 10.1534/genetics.107.080622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haldane JBS, Waddington CH. Inbreeding and linkage. Genetics. 1931;16:357–374. doi: 10.1093/genetics/16.4.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu SC, Kowalski SP, Lan TH, Feldmann KA, Paterson AH. Genome-wide high-resolution mapping by recurrent intermating using Arabidopsis thaliana as a model. Genetics. 1996;142:247–258. doi: 10.1093/genetics/142.1.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Winkler CR, Jensen NM, Cooper M, Podlich DW, Smith OS. On the determination of recombination rates in intermated recombinant inbred populations. Genetics. 2003;164:741–745. doi: 10.1093/genetics/164.2.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bernardo R. Should maize doubled haploids be induced among F(1) or F(2) plants? Theor Appl Genet. 2009;119:255–262. doi: 10.1007/s00122-009-1034-1. [DOI] [PubMed] [Google Scholar]
- 42.Darvasi A, Soller M. Advanced intercross lines, an experimental population for fine genetic mapping. Genetics. 1995;141:1199–1207. doi: 10.1093/genetics/141.3.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Forster BP, Heberle-Bors E, Kasha KJ, Touraev A. The resurgence of haploids in higher plants. Trends Plant Sci. 2007;12:368–375. doi: 10.1016/j.tplants.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 44.Abràmoff MD, Magalhães PJ, Ram SJ. Image processing with ImageJ. Biophotonics. 2004;11:36–42. [Google Scholar]
- 45.Broman KW, Wu H, Sen S, Churchill GA. R/qtl: QTL mapping in experimental crosses. Bioinformatics. 2003;19:889–890. doi: 10.1093/bioinformatics/btg112. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.