Abstract
The SecA ATPase associates with the SecY complex to push preproteins across the bacterial membrane. Because a single SecY is sufficient to create the conducting channel, the function of SecY oligomerization remains unclear. Here, we have analyzed the translocation reaction using nanodiscs. We show that one SecY copy is sufficient to bind SecA and the preprotein, but only the SecY dimer together with acidic lipids supports the activation of the SecA translocation ATPase. In discs, the dimer is predominantly arranged in a back-to-back manner and remains active even if a constituent SecY copy is defective for SecA binding. In membrane vesicles and in intact cells, the coproduction of two inactive SecYs, one for channel gating and the other for SecA binding, recreates a functional translocation unit. These results indisputably argue that the SecY dimer is crucial for the activation of SecA, which is necessary for preprotein transport.
Keywords: SecYEG channel, membrane transporters, nanolipoparticles
The Sec channel is a macromolecular assembly essential to catalyze transport of preproteins carrying an N-terminal signal sequence. In bacteria, the core of the complex is a conserved multispanning membrane protein (SecY) associated with two smaller ones (SecE and SecG) (for review see refs. 1 and 2). Depending on the hydrophobicity of the signal sequence, protein translocation is driven by the ribosome during polypeptide elongation or by SecA, which is an ATPase that pushes preproteins in a stepwise manner across the membrane. Thermodynamic analysis has shown that docking of the signal sequence to the channel lowers the activation energy barrier of protein transport (3). This docking event stimulates the SecA ATPase, which, under optimal conditions, leads to the net movement of 20–30 amino acids of the preprotein across the membrane (4). This SecA ATPase activity has been defined as the SecA translocation ATPase (5) because it is stimulated approximately six- to ninefold in the presence of a preprotein substrate (3, 6). The SecA translocation ATPase also depends on acidic lipids in the membrane, such as phosphatidylglycerol (PG) (7, 8).
The atomic structure of the SecY complex, alone or associated with SecA, has revealed the location of the protein-conducting channel at the center of the SecY subunit (9, 10). This finding has been confirmed by thiol-cross-linking (11, 12) and by cryoelectron microscopy analysis of the eukaryotic Sec complex bound to a translating ribosome (13). It is thus well-established that a single SecY copy is sufficient to form the translocation pathway, yet it remains mysterious that the channel exists as oligomers (14–17).
An earlier study employing a covalently linked SecYEG dimer showed that a preprotein can be transported across a defective channel provided a functional SecY copy is fused to it (18). It was proposed that each copy has a different role, one serving as a docking site for SecA and the other as a translocation channel [a model termed fraternal twins (18, 19)]. In possible support to the model, a photo-cross-linking analysis in intact cells showed that SecA can simultaneously contact two SecYs (20). More recently, a single molecule analysis in proteoliposomes indicated that a single SecY was sufficient to bind the preprotein, although the dimer was necessary to support significant transport (21). These earlier studies have indicated the importance of the dimer, but the exact role of each copy requires additional support. Protein translocation taking place at one SecY copy might be facilitated by the second, or translocation may strictly depend on the dimer. The question is further complicated by the dynamic dimeric state of SecA and whether one or two SecA molecules bind to the channel (22–25).
To help understand this complex dynamic quaternary structure, we have reconstituted SecY in supported nanoscale lipid bilayers termed nanodiscs. We show that the SecY monomer is sufficient to bind SecA and the signal sequence, yet the activation of SecA occurs only when a second SecY and acidic lipids are present in the disc. The two copies are predominantly arranged in a back-to-back manner and create a binding site for one SecA only. Consistent with the fraternal twin model, the SecY dimer can activate the SecA ATPase provided SecA can bind to the assembly. To confirm the involvement of the dimer in the cell context, we combined a mutant defective for channel opening and a mutant defective for SecA binding. When coproduced together, these otherwise inactive SecY channels created a functional assembly. These results strongly argue that two SecY copies are necessary for preprotein transport.
Results
Capture of the SecY Dimer in Nanodiscs and Binding of SecA.
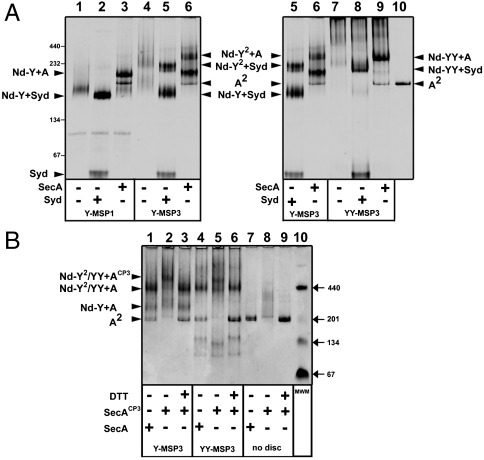
We previously described the incorporation of the SecY monomer into discs using the membrane scaffold protein 1 (MSP1, referred to as Nd-Y) (23). We also reported that SecA and Syd formed a tight complex with the Nd-Y particles (23, 26) (Fig. 1A). Syd is a small protein that binds to the SecY cytosolic loops during assembly of the channel (27). Here, we found that Syd greatly facilitated the electrophoretic mobility of the discs, which helped the analysis of the bands on native gel (Fig. 1A, compare lane 1 to lane 2). Because the SecY channel form oligomers in detergent solution (14), the reconstitution was carried out with the longer scaffold protein, MSP3, which extends the diameter of the disc from approximately 9.7 to approximately 12.1 nm (28). In that case, two particle populations were obtained (Fig. 1A, lane 4) and each was able to associate with SecA and Syd (lanes 5 and 6). To show that the high molecular mass discs contained two SecY complexes (termed Nd-Y2), the reconstitution was performed using a covalently linked SecY dimer (termed Nd-YY; Fig. 1A, lanes 7–9), made from two secY genes that were fused together (29). To determine the stoichiometry of the SecY dimer with SecA, SecA was stabilized as a dimer with a pair of intermolecular disulphide bridges (termed SecACP3; 23). This linked SecA dimer formed a complex with Nd-Y2 (Fig. 1B, lane 2) and Nd-YY (Fig. 1B, lane 5), but compared to native SecA, each assembly had a higher molecular mass (Fig. 1B, compare lanes 1–3 and lanes 4–6). Thus, one SecY forms a binding site for one SecA, and two SecY still form a binding site for one SecA.
