Abstract
The gene encoding the small subunit rRNA serves as a prominent tool for the phylogenetic analysis and classification of Bacteria and Archaea owing to its high degree of conservation and its fundamental function in living organisms. Here we show that the 16S rRNA genes of not-yet-cultivated large sulfur bacteria, among them the largest known bacterium Thiomargarita namibiensis, regularly contain numerous self-splicing introns of variable length. The 16S rRNA genes can thus be enlarged to up to 3.5 kb. Remarkably, introns have never been identified in bacterial 16S rRNA genes before, although they are the most frequently sequenced genes today. This may be caused in part by a bias during the PCR amplification step that discriminates against longer homologs, as we show experimentally. Such length heterogeneity of 16S rRNA genes has so far never been considered when constructing 16S rRNA-based clone libraries, even though an elongation of rRNA genes due to intervening sequences has been reported previously. The detection of elongated 16S rRNA genes has profound implications for common methods in molecular ecology and may cause systematic biases in several techniques. In this study, catalyzed reporter deposition–fluorescence in situ hybridization on both ribosomes and rRNA precursor molecules as well as in vitro splicing experiments were performed and confirmed self-splicing of the introns. Accordingly, the introns do not inhibit the formation of functional ribosomes.
Keywords: group I and II introns, LAGLIDADG homing endonuclease, length heterogeneity bias, microbial diversity, small ribosomal subunit
Studies on the microbial diversity in natural environments or enrichment cultures usually involve the analysis of the genes encoding the small subunit rRNA (16S rRNA in Bacteria and Archaea and 18S rRNA in Eukarya). This gene was selected as a phylogenetic marker because it was considered to be universally present, consistent in size and function, and not subject to horizontal transfer (reviewed in ref. 1). Current methods assessing the diversity of 16S rRNA genes, like the polymerase chain reaction (PCR) and the generation of 16S clone libraries, take advantage in particular of the constant size of the gene (2). In the past decades, these techniques have become fundamental tools in molecular ecology, and the 16S rRNA gene is the most frequently sequenced and analyzed gene today. The universality of this gene has never been questioned, even though in some studies unusual 16S rRNA genes, which contained, e.g., intervening sequences, were encountered (3–7).
In earlier studies, it was observed that clone libraries generated from environments visibly dominated by large sulfur bacteria frequently lacked the 16S rRNA gene sequences of these bacteria (8–13). Large sulfur bacteria occur at high densities predominantly in marine, organically rich coastal sediments (14–16), influencing the local sulfur (17), nitrogen (18), and phosphorous (19) cycle. The present study reports the frequent occurrence of long introns in the 16S rRNA genes of large sulfur bacteria, which explains their discrimination in PCR-based diversity assessments.
Results
Inserted Sequences in rRNA Genes.
By comparative sequence analysis, we identified insertions of variable lengths (290−942 nucleotides) at up to four distinct sites within the 16S rRNA genes of large sulfur bacteria (Fig. 1A). Corresponding to these insertion sites, the inserted elements were classified as S795, S1078, S1396, and S1495 (according to Escherichia coli position numbering). Inserted sequences at the same site generally shared high sequence identities not only when found within one species (92−100%, Table S1), but also when inserted into different species and genera (usually 85−100% identical, except for S1495 with 46.8%, Table S1). In contrast, the four different sequence types shared only low identities among each other (40% or less), and the four insertion sites in the 16S rRNA gene did not show any sequence homology. In addition, the distribution of the inserted sequences varied between the different phylogenetic groups within the Beggiatoaceae (Fig. 1B) and also between individuals in a given species (details in Fig. S1). S1396 was the most frequent sequence in the group of large sulfur bacteria. Its phylogeny within a species reflected the phylogeny derived from the 16S rRNA gene sequence, but differed compared to the 16S-based relationship among higher taxa (details in Fig. S2). In eukaryotes, introns were detected in mitochondrial, chloroplast, and nuclear rRNA genes (20). Among them, introns were located at identical (1078) and nearby (793, 1391, and 1502) positions (refs. 21, 22) as the elements detected here in bacterial 16S rRNA genes. This finding is in accordance with the assumption that certain structural features or particular sequences in the rRNA gene attract introns, thus creating insertion hot spots in Bacteria, Archaea, and Eukarya (21).
Fig. 1.
Introns in the 16S rRNA genes of the large sulfur bacteria. (A) Four introns were inserted in the positions 795, 1078, 1396, and 1495 (according to E. coli numbering) in the gene for the small (“S”) ribosomal subunit (16S rDNA). (B) Multifurcation tree based on nearly full-length 16S rRNA gene sequences of members of the family Beggiatoaceae showing the occurrence of introns in the different genera and species. To date introns have been located in 16S rRNA genes of the genera Thiomargarita, “Candidatus Marithioploca,” “Candidatus Thiopilula,” and “Candidatus Thiophysa.”
