Abstract
The innate immune system is an ancient and broad-spectrum defense system found in all eukaryotes. The detection of microbial elicitors results in the up-regulation of defense-related genes and the elicitation of inflammatory and apoptotic responses. These innate immune responses are the front-line barrier against disease because they collectively suppress the growth of the vast majority of invading microbes. Despite their critical role, we know remarkably little about the diversity of immune elicitors. To address this paucity, we reasoned that hosts are more likely to evolve recognition to “core” pathogen proteins under strong negative selection for the maintenance of essential cellular functions, whereas repeated exposure to host–defense responses will impose strong positive selective pressure for elicitor diversification to avoid host recognition. Therefore, we hypothesized that novel bacterial elicitors can be identified through these opposing forces of natural selection. We tested this hypothesis by examining the genomes of six bacterial phytopathogens and identifying 56 candidate elicitors that have an excess of positively selected residues in a background of strong negative selection. We show that these positively selected residues are atypically clustered, similar to patterns seen in the few well-characterized elicitors. We then validated selected candidate elicitors by showing that they induce Arabidopsis thaliana innate immunity in functional (virulence suppression) and cellular (callose deposition) assays. These finding provide targets for the study of host–pathogen interactions and applied research into alternative antimicrobial treatments.
Keywords: evolutionary genomics, pathogen-associated molecular pattern/microbe-associated molecular pattern, plant pathogen
The innate immune system is an ancient, robust, and broad-spectrum defense system that protects eukaryotes against invading microbes (1, 2). This front-line defense system depends on the detection of microbial elicitors that induces a cascade of defense responses that may include MAP kinase activity, the up-regulation of antimicrobial compounds, elicitation of inflammatory responses, the production of cytokines or mucins, apoptosis, nitric oxide-mediated responses, and the transcription of defense related genes (3–5). In plant–pathogen systems, the innate immune system can be stimulated by a variety of elicitors, including (i) the external recognition of conserved microbial epitopes called microbe-associated molecular patterns (MAMPs) via pattern recognition receptors (PRRs); (ii) by damage-associated molecular patterns (DAMPs) that are endogenous molecules generated during pathogen attack; (iii) by certain toxins and pore-forming molecules such as the fungal toxin fumonisin B1 (6) and the membranotropic peptide alamethicin (7); or (iv) by the internal recognition of pathogen effectors or the functions of these effectors by plant resistance (R) proteins (8, 9). Although there appears to be substantial overlap in the signaling and response to these stimuli, the last of these mechanism, termed effector-trigger immunity (ETI), gives rise to a much more dramatic response than the former mechanisms. The immune response elicited by MAMPs is called MAMP-triggered immunity (MTI), although this term is also typically used to describe the response to the second and third set of elicitors (8, 10).
Despite their critical role in immunity, we know remarkably little about the range and diversity of MTI elicitors. Most studies have focused on a limited number of MAMPs such as lipopolysaccharides, peptidoglycans, β-glucans, flagellin, cold shock protein, and elongation factor Tu (EF-Tu) (3, 11, 12), all of which have been identified after observing host–immune responses. Surprisingly, no method as yet been proposed to systematically identify immunity elicitors.
The ability to develop a systematic approach to discovering MTI elicitors requires the presence of signals or properties that are characteristic of and specific to these elicitors. Fortunately, this is the case for proteinaceous elicitors. Organisms are most able and likely to evolve recognition to microbial epitopes that are indispensable, evolutionarily conserved, and broadly distributed. Thus, proteinaceous MTI elicitors are likely to be found among the subset of microbial proteins that are selectively constrained (i.e., under strong negative selection) due to some vital cellular function. However, pathogens can only mount successful infections if they are able to avoid or disrupt the host–immune response. Consequently, successful pathogens have evolved a number of strategies to overcome MTI, such as delivering virulence proteins (e.g., type III secreted effectors) that specifically disrupt or suppress the innate immune response (13, 14), or by the diversification of the elicitors themselves (12, 13) due to host-imposed positive selection (15–17).