Fig. 1.
The SecY dimer in nanodiscs binds one SecA. (A) The SecY monomer (Y), the dimer (Y2), and the fused dimeric version (YY) were reconstituted in nanodiscs (Nd) with the indicated MSP. The nanodiscs (3 μg each) containing one SecY (labeled Nd-Y), two SecY (labeled Nd-Y2), or the fused dimer (labeled Nd-YY) were incubated with SecA (labeled SecA2) and Syd (2 μg each) followed by native-PAGE and Coomassie blue staining. Syd is a small SecY binding protein that verifies the proper assembly of the SecY complex in some Gram-negative bacteria (45). The analysis was on the same gel but to facilitate figure labeling and to compare the relative migration, lanes 5 and 6 on Left are duplicated on Right. Expected molecular masses: Nd-Y, 124 kDa; Nd-Y2 or Nd-YY, 216 kDa; Syd, 23 kDa. (B) The nonfused or fused SecY dimer (3 μg of Nd-Y2 and Nd-YY, respectively) was incubated with the disulfide-linked SecA dimer (SecACP3, 4 μg). In lane 3 and 6, the sample was incubated with 1 mM DTT (37 °C for 2 min) before loading on the gel. Native SecA is dimeric in solution (∼204 kDa) and migrates next to the 201 kDa marker (lanes 7 and 9). On its own, the cross-linked SecA dimer migrates as a smear (lane 8). Expected molecular masses (not including lipids): Nd-Y2/YY + SecA, 318 kDa and Nd-Y2/YY + SecACP3, 418 kDa.
The SecY Dimer Supports the SecA Translocation ATPase.
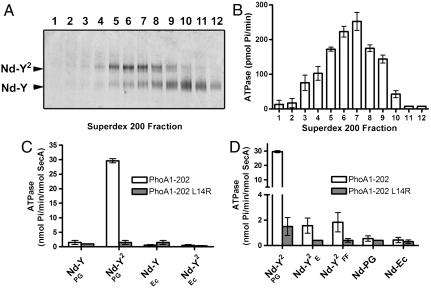
The SecY dimer was assembled in nanodiscs in the presence of PG lipids and purified by gel-filtration chromatography. The fractions were analyzed by native-PAGE (Fig. 2A, Fig. S1A for chromatogram) and equal protein amounts of each fraction were incubated with SecA, ATP, and the preprotein substrate PhoA1-202 (alkaline phosphatase residues 1-202; Fig. 2B). A significant ATPase stimulation occurred with fractions 5–9, which were those enriched for the SecY dimer. Dynamic light scattering analysis indicated that more than 99.8% of the total mass of fraction 7 consisted of monodisperse particles with a diameter of approximately 15 nm (Fig. S1B). In contrast, little ATPase activity was detected with fractions enriched for the SecY monomer (fraction 10–12), indicating that a single SecY is insufficient to support the preprotein-dependent SecA translocation ATPase. Quantification of the ATPase rate showed that the SecY dimer stimulated SecA approximately 20-fold more than its monomeric counterpart (Fig. 2C). Control experiments showed that the SecA ATPase activity was not triggered when the preprotein had a defective signal sequence (PhoA1-202-L14R; Fig. 2C), nor when the SecYEG complex was reconstituted with a crude E. coli lipids extract, which contain high amount of neutral lipids (∼70% neutral; Fig. 2C and Discussion). Moreover, the SecY dimer carrying the mutation R357E in the large SecY cytosolic loop (referred to as 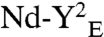 ), or the mutations I82F/I187F in the SecY pore ring (referred to as
), or the mutations I82F/I187F in the SecY pore ring (referred to as 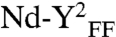 ), failed to support the SecA translocation ATPase (Fig. 2D). These mutations, described below, affect SecY activity in vivo (Fig. S2A) and in membrane vesicles (Fig. S2B). Together, the results showed without ambiguity that activation of the SecA translocation ATPase depends on the SecY dimer, a functional signal sequence and acidic lipids. The same dependencies define the SecA translocation ATPase in the membrane (7).
), failed to support the SecA translocation ATPase (Fig. 2D). These mutations, described below, affect SecY activity in vivo (Fig. S2A) and in membrane vesicles (Fig. S2B). Together, the results showed without ambiguity that activation of the SecA translocation ATPase depends on the SecY dimer, a functional signal sequence and acidic lipids. The same dependencies define the SecA translocation ATPase in the membrane (7).
Fig. 2.