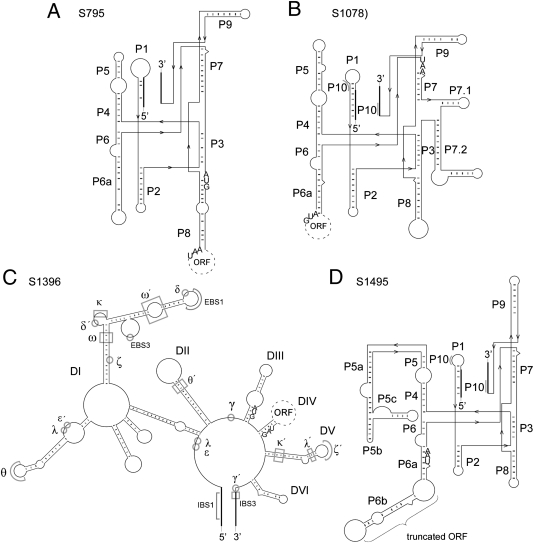
The inserted sequences showed several characteristics of known group I and II introns, which are frequently found in all three domains of life (20). Group I and II introns can be classified into subgroups based on specific RNA folding patterns (Fig. 2; details in Fig. S3). Intron S795 belonged to the group I introns of the ID subgroup (Fig. 2A); S1078 was likewise a group I intron, but of the subgroup IA3 (Fig. 2B); also belonging to the group I introns, S1495 was a IC1 intron (Fig. 2D) (23); and the only group II intron identified so far in large sulfur bacteria was S1396, which belonged to the subgroup IIC (Fig. 2C) (24–27). Furthermore, S1396 showed parallels to a functional type of group II introns described recently by Li et al. (25). Special features include the presence of several additional nucleotides inserted at the 5′-end and the possession of a gene for a LAGLIDADG homing endonuclease [which is more commonly found in group I introns (20)] instead of a reverse transcriptase (25), which were traits likewise found in S1396.
Fig. 2.
RNA folding structures of the four introns detected in the 16S rRNA genes of large sulfur bacteria. Thin lines represent intron regions and thick lines represent exon regions. If present, the start and stop codons of the ORFs are presented. In addition, details about intramolecular conserved structures are given. Folding criteria were extracted from ref. 23 for all group I introns and from refs. 24–27 for the group IIC intron (Fig. S3). (A) Intron S795; a group ID intron with ORF in domain P8 originating from clone sequence AMV001 [accession no. FR690921 (28)]. (B) Intron S1078; a group IA3 intron with OFR in domain P6 originating from clone sequence NAM032 [accession no. FR690922 (28)]. (C) Intron S1396; a group IIC intron with ORF in domain DIV originating from clone NAM056 [accession no. FR690945 (28)]. (D) Intron S1495; a group IC1 intron with truncated ORF in P6 (no start codon detectable) originating from clone sequence AMV001 [accession no. FR690921 (28)].
Most of the introns investigated here (96 of 131) contained an ORF coding for an intron encoded protein (IEP). Translated amino acid sequences had 35−42% identity to site-specific endonucleases of the LAGLIDADG superfamily (29) described from different mitochondria and chloroplast genomes (30–33). The group I intron IEPs are double-motif LAGLIDADG endonucleases, and the group II intron IEPs contain a single LAGLIDADG motif (Fig. S4) (29, 34). Generally, this family of homing endonucleases is encoded by group I introns (20), but can be occasionally found in group II introns (25, 35, 36). The remaining 35 introns either lacked an ORF or had a truncated ORF. Truncated ORFs were found in 2 of the 18 introns S1495 that were isolated, either lacking a start codon (clone AMV001) or containing only a reduced ORF that represented a sequence of 35 amino acids (clone NAM035), which did not reveal any hits in the public databases.
Ligation of Exons During in Vitro Splicing.
The capability of self-splicing and ligation was demonstrated for several bacterial introns (37–40) and was tested accordingly on the introns investigated here (Fig. 3 A and B). All four introns—S795 (clone sequence AMV001, with ORF), S1078 (clone sequence NAM032, with ORF), S1396 (clone sequence NAM056 with ORF), and S1495 (clone sequence AMV001, with truncated ORF)—showed self-splicing potential in transcription buffer during RNA synthesis (Fig. 3A). Ligation of the exons was demonstrated by RT-PCR (Fig. 3B), and sequencing confirmed ligation in all four cases.
Fig. 3.
Self-splicing experiments showed in vitro splicing abilities of RNA introns (A and B), and CARD-FISH hybridization of the 16S rRNA in the native ribosome demonstrated the absence of RNA introns from native ribosomes (C–F). (A) After transcription into RNA, the intron folds and splices itself, thereby ligating the two exons. The three bands in each row indicate the presence of the not-yet-spliced full-length construct “f,” the ligated exons “e,” and the excised intron “i.” (B) Bulk RNA was subject to RT-PCR with primers on both exon ends. The resulting PCR products were the full-length construct “f” and the ligated exons “e.” Bands of intermediate sizes in lane S795 result from incomplete splicing of the intron. (C) Probe VSO1284 was specific for the 16S rRNA of large sulfur bacteria and revealed strong signals for nearly all cells examined. (D) Hybridization of intron regions using a single probe (here S1396-179) never revealed detectable signals. (E) A probe bridging the intron insertion site (here Thm1389) resulted in strong fluorescent signals, whereas (F) a probe binding the exon1/intron boundary sequence (here Thm1397-INT) never showed any signal. These results strongly indicate that the introns are not present in the mature ribosome. (Scale bar, 100 μm.)
Hybridization of Native Ribosomes and Precursor rRNA.
Among individual cells of large sulfur bacteria, a heterogeneous distribution of introns was observed (Fig. S1), except for chain-forming Thiomargarita namibiensis, which always contained introns S1078 and S1396, and chain-forming “Candidatus Thiomargarita nelsonii,” containing either S1396 or no intron. Whole fixed cells of both of these chain-forming types were hybridized by catalyzed reporter deposition–fluorescence in situ hybridization (CARD-FISH) (41) to demonstrate the transcription of the analyzed sequences [16S, internal transcribed spacer (ITS), and intron regions] into RNA. A DNA oligonucleotide that targeted position 1284–1303 of the 16S rRNA in native ribosomes (VSO1284, Table S2) hybridized to nearly all cells, yielding strong signals indicating an overall presence of target molecules (Fig. 3C). In contrast, the application of oligonucleotides targeting the respective intron sequences yielded no signal above background (Fig. 3D; probe S1396-179, Table S2), indicating that introns were absent from mature 16S rRNA. Furthermore, application of a probe that bridged the intron insertion sites revealed strong fluorescent signals (Fig. 3E; probe Thm1389, Table S2). Finally, a probe that targeted the exon1/intron boundary sequence showed no detectable hybridization signal (Fig. 3F; probe Thm1397-INT, Table S2).