Elicitor diversification may seem contrary to the previous description of elicitors as evolutionary conserved molecules. Nevertheless, selective constraints act most directly and intensively on the specific regions that are critical for the structure and function of the protein, and do not preclude the accumulation of genetic variation elsewhere in the molecule. If this diversification occurs in the region that induces MTI, then host recognition may be disrupted while the endogenous protein activity and function are maintained. However, of course MTI avoidance via elicitor diversification will be a viable evolutionary option only if the elicitor is specifically recognized by a receptor, such as the case of MAMPs and their cognate PRRs. The effect of MAMP diversification on immune induction was shown in studies of flagellin genetic variation both within and among species of plant-associated bacteria, which found considerable variation in the defense elicitation activity of different flagellin alleles and flg22 epitopes (18–22), and the natural genetic variation uncovered in the elf18 epitope of EF-Tu from a range of plant-associated Proteobacteria that dramatically differ in their ability to elicit a defense-associated oxidative burst in Arabidopsis (13).
MTI elicitors may therefore be characterized as proteins that are encoded by ubiquitous and conserved genes (i.e., components of the core genome) that have discrete regions containing potentially high rates of nonsynonymous (amino acid changing) substitutions. Thus, we hypothesized that elicitors should be encoded by core genes that are under very strong negative selection as a whole to preserve their critical function, but which have distinct regions under very strong positive selection to avoid host recognition. We exploited these paradoxical signatures of positive and negative selection to identify MTI elicitors in six phytopathogenic bacteria.
Results
Detection of Positive Selection in the Core Phytopathogen Genome.
We screened for novel elicitors by examining the core genome of six plant pathogenic Gram-negative bacteria for proteins exhibiting patterns of both strong negative selection for the maintenance of core functions and strong positive selection for the avoidance of host–immune recognition. The core genome was operationally defined as the orthologous gene set common among the finished (closed) genomes of Pseudomonas syringae pv. tomato DC3000 (PtoDC3000), P. syringae pv. phaseolicola 1448A, P. syringae pv. syringae B728A, Xanthomonas campestris pv. campestris 33913, X. campestris pv. campestris 8004, and X. campestris pv. vesicatoria 85-10. In all, 1,322 orthologous core genes were identified by using the reciprocal smallest distance algorithm, which is less vulnerable to ortholog exclusion because of paralog identification in one BLAST direction (23). Alignments were constructed in ClustalX by using an exhaustive parameter search method that identified the best global alignment, and selection analyses were performed by using the codeml program in the PAML 3.15 application suite (24, 25).
We first measured the ratio of nonsynonymous (Ka) substitutions to synonymous (Ks) substitutions (Ka/Ks = ω) in the core gene set at the whole gene level by using the codeml M0 model and found an average ω = 0.043 ± 0.001 (SD) across the core genome with a median of 0.038 and a range of 0.002–0.183. These very low ω ratios were expected for a functionally constrained core genome under strong negative selection. We then applied codeml models that permit the identification of positive selection acting at individual amino acid residues along the protein sequence. Overall, 35% of the core genes were found to have at least one positively selected residue, and a likelihood ratio test (LRT) rejected the neutral and negative selection null model (M7) in favor of the positive selection model (M8) for 56 proteins [P < 0.01, corresponding to a false discovery rate (FDR) of 10%; Table S1]. Bayes Empirical Bayes (BEB) estimates from model M8 were then used to identify the putatively positively selected sites for each candidate (26).
An important validation of our approach was the identification of the well-characterized MAMP EF-Tu (PSPTO0624) as a statistically significant candidate, with site-specific ω ratios as high as 5.0. Flagellin, which is the best characterized P. syringae MAMP, also had multiple positively selected residues; nevertheless, we could not reject the null hypothesis for this locus at the P = 0.01 level because the intensity of selection on these residues is substantially weaker (ω < 1.5) than that observed for many other candidates including EF-Tu.