Two SecY copies are necessary to activate the SecA translocation ATPase. (A) The SecY complex reconstituted in nanodiscs with MSP3 and PG lipids was purified by gel-filtration chromatography (Fig. S1). Fractions were supplemented with Syd (1 μg) to facilitate analysis by native-PAGE. (B) The same fractions were incubated with SecA (0.2 μM), PhoA1-202 (0.8 μM), and ATP (1 mM) for 30 min at 37 °C. The release of inorganic phosphate was determined by colorimetric assay. Error bars were derived from three independent measurements. (C) Nanodiscs were prepared with the indicated lipids (PG, dioleoylphosphatidylglycerol; Ec, E. coli total lipid extract). Monomeric and dimeric populations were separated by gel-filtration chromatography as in A. The SecA translocation ATPase was determined after plotting the initial ATPase activity rates (Fig. S6 for kinetics) obtained in the presence of nanodiscs and PhoA1-202 or PhoA1-202-L14R (0.8 μM). (D) The SecY complex carrying the mutation R357E (termed YE) or I82F/I187F (termed YFF) was reconstituted into discs. The population of the discs containing the dimer (labeled 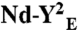 and
and 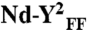 , respectively) was isolated by size-exclusion chromatography and tested for translocation ATPase as in C. Nanodiscs containing only PG lipids (labeled Nd-PG) or E. coli total lipids (labeled Nd-Ec) do not support the SecA translocation ATPase activity.
, respectively) was isolated by size-exclusion chromatography and tested for translocation ATPase as in C. Nanodiscs containing only PG lipids (labeled Nd-PG) or E. coli total lipids (labeled Nd-Ec) do not support the SecA translocation ATPase activity.
The SecA Translocation ATPase Depends on Two SecYs but only One Copy Needs to Bind SecA.
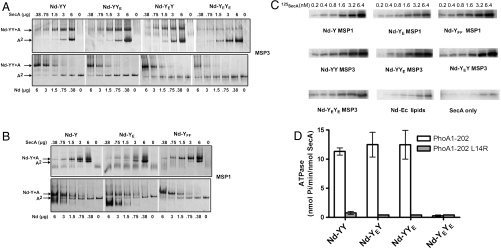
To understand the role of oligomerization, the mutation R357E was introduced on one or the other copy of the covalently linked SecY dimer (termed YYE, YEY, and YEYE, respectively). The R357E mutation strongly reduced the binding of SecA to the channel (23) and caused a severe translocation defect (30). Here, the association of SecA to the mutant channels reconstituted in discs was monitored by native-PAGE (Fig. 3 A and B) and affinity pull-down (Fig. 3C). When present on both copies, the R357E mutation reduced the binding of SecA approximately two- to threefold (Fig. S3) and abolished the SecA translocation ATPase (Fig. 3D). In contrast, the binding of SecA (Fig. 3A) and the translocation ATPase activity (Fig. 3D) were unaffected when the heterodimer contained a wild-type SecY copy (Nd-YY, Nd-YYE, or Nd-YEY). The control experiment showed that the SecYE monomer on its own had a weak affinity for SecA compared to the wild-type complex (Fig. 3B) (23). Together, these results confirmed that the SecA translocation ATPase depends on two SecY copies, although only one is needed for the binding of SecA.
Fig. 3.
The dimer remains active when an SecY copy is defective for SecA binding. (A) A fixed amount of nanodisc containing the indicated fused SecY dimer (3 μg; MSP3 and E. coli lipids) was incubated with an increasing amount of SecA. Alternatively, a fixed amount of SecA (2 μg) was incubated with an increasing amount of discs. Complexes were analyzed by native-PAGE and Coomassie blue staining. Discs containing the fused SecY dimer with the mutation R357E on the first or the second copy are labeled Nd-YEY and Nd-YYE, respectively. (B) As in A, but using the indicated monomeric SecY complex reconstituted with MSP1 and E. coli lipids. (C) The indicated nanodiscs (10 μg each, same series as in Fig. 3 A and B) were incubated with 125I-labeled SecA. The complex was isolated via Ni2+-NTA affinity pull-down (500 μL reaction in TSG buffer, 50 mM Tris·HCl pH 7.9, 100 mM NaCl, 5% glycerol). Nanodiscs and bound SecA were eluted in TSG buffer containing 500 mM imidazole, followed by SDS-PAGE and autoradiography. Nd-Ec lipids refers to nanodiscs made with E. coli total lipid extract. (D) Nanodiscs containing the fused SecY dimer were purified by sucrose density ultracentrifugation (see Fig. S7 for analysis). The kinetics of the measured translocation ATPase are presented in Fig. S6.
The SecY Monomer Suffices to Bind the Signal Sequence.
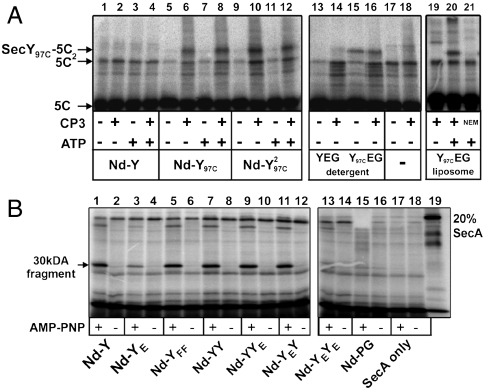
We next tested if the SecY oligomeric state is important for preprotein binding. Previous cross-linking analysis showed that the signal sequence binds near the amino-acyl position 97, close the SecY channel pore (18) (Fig. S4A). To test whether one or two SecY copies were necessary for binding the signal sequence, PhoA1-202 carrying a cysteine residue at position 5 (PhoA1-202-5C) was incubated with SecY97C reconstituted in nanodiscs. Under oxidizing conditions, PhoA1-202-5C formed a disulphide bond with SecY97C, whether SecY was monomeric or dimeric in the disc (Fig. 4A, lanes 5–12). The cross-link was specific because it did not occur with wild-type SecY, which contains two cysteine residues located away from the signal sequence binding site (Fig. 4A, lanes 1–4). However, unlike SecY97C in proteoliposomes, the interaction between the signal sequence and SecY97C in the disc or in detergent did not depend on SecA and ATP (Fig. 4A, lanes 19–21). The position 97C in SecY therefore seems less accessible to the signal sequence when the channel is embedded in the lipid bilayer. To further assess the conformation of the SecY channel in the disc, we tested the capacity to support SecA membrane insertion. Upon binding of a nonhydrolyzable ATP analog, a 30 kDa domain of SecA becomes protease-resistant (31). Formation of this 30 kDa domain was supported by the nanodisc when at least one SecY was able to bind SecA (Fig. 4B). Thus, although the dimer is necessary to activate SecA, the SecY monomer was sufficient to bind the signal sequence and to support SecA insertion.