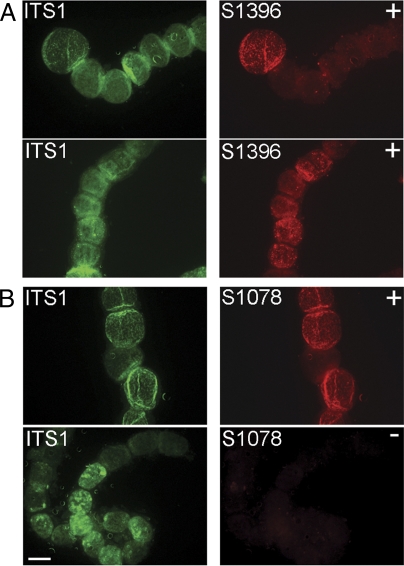
To exclude that the intron-containing 16S rRNA genes are pseudogenes, we applied a more sensitive CARD-FISH assay and tested whether the introns could be hybridized as part of the rRNA precursor (Fig. 4). Therefore, specific regions of the precursor (ITS, S1078, or S1396) were targeted simultaneously with three or four probes (Table S2), hybridized at a lower temperature (35 °C instead of 46 °C), and the RNA secondary structure was loosened by the addition of helper oligonucleotides (42).
Fig. 4.
CARD-FISH on the rRNA precursor of large sulfur bacteria. Hybridization results for the ITS region are shown on the left (green) and for the intron region on the right (red). Successful hybridization of introns in those cells that simultaneously showed an ITS-specific signal indicates that introns are transcribed as part of the rRNA precursor. (A) Simultaneous hybridization of the ITS region and intron S1396 in Thiomargarita namibiensis (Upper panels) and “Ca. T. nelsonii” (Lower panels) revealed positive fluorescent signals. Sequencing 16S rRNA genes of individual cells of these species implied that intron S1396 should be present in both species (“+”). (B) Simultaneous hybridization of the ITS region and intron S1078, which is expected to be present only in T. namibiensis (Upper panel, “+”) and not in “Ca. T. nelsonii” (Lower panel, “−”). (Scale bar, 100 μm.)
Both ITS- and intron-specific signals were obtained with the altered hybridization conditions, which indicated that the rRNA precursor could be detected. Intracellular precursor concentrations are generally low, but can vary highly among individual cells depending on their metabolic state (43). This fluctuation was also observed in this study as generally few of the analyzed cells showed detectable signals with the altered protocol. Double-hybridization experiments targeting successively the intron and the ITS region revealed that all cells having intron-specific signals simultaneously showed ITS-specific signals (Fig. 4 A and B, “ITS” and “+” panels). In “Ca. T. nelsonii” cells, intron S1078 is not present, and indeed only an ITS-specific signal was detectable (Fig. 4B, “ITS” and “−” panels). These findings confirmed that intron-containing 16S rRNA genes were transcribed and could be specifically hybridized in the rRNA precursor.
Frequently, ITS signals were detected although intron signals were not, but never vice versa. This indicated that introns of the 16S rRNA might be excised and/or degraded more efficiently than the ITS region, whereas the latter might be excised and degraded in a later step. We can conclude from the CARD-FISH results that the formation of native ribosomes in large sulfur bacteria is not affected by the presence of intron sequences in their rDNA because they are removed during rRNA maturation processes.
Length-Heterogeneity PCR Bias.
Length deviation in target sequences can cause a PCR bias that preferentially amplifies shorter over longer homologs (44, 45). We assumed that this bias could be the potential reason for intron-containing 16S rRNA genes of large sulfur bacteria being frequently missing from universal clone libraries (8–13). Equal amounts of cloned intron-containing (∼2,350 nt) and intron-lacking (∼1,500 nt) 16S rRNA genes were combined and subject to a universal PCR approach. Indeed, we observed a systematic discrimination of the longer gene over the homolog being shorter by 850 nt (Fig. S5, lane 3). Simulating a more realistic approach, which assumed that intron-containing genes are less frequent in a natural sample than intron-lacking genes, the longer homolog was provided less concentrated (ratios of 1:10, 1:100, and 1:1,000). In all these approaches, again only the shorter gene was amplified (Fig. S5, lanes 4–6), which further demonstrated the strong effect of the length-heterogeneity bias. This bias can be explained by the kinetics of the enzymatic reaction during the PCR amplification step (44) and may affect those reactions containing homologs that exceed a certain length deviation and/or that are combined in unfavorable concentrations.
Discussion
In this study we demonstrate that the 16S rRNA genes of large sulfur bacteria can contain self-splicing introns, up to three being present in a single gene and extending its length to more than 3,500 nucleotides. Introns are generally rare in bacteria and have never been encountered previously in bacterial 16S rRNA genes. So, it was quite surprising to even identify multiple introns inserted into these bacterial 16S rRNA genes.