Although we identified 56 core bacterial proteins exhibiting significant signatures of positive selection, it is unlikely that all of these candidates are MTI elicitors. Bacteria are exposed to a variety of selective pressures that can give rise to similar patterns of positive selection among the core genome. For example, some of these proteins may serve as bacteriophage receptors, selecting for increased genetic variation to avoid phage predation. To distinguish between molecular signatures of selection arising from interactions unrelated to the evasion of host–plant defenses, we performed an additional evolutionary analysis on the 56 candidate elicitors from seven soil-inhabiting, nonpathogenic Pseudomonas strains (P. fluorescens strains Pf-5, Pf0-1, SBW25 and P. putida strains F1, GB-1, KT2440, and W619). We hypothesized that nonpathogenic soil microbes will not be exposed to the same host-imposed selective pressures as plant-associated pathogenic microbes, and, therefore, we would not expect to see the same pattern of diversification on these candidate MTI elicitors. Only eight of the 56 candidates show statistically significant evidence for positive selection among nonpathogenic strains (P < 0.01, LRT between M7 and M8; Table S1), suggesting these proteins are less likely to be targets for plant recognition.
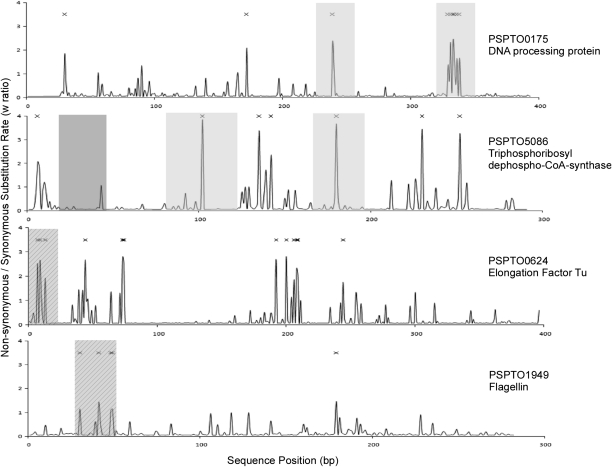
A number of strong MTI elicitors are short peptides encoded within proteinaceous MAMPs. For example, the primary EF-Tu epitope is an 18-aa peptide (elf18), whereas the 22-aa flg22 peptide is the primary flagellin MAMP epitope (6, 7, 13). Our analysis uncovered three positively selected residues within the elf18 epitope and four within the flg22 epitope (Fig. 1). Interestingly, Lacombe and colleagues (13) tested synthetic elf18 epitopes derived from several genera of phytopathogenic bacteria and showed that they have variable defense elicitation activities in Arabidopsis. One of the synthetic peptides tested (class b in figure 1 of ref. 13) carries substitutions at two residues under very strong positive selection (P = 0.843 and 0.945, respectively, for positions 7 and 9). This peptide reduces oxidative burst elicitation activity by half in Arabidopsis. Another peptide tested carries a substitution found only in PtoDC3000 (group G in figure 1 of ref. 13) that is also under positive selection, albeit weaker (P = 0.604), and that also reduces elicitation activity by half. None of the other elf18 peptides tested carried positively selected substitutions or significantly modulate elicitation activity.
Fig. 1.
Estimation of the nonsynonymous (Ka) to synonymous substitution (KS) ratio (ω) at each amino acid position using the Bayes Empirical Bayes (BEB) estimates from PAML/codeml model M8. Crosses above the sequence denote sites with moderate to high probability of positive selection (P > 0.50). Solid gray shading shows regions for which candidate peptides were synthesized for functional analysis, whereas the dark gray shading in PSPTO5086 indicates the region used for synthesizing the negative candidate peptide. Hatched shading indicates the region in EF-Tu (elf18) and flagellin (Flg22) previously identified as an elicitor of innate immunity in A. thaliana.
These results led us to hypothesize that not only will MTI elicitors have an excess of amino acids under strong positive selection relative to other core genes, but that these positively selected amino acids will be clustered into islands that correspond to the specific elicitor epitopes. To assess this hypothesis, we determined the number of candidate elicitor and core genome loci that had multiple positively selected residues clustered within a 30-aa window. We only considered loci with two or more positively selected residues to control for biases imposed by a general excess of positively selected sites among the candidates. A total of 210 core genome loci had two or more positively selected sites. Forty-five of these loci were from candidate elicitors, and 165 were noncandidate core genes. Out of this group, 62% of the candidate elicitors carried clusters of positively selected sites, compared with 46% of noncandidates, indicating a significant overrepresentation of clusters among the candidates (P = 0.018, Fisher's exact test). Each candidate elicitor in this set carried an average of 1.30 ± 1.68 (SD) clusters, whereas core genes had 0.64 ± 1.02 (SD) clusters (P = 0.0014, unpaired t test). If we consider the entire core genome dataset, rather than restricting it to those loci with two or more positively selected residues, 50% of all candidate elicitors had clusters of positively selected residues, whereas this pattern was found in only 6% of the entire core genome (P < 0.0001, Fisher's exact test).