Fig. 4.
The SecY monomer suffices to bind the signal sequence. (A) The 125I-labeled PhoA1-202 (∼25,000 c.p.m., 200 ng) bearing a unique cysteine residue at position 5 of the signal sequence (labeled 5C) was incubated with the indicated nanodiscs (3 μg each) in TL buffer for 5 min at 37 °C in the presence of SecA (0.1 μM) and ATP (1 mM). The oxidation of the cysteines was started with CP3 (0.2 mM for 5 min at room temperature) and terminated with NEM (10 mM). The cross-link products were analyzed by 12% SDS-PAGE and autoradiography. The same cysteine cross-linking experiment was performed with the purified SecY97CEG complex in detergent solution (3 μg in TL buffer +0.03% dodecyl maltoside) or reconstituted in liposomes (3 μg at a protein∶lipid molar ratio of 1∶2,000). The cysteine cross-link between SecY97C and PhoA1-202-5C is labeled SecY97C-5C. A fraction of PhoA1-202-5C forms cysteine linked dimers (labeled 5C2). (B) The 125I-labeled SecA (∼50,000 c.p.m., 0.02 μM) was incubated with the indicated nanodiscs (3 μg each), PhoA1-202 (0.8 μM), and AMPPNP (1 mM) for 10 min at 37 °C. Trypsinolysis (0.08 μg/mL trypsin) was for 15 min on ice. Samples were precipitated with trichloroacetic acid (17% final) and analyzed by SDS-PAGE and autoradiography. Nanodiscs bearing the monomeric SecY complexes (Nd-Y, Nd-YE, and Nd-YFF) were reconstituted with MSP1 and PG lipid. Nanodiscs bearing the covalently linked SecY dimer were prepared as in Fig. S7.
The Back-to-Back Dimer is the Predominant Formation in Discs.
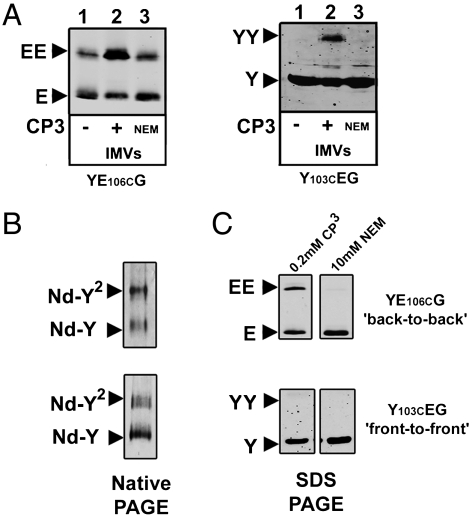
Two distinct dimeric arrangements have been observed in the lipid bilayer and in detergent solution: the front-to-front where the SecY lateral gates are facing each other and the back-to-back with the SecE subunits at the interface (20, 21, 32–34). The nanodiscs allowed to probe the orientation of the SecY dimer in a restricted environment. To differentiate the two conformations (Fig. S4B for representation), a cysteine residue was introduced at position 106 on SecE (YE106CG) and 103 on SecY (Y103CEG). Upon oxidation, SecE-SecE and SecY-SecY disulfide linkages were detected in the cell membrane (Fig. 5A) suggesting that both dimeric arrangements exist, although artifacts related to overproduction are possible. In detergent, SecE-SecE cross-links were detected but this linkage appeared only upon dissociation of the SecYEG complex by temperature or SDS (Fig. S5). In nanodiscs (Fig. 5B) SecE-SecE cross-links were detected, but in the same conditions SecY-SecY cross-links were not (Fig. 5C). For some reason, the SecY103C mutant formed less dimers in nanodiscs than the SecE106C version, but the amount of material analyzed by Western blotting (∼3 μg) should have been sufficient to detect even trace amounts of SecY-SecY cross-links.
Fig. 5.
Orientation of the SecY copies within the disc. (A) Inner membrane vesicles (2 μg) enriched for the SecY complex carrying the mutation SecE-L106C or SecY-A103C were incubated for 5 min at room temperature followed by addition of 0.2 mM CP3 (2 min) then NEM (10 mM). Samples were analyzed by 12% SDS-PAGE and Western blotting using a polyclonal antibody against SecE, or monoclonal antibody against SecY. (B) The indicated SecY complexes were purified and reconstituted in nanodiscs (using MSP3 and PG lipids). A sample was analyzed by native-PAGE and Coomassie blue staining. (C) The same samples were oxidized with 0.2 mM CP3 (2 min at 37 °C) followed by NEM treatment (10 mM), 12% SDS-PAGE and immuno-staining with anti-SecE or anti-SecY antibodies. Cysteine cross-linking is not detected when the samples are treated with NEM before addition of CP3.
Complementation Between Two SecY Mutants Restores Protein Translocation.