The four introns are probably analogs because they do not share a common insertion site and their sequences are not related to each other. Thus, every intron type has probably inserted independently into the genome. Phylogenetic analysis of the most frequently observed intron S1396, a group II intron, revealed that it was probably acquired both vertically and horizontally within the family. This feature is well known among group II introns (46–49). Comparative sequence analysis of intron S1396 generally reflects the phylogeny suggested by 16S rRNA sequence analysis at species level (Table S1 and Fig. S2), indicating that the intron coevolved with the host after insertion into its genome. At the family level, the relationship of S1396 between genera is different from the 16S rRNA-derived phylogenetic pattern, suggesting horizontal transfer of this intron between different taxa. This phenomenon is likely caused by the high mobility of these elements, frequently inserting and exiting from genomes (46, 47). Furthermore, introns are preferentially found in conserved regions of the rRNA genes (21, 22), which can explain why the insertion sites of introns analyzed in this study are homologous to locations of introns found in mitochondrial and nuclear rRNA genes of eukaryotes. This is consistent with the hypothesis that introns can be horizontally transferred over large phylogenetic distances (50, 51). All ORFs in the introns described here encoded IEPs typical of a common family of homing endonucleases, suggesting that the introns spread into cognate sites as DNA elements via homing (20). The existence of variants that lack, or have truncated IEPs, is considered to be a consequence of introns saturating all possible insertion sites and of IEPs for intron homing becoming redundant (52).
Introns have been well studied regarding their splicing mechanism from RNA precursors and their methods of propagation in the host genome (35, 46, 53–57). Here we show that introns in large sulfur bacteria also demonstrate self-splicing abilities in vitro (Fig. 3) and removal from precursor rRNA molecules in vivo (Figs. 3 and 4). CARD-FISH on the rRNA precursor targeting the ITS region was previously performed (58, 59), but in this study also introns in the rRNA precursors were hybridized. The successful and specific labeling of the different introns demonstrates their transcription into RNA, strongly indicating that the 16S rRNA genes retrieved by PCR were not pseudogenes. Also, the invariable absence of intron-free PCR products from a cell yielding an intron-containing 16S rRNA gene amplicon further supports the suggestion that the intron-containing gene is functional. The absence of motifs for precursor processing enzymes like RNase III and the in vitro self-splicing activity indicate the autonomous and uncoupled removal of the introns from the precursor in vivo (Figs. 3 and 4), which certainly prevents negative impact on ribosome functionality.
The detection of introns in the 16S rRNA genes of large sulfur bacteria raises questions of why they are retained in the genes of this group of organisms, whether they have a biological function, or whether they are strictly selfish parasitic DNA elements. Currently, we can offer only suggestions for the first of these questions. We presume that introns cause less constraints and thus experience reduced selective pressure in large sulfur bacteria, compared with introns in most ordinary-sized bacteria. This hypothesis is based on the assumption that introns might persist longer in organisms that spend only a small fraction of their energy on replication (60). This energy surplus could be produced either by mitochondria in eukaryotes (60) or by the storage of tremendous amounts of energy reserves as in the group of large sulfur bacteria (16, 18, 19, 61). The replication, transcription, and excision of introns may thus not represent a major bioenergentic expense for large sulfur bacteria. DNA replication is considered to require only about 2% of a cell's energy budget (62), and transcription can be regarded as even less costly. Accordingly, large sulfur bacteria could afford the retention of intron-containing genes (60) owing to their energy surplus in the form of storage compounds (16, 18, 19, 61). Alternatively, it could be speculated that the high frequency of introns in this group of bacteria is somehow connected to the extreme polyploidy in these organisms: nucleic-acid staining in Thiomargarita spp. reveals the presence of several thousand nucleoids per cell (62, 63). In view of the insertion mechanism of introns, initiated by a double-strand break of the DNA and followed by cellular repair mechanisms involving homologous recombination, the presence of multiple alleles within a cell is necessary for efficient repair of the intron insertion site. In fact, the spreading of introns in organisms with several thousand genomes could be extremely efficient as only a few IEPs are required to introduce many double-strand breaks, which could then be repaired by copying intron sequences into new sites. Thus, extreme polyploidy can cause rapid intron spread, while at the same time allowing efficient repair of harmful double-strand breaks in the host.
Considering that the 16S rRNA gene is the most sequenced gene today, it seems surprising that bacterial intron-containing 16S rRNA genes have never been encountered before. Thus, it is tempting to speculate that large sulfur bacteria are the only group of bacteria accumulating introns in their 16S rRNA genes. However, the presence of introns, even multiple ones, has been described for the 23S rRNA genes of several distantly related bacteria (39, 50, 51, 64, 65). This gene bears the same evolutionary importance and conservation as the 16S rRNA gene, which indicates that 16S introns might be at least as frequent as 23S introns. Furthermore, introns in archaeal rRNA genes are very common and have been detected in 16S rRNA genes several times (66, 67). Generally, introns in rRNA may have a better opportunity for self-splicing because they are not subject to translation compared with introns inserted into mRNA (68–70). Also, the high level of sequence conservation of rRNA genes and their frequent occurrence in multiple copies could increase intron mobility among these regions (71). We therefore speculate that introns in bacterial rRNA genes might be more common than previously recognized. Nonetheless, public databases do not contain any entries of bacterial 16S rRNA genes with introns (21, 72, 73). This is either a coincidence or a consequence of biases introduced during the application of techniques and/or the interpretation of results.