Functionally Assaying Candidate Elicitors for Immune Elicitation.
We selected eight candidate elicitors for functional validation (Table 1), based on significant statistical evidence of positive selection under the M7-M8 likelihood ratio test, and BEB estimates identifying one or more positively selected residues (P > 0.9). We then identified candidate elicitor epitopes based on the location of the positively selected sites within these proteins and synthesized three classes of peptides ranging in size from 20 to 40 aa (Table S2). Class a peptides (positive candidates) were designed to encompass at least one positively selected site within a candidate elicitor. Class b peptides (negative candidates) were designed to encompass only neutral or negatively selected regions within a candidate elicitor. Class c peptides (noncandidates) were designed from noncandidates that showed no signs of positive selection. Peptides were also synthesized corresponding to the region in flagellin (PSPTO1949, flg22) with previously demonstrated elicitation activity. Our expectations were that the class a positive candidate peptides should elicit MTI, whereas the class c noncandidate peptides should not elicit MTI. The class b negative candidate peptides were generally expected not to elicit MTI because they were not designed to encompass positively selected sites, although we expected substantial variation in their elicitation ability because of their linkage to selected regions.
Table 1.
Putative MTI elicitor proteins used in validation studies
| Protein ID | Annotation (P. syringae pv. tomato DC3000) | P* | PSS† | Clusters‡ |
| PSPTO2757 | GGDEF domain/EAL domain protein | 6.76e-07 | 10 | 6 |
| PSPTO3468 | Conserved hypothetical protein | 4.20e-05 | 5 | 1 |
| PSPTO3620 | HlyD family secretion protein | 4.23e-05 | 2 | 1 |
| PSPTO4436 | Conserved hypothetical protein | 1.63e-03 | 3 | 1 |
| PSPTO0624 | Translation elongation factor EF-Tu | 1.79e-03 | 8 | 5 |
| PSPTO1253 | ATP-dependent RNA helicase RhlB | 2.89e-03 | 1 | 0 |
| PSPTO5086 | Triphosphoribosyl-dephospho-CoA synthase MdcB | 4.41e-03 | 5 | 1 |
| PSPTO0175 | DNA processing protein DprA, putative | 4.77e-03 | 4 | 1 |
Multiple independent iterations of the M7/M8 codeml analyses were performed on the entire core genome. Only those proteins consistently identified as significant by the likelihood ratio tests (LRT) are presented.
*P value of the LRT between models M7 and M8 of codeml (PAML 3.15).
†Positively selected sites (PSS) estimated by BEB with a P > 0.75.
‡The number of positively selected sites found within a 30-aa window.
Virulence suppression assays are perhaps the best method for monitoring the induction of the immune system because they directly assess whether the application of a candidate elicitor results in the decreased susceptibility. The virulence suppression activity of each candidate was assayed by preinoculating A. thaliana ecotype Col-0 with the peptides followed by an infiltration 24 h later with either PtoDC3000 or PmaM6—two A. thaliana Col-0 pathogens. An example of typical virulence suppression assay is presented in Fig. 2A. In this test, the positive candidates PSPTO0175-1:a, PSPTO0175-2:a, PSPTO1253-1:a, and PSPTO5086-1:a significantly reduced bacterial growth compared with the water control, although not to the same degree as flg22Pto. The positive candidate peptides PSPTO4436-1:a and PSPTO5086-2:a also reduced the growth of PtoDC3000 relative to the mock inoculation, but not significantly. The negative candidate peptide PSPTO5086-3:b showed no effect on bacterial growth compared with a mock inoculation. Interestingly, although the flg22 peptide from Agrobacterium tumefaciens is commonly perceived not to induce immunity in the PtoDC3000–A. thaliana pathosystem, it did significantly reduced the growth of PtoDC3000 in this particular assay. Nevertheless, virulence suppression results with this peptide were highly inconsistent across assays and more typically did not induce a significant immune response. We also tested elicitation activity of pooled candidate peptides and found that pooling three negative peptides had no greater effect than the individually tested peptides and did not significantly decrease susceptibility overall, whereas three pooled positive candidate peptides significantly suppressed virulence, but the effect was not significantly greater than that observed for one of the three positive candidates tested individually (Fig. S1).