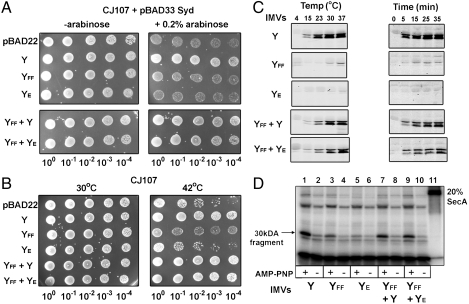
To show the importance of the SecY dimer for the cell physiology, the SecYFF and SecYE mutants were coexpressed in the same cell. The SecYFF mutant had an altered pore ring structure and was defective for channel gating (Fig. S2B) (35), but otherwise interacted with SecA as strongly as the wild-type (Fig. 3B, Fig. 4B). The SecYE mutant was defective for SecA binding (Fig. 3B), but otherwise contained an unaltered translocation pore. Because the molecular basis for the translocation defect of each mutant was different, we could test their ability to complement each other. Neither SecYFF nor SecYE on their own was able to restore the growth of a thermo-sensitive SecY strain (Fig. 6 A and B). In contrast, the coproduction of the mutants restored cell viability in two different backgrounds and temperatures (Fig. 6 A and B). Similarly, the SecYFF and SecYE mutants supported in vitro protein translocation and SecA membrane insertion, but only when both mutants were simultaneously expressed in the membrane (Fig. 6 C and D). Therefore, the two inactive SecY complexes were able to associate together to form a functional channel. In a possible scenario, the SecYFF copy would recruit SecA whereas the neighboring SecYE channel would provide the translocation pathway for the preprotein.
Fig. 6.
Transcomplementation between SecYFF and SecYE mutant channels. (A) Cell growth complementation assays in E. coli CJ107 transformed with the indicated plasmid and pBAD33-Syd were performed at 37 °C as described in Fig. S2A. Cellular growth is observed when the YFF and YE mutant channels (labeled YFF + YE) are coexpressed together in the same strain. (B) The experiment as in A was performed in CJ107 cells (without Syd) incubated at 30 °C and 42 °C. The thermo-sensitive SecY24 complex is compromised at 42 °C. (C) In vitro translocation assay using membranes vesicles enriched with comparable amounts of the indicated SecY complexes (see Fig. S8 for Western blotting). Each assay was performed using 16 ng of fluorescent dye-labeled PhoA1-202 mixed with 100 ng of unlabeled PhoA1-202 (total PhoA1-202 concentration 0.09 μM). Translocation activity was determined at different temperatures (Left) or at 30 °C at different time points (Right). (D) The SecA membrane insertion assay was performed as in Fig. 4B, using 3 μg of membrane vesicles containing the indicated SecY complex.
Discussion
The difficulty of understanding the oligomerization of the SecY complex has led to uncertainty regarding the functional state of the channel. Here, the nanodisc allowed to isolate the SecY dimer, which, unlike the monomer, was able to support the SecA translocation ATPase. The importance of SecY dimerization was also observed in membrane vesicles and in vivo because the coproduction of two inactive SecY subunits, each for a different reason, recreated a functional unit. Together, these results would strongly argue that the SecY dimer is crucial for the activation of SecA and subsequent preprotein transport. Yet, a recent analysis concluded that a single SecY channel suffices to support SecA-driven protein translocation (36). In that study, the SecYEG complex was incorporated into giant liposomes at extremely dilute protein concentration. Using single molecule fluorescence spectroscopy, it was found that the SecY monomer supported SecA binding and formation of a preprotein translocation intermediate. Remarkably, the preprotein was not detected with the SecY dimer. In contrast, in another single molecule analysis at even lower protein to lipid ratio, the monomer was found insufficient to support protein translocation (21). The opposed conclusions reached in these earlier liposome assays may simply highlight the difficulty of controlling or measuring the SecY oligomeric state in the fluidic lipid environment.
In nanodiscs, the monomer was sufficient to bind SecA and the preprotein signal sequence (Fig. 3B, Fig. 4A). These observations were consistent with earlier disulphide cross-links and confocal microscopy analysis that probed the interaction of the preprotein with the channel (11, 21), and also with the crystal structure of SecA bound to the SecY complex (10). The results in nanodiscs also showed that only the dimer was able support the preprotein-dependent SecA ATPase. Why and how this dimeric assembly is necessary to activate SecA will require further investigation. The mechanism of the SecA translocation ATPase itself is still unclear. Arrhenius plots have indicated that the docking of the signal sequence onto the channel lowers the SecA activation energy barrier, a process termed triggering (3). In the membrane, this step would be normally followed by the irreversible engagement of the substrate with the channel and by cycles of ATP hydrolysis coupled to protein transport. The later step was apparently not reproduced in the nanodisc, perhaps as a result of the low kcat supported by the system (29.6 min-1, Fig. 2C), compared to membrane vesicles and proteoliposomes (70 min-1 and 456 min-1, respectively) (3, 37). The number of lipid captured inside the disc (< 40–50 lipids per leaflet given size constraints) may also be insufficient for the signal sequence to interact productively with the channel. This limitation might explain why the preprotein-dependent SecA translocation ATPase was stimulated only two–threefold, compared to the six–ninefold in the membrane (3). In addition, the SecY conformation may be affected by the membrane lateral pressure perhaps absent in the disc. This other limitation may explain why the binding of the signal sequence was not dependent on ATP (Fig. 4A) and why the SecY pore mutant could still facilitate SecA insertion (Fig. 4B). Nevertheless, the fact that the SecA ATPase activity was dependent on a correct signal sequence is strong evidence that the triggering step of the reaction has been recreated in the disc.