The possibility that the 16S rRNA gene may not have a conserved size has not been sufficiently considered in designing and applying techniques for diversity studies, even though elongations of 16S rRNA genes due to intervening sequences have been reported before (3–7). Intervening sequences, in contrast to introns, cannot self-splice from precursor RNA and, upon removal by cellular maturation enzymes, leave the ribosomal RNA fragmented (3–7). In this study, we also showed that true self-splicing introns can be present in bacterial 16S rRNA genes, leading to the conclusion that the 16S rRNA genes of bacteria can indeed be far longer than ∼1,500 nucleotides. Surely, this finding has far-reaching consequences because the gene for the small ribosomal subunit plays a fundamental role in phylogenetic and diversity studies and most currently applied methods to study this gene are based on its consistency.
The present study provides further evidence that the retrieval frequency of rRNA genes should not be considered a reliable measure for the abundance of microbial species. We demonstrate severe size selection of PCR products, i.e., the discrimination of intron-containing 16S rRNA gene sequences in a heterogeneous sample. This has far-reaching consequences for environmental diversity studies because entire phylogenetic groups may be missed. Future genome sequencing as well as the generation of large metagenomic datasets will reveal whether introns in 16S rRNA genes, or elongated 16S rRNA genes in general, might be a more common feature of Bacteria than previously recognized.
Materials and Methods
16S rRNA and Intron Sequences.
The amplification of the 16S rRNA genes including intron sequences has been recently published (28). Comparative sequence analysis of 16S rRNA genes and introns was performed in BioEdit (74) and in the ARB software package (75). RNA folding structures of introns were created manually in Canvas (ACD Systems) using folding criteria assigned by ref. 23 for group I introns and by refs. 24–27 for group II introns (Fig. 2 and Fig. S3). More details can be found in SI Materials and Methods.
CARD-FISH.
Probe 1284 targeting the 16S rRNA of all nonfilamentous large sulfur bacteria (Table S2) was designed in ARB (75). All intron probes, ITS probes, and helper oligonucleotides (Tables S2 and S3) were designed manually. Hybridization of Thiomargarita spp. was performed on slides using a protocol modified from ref. 41. For more details, see SI Materials and Methods.
In Vitro Splicing.
Intron-containing 16S rRNA genes were either cloned into vectors featuring a T7 RNA polymerase promotor or PCR-amplified with a primer containing a 5′-promoter overhang. In vitro transcription was performed using the T7 RNA Polymerase-Plus kit (Ambion), and cDNA was generated with the RevertAid H-Minus First Strand cDNA Sythesis kit (Fermentas). Splicing products were amplified via PCR using High Fidelity PCR Enzyme mix (Fermentas). RNA and DNA products were analyzed by agarose gel electrophoresis. DNA bands were further analyzed by sequencing. Details on the reactions are in SI Materials and Methods.
PCR of Homologous Genes with Different Lengths.
Intron-containing and intron-less 16S rRNA genes were cloned and used for PCR with universal 16S primers (GM3F and GM4R) (76) and the High Fidelity PCR Enzyme mix (Fermentas). The reaction contained either one of the two homologs or a mixture of both in varying ratios. For details on the reactions, see SI Materials and Methods.
Supplementary Material
Acknowledgments
This study was funded by the Max Planck Society.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1120192109/-/DCSupplemental.
References
- 1.Pace NR, Olsen GJ, Woese CR. Ribosomal RNA phylogeny and the primary lines of evolutionary descent. Cell. 1986;45:325–326. doi: 10.1016/0092-8674(86)90315-6. [DOI] [PubMed] [Google Scholar]
- 2.Woese CR. Bacterial evolution. Microbiol Rev. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burgin AB, Parodos K, Lane DJ, Pace NR. The excision of intervening sequences from Salmonella 23S ribosomal RNA. Cell. 1990;60:405–414. doi: 10.1016/0092-8674(90)90592-3. [DOI] [PubMed] [Google Scholar]
- 4.Linton D, Clewley JP, Burnens A, Owen RJ, Stanley J. An intervening sequence (IVS) in the 16S rRNA gene of the eubacterium Helicobacter canis. Nucleic Acids Res. 1994;22:1954–1958. doi: 10.1093/nar/22.11.1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rainey FA, Ward-Rainey NL, Janssen PH, Hippe H, Stackebrandt E. Clostridium paradoxum DSM 7308(T) contains multiple 16S rRNA genes with heterogeneous intervening sequences. Microbiology. 1996;142:2087–2095. doi: 10.1099/13500872-142-8-2087. [DOI] [PubMed] [Google Scholar]
- 6.Ralph D, McClelland M. Intervening sequence with conserved open reading frame in eubacterial 23S rRNA genes. Proc Natl Acad Sci USA. 1993;90:6864–6868. doi: 10.1073/pnas.90.14.6864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Springer N, et al. Occurrence of fragmented 16S rRNA in an obligate bacterial endosymbiont of Paramecium caudatum. Proc Natl Acad Sci USA. 1993;90:9892–9895. doi: 10.1073/pnas.90.21.9892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Angert ER, et al. Molecular phylogenetic analysis of a bacterial community in Sulphur River, Parker Cave, Kentucky. Am Mineral. 1998;83:1583–1592. [Google Scholar]