Fig. 2.
Functional validation of candidate elicitors. (A) Virulence suppression activity of putative elicitor peptides in A. thaliana. A. thaliana Col-0 leaves were infiltrated with treatment peptides, followed by inoculation with the strong pathogen P. syringae pv. tomato DC3000 (PtoDC3000) 24 h later. Peptides that induce innate immunity should suppress the growth of PtoDC3000. Average bacterial growth (±SE) was assayed immediately after inoculation (white bars) and again 1 d later (gray bars). Asterisks indicate that the day 1 growth is significantly different (P < 0.05) than the water control by an unpaired, homoscedastic t test. (B) Representative images analyzed for callose deposition in A. thaliana Col-0 leaves. Callose deposits are indicated by dark spots. Images have been converted to grayscale and inverted. (Scale bar: 200 μm.) (C) Average proportion of the field of view (2.9 mm2) occupied by callose deposits for each treatment peptide (±SE). Significant comparisons with the water control are designated (*P < 0.05 and **P < 0.005).
We used a Fisher's combined probability metaanalysis to objectively assess the overall trend in the virulence suppression data (Table 2). We first evaluated whether peptides behaved consistently between trials, predicting that only the positive peptides would consistently suppress virulence relative to the water control, while both the negative candidates and the noncandidate peptides were expected to either vary inconsistently above and below the water control or show no significant difference. Using a FDR of 5%, we found that 8 of 10 positive candidate peptides showed significant and consistent virulence suppression activity, whereas only 1 of 9 negative candidates or noncandidate peptides showed significant and consistent virulence suppression activity (P = 0.0055; Fisher's exact test). Intriguingly, the only class a candidate peptide that did not show any virulence suppression activity was also recovered as a marginally significant candidate (PSPTO2757-2:a; P = 0.02) in the analysis of the nonpathogenic Pseudomonads.
Table 2.
Virulence suppression metaanalysis
| Peptide* | Fisher's P† | n‡ | Consistency§ | FDR signif¶ |
| Consistent peptides | ||||
| 0624–1:a | 2 × 10−05 | 10 | 0.80 | + |
| 5086–1:a | 0.0002 | 9 | 0.78 | + |
| 4436–2:b | 0.0005 | 4 | 1.00 | + |
| 1253–1:a | 0.0009 | 3 | 1.00 | + |
| 3468–1:a | 0.0020 | 6 | 0.83 | + |
| 0175–1:a | 0.0020 | 3 | 1.00 | + |
| 3620–1:a | 0.0090 | 7 | 1.00 | + |
| 0175–2:a | 0.0209 | 2 | 1.00 | + |
| 4436–1:a | 0.0221 | 4 | 0.75 | + |
| 3468–2:b | 0.1110 | 5 | 0.80 | − |
| 2291–1:c | 0.1523 | 2 | 0.00 | − |
| 2757–2:a | 0.2401 | 1 | 1.00 | − |
| 2757–3:b | 0.4849 | 1 | 1.00 | − |
| Inconsistent peptides | ||||
| 4407–1:c | 0.0003 | 3 | 0.67 | − |
| 0624–3:b | 0.0017 | 3 | 0.67 | − |
| 3620–2:b | 0.0033 | 6 | 0.67 | − |
| 5086–2:a | 0.0170 | 3 | 0.67 | − |
| 4361–1:c | 0.0740 | 3 | 0.33 | − |
| 5086–3:b | 0.1754 | 6 | 0.33 | − |
*Peptide class is denoted by the letter following the colon. Class a, positive candidate peptides from candidate elicitor proteins containing positively selected sites. Class b, negative candidate peptides from candidate elicitor proteins containing no positively selected sites. Class c, noncandidate peptides from noncandidate elicitor proteins.