The results with nanodiscs also showed that acidic lipids contribute directly to the SecA translocation ATPase activity. In the membrane, these lipids are essential for the binding of SecA (6, 7) and for the so-called SecA lipid ATPase, which occur only in liposomes, at low magnesium concentration (micromolar) and in the absence of a preprotein (7). We reported earlier that the SecY monomer in discs with acidic lipids could stimulate an ATPase rate up to approximately 80 min-1 (23). However, in the presence of physiological amount of magnesium (∼1 mM), the ATPase rate supported by the SecY monomer was reduced to less than 2 min-1 (Fig. 2C, Nd-Y/PG). Clearly, acidic lipids facilitate preprotein-dependent SecA activation but only in the context of the SecY dimer. How these lipids and magnesium lower the SecA activation energy barrier also need to be determined. Acidic lipids may have an allosteric effect on SecA, increase the strength of the interaction with SecY (16, 23), or favor the monomerization of the SecA dimer (38). Together, these effects might directly contribute to the triggering step of the translocation reaction.
The organization of the SecY copies in the functional dimer has been controversial. Our results show that both front-to-front and back-to-back conformations exist in the membrane but most likely as a result of protein overproduction. Furthermore, the SecE subunit self-dimerizes when unbound to SecY (Fig. S5) (39), which complicates earlier cysteine cross-link analysis performed on membrane vesicles. In nanodiscs, the majority of the SecY complex was arranged in a back-to-back manner. Because the formation of the disc is a self-assembly process, the back-to-back orientation may be an energetically favorable state preferentially selected during the reconstitution. These results are compatible with previous experiments showing that a disulphide stabilized back-to-back dimer is active in liposomes (21). These results do not exclude, however, that other functional arrangements exist. In fact, the exact orientation of the monomers may not be critical as long as the dimeric assembly satisfies signal sequence binding and SecA activation.
Our knowledge on the SecY channel is considerable, yet why the channel dimerizes and the functional advantage, if any, is still not understood. Dimerization might increase the affinity of the channel for its binding partners (18), recruit acidic lipids essential to activate SecA (16, 21), or facilitate gating through some sort of allosteric communications between monomers (40, 41). Previous site-directed cysteine cross-linking experiments revealed that each SecY copy is separately engaged with SecA and the preprotein during translocation (18). Our results provide additional support for this functional asymmetry because the functional complementation between two SecY mutants, each with different translocation defects, was possible. The details regarding the contribution of each copy remain to be determined, but that a single gene contribute to two different functions, receptor and channel, is perhaps an advantage or an evolutionary adaptation to SecA, which is unique to bacteria.
Materials and Methods
Biological Reagents.
pBAD22-based plasmids encoding for His-tagged SecYEG and covalently linked SecYEG dimers were previously described (29, 42). The cloning and expression of the two SecY complexes but from the same plasmid is described in SI Materials and Methods. The SecYEG complexes were expressed in Escherichia coli BL21 (DE3) and purified by Ni-nitrilotriacetate (NTA) and cation exchange chromatography (26). The reconstitution of the SecY complex in nanodisc is described in SI Materials and Methods. Membrane scaffold proteins (MSP) employed were prepared as described (23, 43): MSP1D1 (referred to as MSP1; 24.6 kDa), MSP1E3D1 (referred to as MSP3; 32.6 kDa), and MSP2N2 (45.5 kDa). The first 202 amino acids of the alkaline phosphatase A (PhoA1-202) were expressed from plasmid pET-23 and purified from inclusion bodies using Ni-NTA chromatography under denaturing conditions (50 mM Tris·HCl pH 7.9, 6 M Urea, 1 mM DTT). The 125I-labeling of SecA was performed with iodogen-coated tubes (Pierce) as previously described (29). DOPG lipids (dioleoylphosphatidylglycerol) and E. coli total lipids were purchased from Avanti Polar Lipids. Cu2+-phenanthroline3 (CP3) and N-ethylmaleimide (NEM) were from Sigma.
Translocation ATPase Measurements.
The purified nanodisc preparations (0–155 nM) were incubated with SecA (0.2 μM) and ATP (1 mM) in TL buffer (50 mM Tris·HCl pH 7.9, 50 mM KCl, 50 mM NaCl, 5 mM MgCl2, 1 mM DTT), and 0.8 μM PhoA1-202 or PhoA1-202-L14R. The release of inorganic phosphate was measured by colorimetric method (Malachite Green) as previously described (23). ATPase rates in the presence or absence of preprotein substrate were measured in intervals over a range of 30 min at each nanodisc concentration (Fig. S6, Fig. S7), followed by fitting the initial rates to a one site quadratic binding equation to determine kcat values as previously described (37). To determine the amount of preprotein-dependent ATPase, the rate of inorganic phosphate release observed in the absence of the substrate was subtracted in each experiment
Other Methods.
Protein concentration was determined using the Bradford reagent (BioRad). Colorless native gels (4–13% linear gradient) and electrophoresis conditions were performed as described (26). The molecular mass markers employed on native gel are, as follows: ferritin, 440 kDa; catalase, 232 kDa; BSA (trimer/dimer/monomer), 201/134/67 kDa. The covalent SecA dimer (SecACP3) was obtained following oxidation with CP3 and size-exclusion chromatography as previously described (23). Dynamic light scattering measurements on the nanodisc particles (0.1 μg/mL) were performed at 25 °C on a Wyatt DynaPro Nanostar equipped with a 661 nm laser beam. Affinity pull-down experiments were performed by binding the nanodiscs onto Ni-NTA beads (GenScript) via a 6-Histidine N-terminal tag on MSP1 and MSP3, followed by incubation with SecA as described in Fig. 3C. In vitro protein translocation experiments were carried out as previously described (44) using 3 μg inner membrane vesicles (IMVs) prepared from E. coli strain KM9 (unc-, see Figs. S2C and S8 for Western blotting) in a 50 μL reaction volume with 0.2 μM SecA, 0.2 mg/mL BSA, 1 mM ATP (10 min, 37 °C), and 125I or fluorescent-labeled PhoA1-202. Dye labeling of PhoA1-202 is described in SI Materials and Methods. In vivo complementation experiments were performed in E. coli conditional lethal strain CJ107 carrying the secY24 mutation (45).