- 9.Edgcomb VP, Kysela DT, Teske A, de Vera Gomez A, Sogin ML. Benthic eukaryotic diversity in the Guaymas Basin hydrothermal vent environment. Proc Natl Acad Sci USA. 2002;99:7658–7662. doi: 10.1073/pnas.062186399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gillan DC, Speksnijder AGCL, Zwart G, De Ridder C. Genetic diversity of the biofilm covering Montacuta ferruginosa (Mollusca, Bivalvia) as evaluated by denaturing gradient gel electrophoresis analysis and cloning of PCR-amplified gene fragments coding for 16S rRNA. Appl Environ Microbiol. 1998;64:3464–3472. doi: 10.1128/aem.64.9.3464-3472.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.López-García P, et al. Bacterial diversity in hydrothermal sediment and epsilonproteobacterial dominance in experimental microcolonizers at the Mid-Atlantic Ridge. Environ Microbiol. 2003;5:961–976. doi: 10.1046/j.1462-2920.2003.00495.x. [DOI] [PubMed] [Google Scholar]
- 12.Sekar R, Mills DK, Remily ER, Voss JD, Richardson LL. Microbial communities in the surface mucopolysaccharide layer and the black band microbial mat of black band-diseased Siderastrea siderea. Appl Environ Microbiol. 2006;72:5963–5973. doi: 10.1128/AEM.00843-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stevens H, Ulloa O. Bacterial diversity in the oxygen minimum zone of the eastern tropical South Pacific. Environ Microbiol. 2008;10:1244–1259. doi: 10.1111/j.1462-2920.2007.01539.x. [DOI] [PubMed] [Google Scholar]
- 14.Gallardo VA. Large benthic microbial communities in sulfide biota under Peru-Chile subsurface countercurrent. Nature. 1977;268:331–332. [Google Scholar]
- 15.Mussmann M, et al. Phylogeny and distribution of nitrate-storing Beggiatoa spp. in coastal marine sediments. Environ Microbiol. 2003;5:523–533. doi: 10.1046/j.1462-2920.2003.00440.x. [DOI] [PubMed] [Google Scholar]
- 16.Schulz HN, et al. Dense populations of a giant sulfur bacterium in Namibian shelf sediments. Science. 1999;284:493–495. doi: 10.1126/science.284.5413.493. [DOI] [PubMed] [Google Scholar]
- 17.Jørgensen BB, Fenchel T. The sulfur cycle of a marine sediment model system. Mar Biol. 1974;24:189–201. [Google Scholar]
- 18.Fossing H, et al. Concentration and transport of nitrate by the mat-forming sulfur bacterium Thioploca. Nature. 1995;374:713–715. [Google Scholar]
- 19.Schulz HN, Schulz HD. Large sulfur bacteria and the formation of phosphorite. Science. 2005;307:416–418. doi: 10.1126/science.1103096. [DOI] [PubMed] [Google Scholar]
- 20.Lambowitz AM, Belfort M. Introns as mobile genetic elements. Annu Rev Biochem. 1993;62:587–622. doi: 10.1146/annurev.bi.62.070193.003103. [DOI] [PubMed] [Google Scholar]
- 21.Jackson SA, Cannone JJ, Lee JC, Gutell RR, Woodson SA. Distribution of rRNA introns in the three-dimensional structure of the ribosome. J Mol Biol. 2002;323:35–52. doi: 10.1016/s0022-2836(02)00895-1. [DOI] [PubMed] [Google Scholar]
- 22.Johansen SD, Haugen P, Nielsen H. Expression of protein-coding genes embedded in ribosomal DNA. Biol Chem. 2007;388(1):679–686. doi: 10.1515/BC.2007.089. [DOI] [PubMed] [Google Scholar]
- 23.Michel F, Westhof E. Modelling of the three-dimensional architecture of group I catalytic introns based on comparative sequence analysis. J Mol Biol. 1990;216:585–610. doi: 10.1016/0022-2836(90)90386-Z. [DOI] [PubMed] [Google Scholar]
- 24.Granlund M, Michel F, Norgren M. Mutually exclusive distribution of IS1548 and GBSi1, an active group II intron identified in human isolates of group B streptococci. J Bacteriol. 2001;183:2560–2569. doi: 10.1128/JB.183.8.2560-2569.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li C-F, Costa M, Bassi G, Lai Y-K, Michel F. Recurrent insertion of 5′-terminal nucleotides and loss of the branchpoint motif in lineages of group II introns inserted in mitochondrial preribosomal RNAs. RNA. 2011;17:1321–1335. doi: 10.1261/rna.2655911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Quiroga C, Roy PH, Centrón D. The S.ma.I2 class C group II intron inserts at integron attC sites. Microbiology. 2008;154:1341–1353. doi: 10.1099/mic.0.2007/016360-0. [DOI] [PubMed] [Google Scholar]
- 27.Toor N, Robart AR, Christianson J, Zimmerly S. Self-splicing of a group IIC intron: 5′ exon recognition and alternative 5′ splicing events implicate the stem-loop motif of a transcriptional terminator. Nucleic Acids Res. 2006;34:6461–6471. doi: 10.1093/nar/gkl820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salman V, et al. A single-cell sequencing approach to the classification of large, vacuolated sulfur bacteria. Syst Appl Microbiol. 2011;34:243–259. doi: 10.1016/j.syapm.2011.02.001. [DOI] [PubMed] [Google Scholar]
- 29.Hensgens LAM, Bonen L, de Haan M, van der Horst G, Grivell LA. Two intron sequences in yeast mitochondrial COX1 gene: Homology among URF-containing introns and strain-dependent variation in flanking exons. Cell. 1983;32:379–389. doi: 10.1016/0092-8674(83)90457-9. [DOI] [PubMed] [Google Scholar]
- 30.Côté V, Mercier JP, Lemieux C, Turmel M. The single group-I intron in the chloroplast rrnL gene of Chlamydomonas humicola encodes a site-specific DNA endonuclease (I-ChuI) Gene. 1993;129(1):69–76. doi: 10.1016/0378-1119(93)90697-2. [DOI] [PubMed] [Google Scholar]