Supplementary Material
Acknowledgments.
We thank J. Montariol for help in purifying the SecYEG complexes. K.D. was supported by the Alexander Bell Canada graduate scholarship. C.S.C was supported by a Postdoctoral Fellowship from the Natural Sciences and Engineering Research Council. F.D. is a Canada Research Chair Tier II. S.G.S. is supported by National Institutes of Health Grant GM33775. This work was supported by the Canadian Institutes of Health Research.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. D.B.O. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1117783109/-/DCSupplemental.
References
- 1.Dalal K, Duong F. The SecY complex: Conducting the orchestra of protein translocation. Trends Cell Biol. 2011;21:506–514. doi: 10.1016/j.tcb.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 2.du Plessis DJ, Nouwen N, Driessen AJ. The sec translocase. Biochim Biophys Acta. 2011;1808:851–865. doi: 10.1016/j.bbamem.2010.08.016. [DOI] [PubMed] [Google Scholar]
- 3.Gouridis G, Karamanou S, Gelis I, Kalodimos CG, Economou A. Signal peptides are allosteric activators of the protein translocase. Nature. 2009;462:363–367. doi: 10.1038/nature08559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schiebel E, Driessen AJ, Hartl FU, Wickner W. Delta mu H+ and ATP function at different steps of the catalytic cycle of preprotein translocase. Cell. 1991;64:927–939. doi: 10.1016/0092-8674(91)90317-r. [DOI] [PubMed] [Google Scholar]
- 5.Cunningham K, Wickner W. Specific recognition of the leader region of precursor proteins is required for the activation of translocation ATPase of Escherichia coli. Proc Natl Acad Sci USA. 1989;86:8630–8634. doi: 10.1073/pnas.86.22.8630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hendrick JP, Wickner W. SecA protein needs both acidic phospholipids and SecY/E protein for functional high-affinity binding to the Escherichia coli plasma membrane. J Biol Chem. 1991;266:24596–24600. [PubMed] [Google Scholar]
- 7.Lill R, Dowhan W, Wickner W. The ATPase activity of SecA is regulated by acidic phospholipids, SecY, and the leader and mature domains of precursor proteins. Cell. 1990;60:271–280. doi: 10.1016/0092-8674(90)90742-w. [DOI] [PubMed] [Google Scholar]
- 8.de Vrije T, de Swart RL, Dowhan W, Tommassen J, de Kruijff B. Phosphatidylglycerol is involved in protein translocation across Escherichia coli inner membranes. Nature. 1988;334:173–175. doi: 10.1038/334173a0. [DOI] [PubMed] [Google Scholar]
- 9.Van den Berg B, et al. X-ray structure of a protein-conducting channel. Nature. 2004;427:36–44. doi: 10.1038/nature02218. [DOI] [PubMed] [Google Scholar]
- 10.Zimmer J, Nam Y, Rapoport TA. Structure of a complex of the ATPase SecA and the protein-translocation channel. Nature. 2008;455:936–943. doi: 10.1038/nature07335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cannon KS, Or E, Clemons WM, Jr, Shibata Y, Rapoport TA. Disulfide bridge formation between SecY and a translocating polypeptide localizes the translocation pore to the center of SecY. J Cell Biol. 2005;169:219–225. doi: 10.1083/jcb.200412019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bauer BW, Rapoport TA. Mapping polypeptide interactions of the SecA ATPase during translocation. Proc Natl Acad Sci USA. 2009;106:20800–20805. doi: 10.1073/pnas.0910550106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Becker T, et al. Structure of monomeric yeast and mammalian Sec61 complexes interacting with the translating ribosome. Science. 2009;326:1369–1373. doi: 10.1126/science.1178535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bessonneau P, Besson V, Collinson I, Duong F. The SecYEG preprotein translocation channel is a conformationally dynamic and dimeric structure. EMBO J. 2002;21:995–1003. doi: 10.1093/emboj/21.5.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scheuring J, et al. The oligomeric distribution of SecYEG is altered by SecA and translocation ligands. J Mol Biol. 2005;354:258–271. doi: 10.1016/j.jmb.2005.09.058. [DOI] [PubMed] [Google Scholar]
- 16.Gold VA, et al. The action of cardiolipin on the bacterial translocon. Proc Natl Acad Sci USA. 2010;107:10044–10049. doi: 10.1073/pnas.0914680107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rusch SL, Kendall DA. Oligomeric states of the SecA and SecYEG core components of the bacterial sec translocon. Biochim Biophys Acta. 2007;1768:5–12. doi: 10.1016/j.bbamem.2006.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Osborne AR, Rapoport TA. Protein translocation is mediated by oligomers of the SecY complex with one SecY copy forming the channel. Cell. 2007;129:97–110. doi: 10.1016/j.cell.2007.02.036. [DOI] [PubMed] [Google Scholar]
- 19.Duong F. Cell biology: Fraternal twins. Nature. 2007;446:741–743. doi: 10.1038/446741a. [DOI] [PubMed] [Google Scholar]
- 20.Das S, Oliver DB. Mapping of the SecA{middle dot}SecY and SecA{middle dot}SecG interfaces by site-directed in vivo photocross-linking. J Biol Chem. 2011;286:12371–12380. doi: 10.1074/jbc.M110.182931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deville K, et al. The oligomeric state and arrangement of the active bacterial translocon. J Biol Chem. 2011;286:4659–4669. doi: 10.1074/jbc.M110.175638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Or E, Boyd D, Gon S, Beckwith J, Rapoport T. The bacterial ATPase SecA functions as a monomer in protein translocation. J Biol Chem. 2005;280:9097–9105. doi: 10.1074/jbc.M413947200. [DOI] [PubMed] [Google Scholar]