- 31.Saiki RK, et al. Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science. 1985;230:1350–1354. doi: 10.1126/science.2999980. [DOI] [PubMed] [Google Scholar]
- 32.Sethuraman J, Majer A, Friedrich NC, Edgell DR, Hausner G. Genes within genes: Multiple LAGLIDADG homing endonucleases target the ribosomal protein S3 gene encoded within an rnl group I intron of Ophiostoma and related taxa. Mol Biol Evol. 2009;26:2299–2315. doi: 10.1093/molbev/msp145. [DOI] [PubMed] [Google Scholar]
- 33.Wakasugi T, et al. Complete nucleotide sequence of the chloroplast genome from the green alga Chlorella vulgaris: The existence of genes possibly involved in chloroplast division. Proc Natl Acad Sci USA. 1997;94:5967–5972. doi: 10.1073/pnas.94.11.5967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dalgaard JZ, et al. Statistical modeling and analysis of the LAGLIDADG family of site-specific endonucleases and identification of an intein that encodes a site-specific endonuclease of the HNH family. Nucleic Acids Res. 1997;25:4626–4638. doi: 10.1093/nar/25.22.4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haugen P, Simon DM, Bhattacharya D. The natural history of group I introns. Trends Genet. 2005;21(2):111–119. doi: 10.1016/j.tig.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 36.Toor N, Zimmerly S. Identification of a family of group II introns encoding LAGLIDADG ORFs typical of group I introns. RNA. 2002;8:1373–1377. doi: 10.1017/s1355838202023087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Adamidi C, Fedorova O, Pyle AM. A group II intron inserted into a bacterial heat-shock operon shows autocatalytic activity and unusual thermostability. Biochemistry. 2003;42:3409–3418. doi: 10.1021/bi027330b. [DOI] [PubMed] [Google Scholar]
- 38.Chien MF, Tosa S, Huang CC, Endo G. Splicing of a bacterial group II intron from Bacillus megaterium is independent of intron-encoded protein. Microbes Environ. 2009;24(1):28–32. doi: 10.1264/jsme2.me08540. [DOI] [PubMed] [Google Scholar]
- 39.Nesbø CL, Doolittle WF. Active self-splicing group I introns in 23S rRNA genes of hyperthermophilic bacteria, derived from introns in eukaryotic organelles. Proc Natl Acad Sci USA. 2003;100:10806–10811. doi: 10.1073/pnas.1434268100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ferat JL, Le Gouar M, Michel F. A group II intron has invaded the genus Azotobacter and is inserted within the termination codon of the essential groEL gene. Mol Microbiol. 2003;49:1407–1423. doi: 10.1046/j.1365-2958.2003.03649.x. [DOI] [PubMed] [Google Scholar]
- 41.Pernthaler A, Pernthaler J, Amann R. Fluorescence in situ hybridization and catalyzed reporter deposition for the identification of marine bacteria. Appl Environ Microbiol. 2002;68:3094–3101. doi: 10.1128/AEM.68.6.3094-3101.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fuchs BM, Glöckner FO, Wulf J, Amann R. Unlabeled helper oligonucleotides increase the in situ accessibility to 16S rRNA of fluorescently labeled oligonucleotide probes. Appl Environ Microbiol. 2000;66:3603–3607. doi: 10.1128/aem.66.8.3603-3607.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Britschgi TB, Cangelosi GA. Detection of rifampin-resistant bacteria using DNA probes for precursor rRNA. Mol Cell Probes. 1995;9(1):19–24. doi: 10.1016/s0890-8508(95)90932-x. [DOI] [PubMed] [Google Scholar]
- 44.Cardinale M, et al. Comparison of different primer sets for use in automated ribosomal intergenic spacer analysis of complex bacterial communities. Appl Environ Microbiol. 2004;70:6147–6156. doi: 10.1128/AEM.70.10.6147-6156.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jeffreys AJ, Wilson V, Neumann R, Keyte J. Amplification of human minisatellites by the polymerase chain reaction: Towards DNA fingerprinting of single cells. Nucleic Acids Res. 1988;16:10953–10971. doi: 10.1093/nar/16.23.10953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dai L, Zimmerly S. Compilation and analysis of group II intron insertions in bacterial genomes: Evidence for retroelement behavior. Nucleic Acids Res. 2002;30:1091–1102. doi: 10.1093/nar/30.5.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Toro N. Bacteria and Archaea Group II introns: Additional mobile genetic elements in the environment. Environ Microbiol. 2003;5(3):143–151. doi: 10.1046/j.1462-2920.2003.00398.x. [DOI] [PubMed] [Google Scholar]
- 48.Tourasse NJ, Kolstø AB. Survey of group I and group II introns in 29 sequenced genomes of the Bacillus cereus group: Insights into their spread and evolution. Nucleic Acids Res. 2008;36:4529–4548. doi: 10.1093/nar/gkn372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zimmerly S, Hausner G, Wu Xc. Phylogenetic relationships among group II intron ORFs. Nucleic Acids Res. 2001;29:1238–1250. doi: 10.1093/nar/29.5.1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Haugen P, et al. Cyanobacterial ribosomal RNA genes with multiple, endonuclease-encoding group I introns. BMC Evol Biol. 2007;7:159. doi: 10.1186/1471-2148-7-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Raghavan R, Miller SR, Hicks LD, Minnick MF. The unusual 23S rRNA gene of Coxiella burnetii: Two self-splicing group I introns flank a 34-base-pair exon, and one element lacks the canonical omegaG. J Bacteriol. 2007;189:6572–6579. doi: 10.1128/JB.00812-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Goddard MR, Burt A. Recurrent invasion and extinction of a selfish gene. Proc Natl Acad Sci USA. 1999;96:13880–13885. doi: 10.1073/pnas.96.24.13880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Edgell DR, Belfort M, Shub DA. Barriers to intron promiscuity in bacteria. J Bacteriol. 2000;182:5281–5289. doi: 10.1128/jb.182.19.5281-5289.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kruger K, et al. Self-splicing RNA: Autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell. 1982;31(1):147–157. doi: 10.1016/0092-8674(82)90414-7. [DOI] [PubMed] [Google Scholar]