- 23.Alami M, Dalal K, Lelj-Garolla B, Sligar SG, Duong F. Nanodiscs unravel the interaction between the SecYEG channel and its cytosolic partner SecA. EMBO J. 2007;26:1995–2004. doi: 10.1038/sj.emboj.7601661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kusters I, et al. Quaternary structure of SecA in solution and bound to SecYEG probed at the single molecule level. Structure. 2011;19:430–439. doi: 10.1016/j.str.2010.12.016. [DOI] [PubMed] [Google Scholar]
- 25.Wowor AJ, Yu D, Kendall DA, Cole JL. Energetics of SecA dimerization. J Mol Biol. 2011;408:87–98. doi: 10.1016/j.jmb.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dalal K, Duong F. Reconstitution of the SecY translocon in nanodiscs. Methods Mol Biol. 2010;619:145–156. doi: 10.1007/978-1-60327-412-8_9. [DOI] [PubMed] [Google Scholar]
- 27.Dalal K, et al. Structure, binding, and activity of syd, a SecY-interacting protein. J Biol Chem. 2009;284:7897–7902. doi: 10.1074/jbc.M808305200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ritchie TK, et al. Chapter 11—reconstitution of membrane proteins in phospholipid bilayer nanodiscs. Methods Enzymol. 2009;464:211–231. doi: 10.1016/S0076-6879(09)64011-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Duong F. Binding, activation and dissociation of the dimeric SecA ATPase at the dimeric SecYEG translocase. EMBO J. 2003;22:4375–4384. doi: 10.1093/emboj/cdg418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mori H, Ito K. An essential amino acid residue in the protein translocation channel revealed by targeted random mutagenesis of SecY. Proc Natl Acad Sci USA. 2001;98:5128–5133. doi: 10.1073/pnas.081617398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Economou A, Wickner W. SecA promotes preprotein translocation by undergoing ATP-driven cycles of membrane insertion and deinsertion. Cell. 1994;78:835–843. doi: 10.1016/s0092-8674(94)90582-7. [DOI] [PubMed] [Google Scholar]
- 32.Kaufmann A, Manting EH, Veenendaal AK, Driessen AJ, van der Does C. Cysteine-directed cross-linking demonstrates that helix 3 of SecE is close to helix 2 of SecY and helix 3 of a neighboring SecE. Biochemistry. 1999;38:9115–9125. doi: 10.1021/bi990539d. [DOI] [PubMed] [Google Scholar]
- 33.Breyton C, Haase W, Rapoport TA, Kuhlbrandt W, Collinson I. Three-dimensional structure of the bacterial protein-translocation complex SecYEG. Nature. 2002;418:662–665. doi: 10.1038/nature00827. [DOI] [PubMed] [Google Scholar]
- 34.van der Sluis EO, Nouwen N, Driessen AJ. SecY-SecY and SecY-SecG contacts revealed by site-specific crosslinking. FEBS Lett. 2002;527:159–165. doi: 10.1016/s0014-5793(02)03202-7. [DOI] [PubMed] [Google Scholar]
- 35.Dalal K, Bao H, Duong F. Modulation of the SecY channel permeability by pore mutations and trivalent cations. Channels (Austin) 2010;4:83–86. doi: 10.4161/chan.4.2.10533. [DOI] [PubMed] [Google Scholar]
- 36.Kedrov A, Kusters I, Krasnikov VV, Driessen AJ. A single copy of SecYEG is sufficient for preprotein translocation. EMBO J. 2011;30:4387–4397. doi: 10.1038/emboj.2011.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robson A, Gold VA, Hodson S, Clarke AR, Collinson I. Energy transduction in protein transport and the ATP hydrolytic cycle of SecA. Proc Natl Acad Sci USA. 2009;106:5111–5116. doi: 10.1073/pnas.0809592106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Or E, Navon A, Rapoport T. Dissociation of the dimeric SecA ATPase during protein translocation across the bacterial membrane. EMBO J. 2002;21:4470–4479. doi: 10.1093/emboj/cdf471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matsuo E, Mori H, Ito K. Interfering mutations provide in vivo evidence that Escherichia coli SecE functions in multimeric states. Mol Genet Genomics. 2003;268:808–815. doi: 10.1007/s00438-003-0803-9. [DOI] [PubMed] [Google Scholar]
- 40.Bostina M, Mohsin B, Kuhlbrandt W, Collinson I. Atomic model of the E. coli membrane-bound protein translocation complex SecYEG. J Mol Biol. 2005;352:1035–1043. doi: 10.1016/j.jmb.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 41.Tam PC, Maillard AP, Chan KK, Duong F. Investigating the SecY plug movement at the SecYEG translocation channel. EMBO J. 2005;24:3380–3388. doi: 10.1038/sj.emboj.7600804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Collinson I, et al. Projection structure and oligomeric properties of a bacterial core protein translocase. EMBO J. 2001;20:2462–2471. doi: 10.1093/emboj/20.10.2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Denisov IG, Grinkova YV, Lazarides AA, Sligar SG. Directed self-assembly of monodisperse phospholipid bilayer nanodiscs with controlled size. J Am Chem Soc. 2004;126:3477–3487. doi: 10.1021/ja0393574. [DOI] [PubMed] [Google Scholar]
- 44.Dalal K, Duong F. The SecY complex forms a channel capable of ionic discrimination. EMBO Rep. 2009;10:762–768. doi: 10.1038/embor.2009.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shimoike T, et al. Product of a new gene, syd, functionally interacts with SecY when overproduced in Escherichia coli. J Biol Chem. 1995;270:5519–5526. doi: 10.1074/jbc.270.10.5519. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.