- 55.Lambowitz AM, Zimmerly S. Mobile group II introns. Annu Rev Genet. 2004;38:1–35. doi: 10.1146/annurev.genet.38.072902.091600. [DOI] [PubMed] [Google Scholar]
- 56.Michel F, et al. Activation of the catalytic core of a group I intron by a remote 3′ splice junction. Genes Dev. 1992;6:1373–1385. doi: 10.1101/gad.6.8.1373. [DOI] [PubMed] [Google Scholar]
- 57.Peebles CL, et al. A self-splicing RNA excises an intron lariat. Cell. 1986;44:213–223. doi: 10.1016/0092-8674(86)90755-5. [DOI] [PubMed] [Google Scholar]
- 58.Oerther DB, Pernthaler J, Schramm A, Amann R, Raskin L. Monitoring precursor 16S rRNAs of Acinetobacter spp. in activated sludge wastewater treatment systems. Appl Environ Microbiol. 2000;66:2154–2165. doi: 10.1128/aem.66.5.2154-2165.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schmid M, Schmitz-Esser S, Jetten M, Wagner M. 16S-23S rDNA intergenic spacer and 23S rDNA of anaerobic ammonium-oxidizing bacteria: Implications for phylogeny and in situ detection. Environ Microbiol. 2001;3:450–459. doi: 10.1046/j.1462-2920.2001.00211.x. [DOI] [PubMed] [Google Scholar]
- 60.Darnell JE, Doolittle WF. Speculations on the early course of evolution. Proc Natl Acad Sci USA. 1986;83:1271–1275. doi: 10.1073/pnas.83.5.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Preisler A, et al. Biological and chemical sulfide oxidation in a Beggiatoa inhabited marine sediment. ISME J. 2007;1:341–353. doi: 10.1038/ismej.2007.50. [DOI] [PubMed] [Google Scholar]
- 62.Lane N, Martin W. The energetics of genome complexity. Nature. 2010;467:929–934. doi: 10.1038/nature09486. [DOI] [PubMed] [Google Scholar]
- 63.Schulz HN. The genus Thiomargarita. In: Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E, editors. In The Prokaryotes. New York: Springer; 2006. pp. 1156–1163. [Google Scholar]
- 64.Everett KDE, Kahane S, Bush RM, Friedman MG. An unspliced group I intron in 23S rRNA links Chlamydiales, chloroplasts, and mitochondria. J Bacteriol. 1999;181:4734–4740. doi: 10.1128/jb.181.16.4734-4740.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Seshadri R, et al. Complete genome sequence of the Q-fever pathogen Coxiella burnetii. Proc Natl Acad Sci USA. 2003;100:5455–5460. doi: 10.1073/pnas.0931379100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Burggraf S, Larsen N, Woese CR, Stetter KO. An intron within the 16S ribosomal RNA gene of the archaeon Pyrobaculum aerophilum. Proc Natl Acad Sci USA. 1993;90:2547–2550. doi: 10.1073/pnas.90.6.2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Itoh T, Suzuki K, Nakase T. Occurrence of introns in the 16S rRNA genes of members of the genus Thermoproteus. Arch Microbiol. 1998;170(3):155–161. doi: 10.1007/s002030050628. [DOI] [PubMed] [Google Scholar]
- 68.Cavalier-Smith T. Intron phylogeny: A new hypothesis. Trends Genet. 1991;7(5):145–148. [PubMed] [Google Scholar]
- 69.Doolittle WF. The origins of introns. Curr Biol. 1991;1(3):145–146. doi: 10.1016/0960-9822(91)90214-h. [DOI] [PubMed] [Google Scholar]
- 70.Martin W, Koonin EV. Introns and the origin of nucleus-cytosol compartmentalization. Nature. 2006;440:41–45. doi: 10.1038/nature04531. [DOI] [PubMed] [Google Scholar]
- 71.Thompson AJ, Herrin DL. A chloroplast group I intron undergoes the first step of reverse splicing into host cytoplasmic 5.8S ribosomal RNA: Implications for intron-mediated RNA recombination, intron transposition and 5.8S ribosomal RNA structure. J Mol Biol. 1994;236:455–468. doi: 10.1006/jmbi.1994.1157. [DOI] [PubMed] [Google Scholar]
- 72.Cannone JJ, et al. The Comparative RNA Web (CRW) Site: An online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics. 2002;3:2. doi: 10.1186/1471-2105-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Simon DM, et al. Group II introns in eubacteria and archaea: ORF-less introns and new varieties. RNA. 2008;14:1704–1713. doi: 10.1261/rna.1056108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hall TA. 1999. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser, no. 41: 95–98.
- 75.Ludwig W, et al. ARB: A software environment for sequence data. Nucleic Acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Muyzer G, Teske A, Wirsen CO, Jannasch HW. Phylogenetic relationships of Thiomicrospira species and their identification in deep-sea hydrothermal vent samples by denaturing gradient gel electrophoresis of 16S rDNA fragments. Arch Microbiol. 1995;164(3):165–172. doi: 10.1007/BF02529967. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.