†Fisher's combined probability test based on individual t tests between test peptide and water control.
‡Sample size.
§Proportion of replicate assays that showed virulence suppression relative to water control.
¶Significant metaanalysis based on a 5% FDR.
There are numerous molecular and cellular assays for the induction of innate immunity in plants, including testing for the activation of protein kinases involved in signaling, measuring the release of reactive oxygen species, and quantifying the induction of defense-related genes. We elected to test the candidate elicitors ability to induce the formation of cell wall papillae via the well-established callose formation assay (27). Previous work has shown that the flg22 flagellin MAMP from the tomato and A. thaliana pathogen PtoDC3000 (flg22Pto) induces significant callose deposition in A. thaliana (28). An example of typical callose deposition assay is presented in Fig. 2B. In this test, we used six positive candidate peptides, a negative candidate peptide, flg22Pto as a positive control, BSA as a nonspecific protein control, and water as a negative control. Peptides were infiltrated into A. thaliana, and leaves were stained with aniline blue 24 h after infiltration. Callose deposition was monitored via epifluorescent microscopy and analyzed with ImageJ (29) by quantifying the proportion of the area occupied by callose, excluding vasculature deposits. All candidate elicitors exhibited significantly higher callose deposition than water or the negative candidate peptide PSPTO5086-1b (P < 0.05; unpaired t test) (Fig. 2C). The BSA treatment induced no significant callose deposition. None of the candidate peptides induced callose deposition as strongly as flg22Pto, but this result is not surprising as flg22 is the strongest MAMP known in this pathosystem.
A callose deposition metaanalysis (Table S3) found that eight of nine positive candidates peptides resulted in significantly greater callose than the water control by using an FDR of 0.05% (the low FDR is necessary due to the highly sensitive nature of the assay), whereas only two of five negative candidate peptides acted in the same manner (P = 0.095; Fisher's exact test). One possible explanation for the weaker overall pattern of significance relative to the virulence suppression assay is that this assay measures a specific cellular reaction as opposed to nonspecific immune response, and we do not know the potential spectrum of cell wall-associated responses induced by MTI elicitors.
Discussion
MTI is arguably the single most important mechanism maintaining plant health. Nevertheless, we know remarkably little about the breadth and diversity of the molecules that elicit this response and how their natural genetic variation influences disease and defense. The common perception of MTI elicitors as broadly distributed and highly conserved molecules that induce innate immunity in a nonhost-specific manner is largely true, but very much an oversimplification (11, 12). There is clear evidence of natural genetic variation in both MAMPs (9, 13, 14) and their cognate PRRs (22) affecting defense induction and the effectiveness of the immune response. Proteinaceous elicitors must play by the same evolutionary rules as other proteins, so although hosts will more readily evolve recognition to highly conserved molecules, this recognition will, in turn, drive their diversification until it impacts the overall fitness of the microbe. Likewise, the same evolutionary pressures will drive hosts to continually evolve recognition to new microbial epitopes, resulting in changes in the relative “effectiveness” of specific elicitors as the frequency of these elicitors and PRR alleles with different affinities shift over time.
We have identified 55 unique candidate MTI elicitors (not including the previously identified EF-Tu). Most of the tested candidates prime the innate immune system and induce callose formation, which therefore warrants their designation as unique MTI elicitors. It is likely that these elicitors are true MAMPs. However, validation of this designation requires the identification of the cognate PRR, because it is formally possible that some of these candidates may be mimicking DAMPs endogenously produced by the host upon pathogen attack (30). Nevertheless, as Boller and Felix (8) point out, many well-defined MAMPs have unknown receptors. Examples include bacterial cold shock protein, superoxide dismutase, harpins, a secreted transglutaminase, strerol-binding elicitins, and a cellulose-binding lectin from oomycetes. It is also extremely difficult to understand why proteins would carry the specific selective signatures used in this screen if they did not interact with specific host receptors.
Although the peptides chosen for this study differ quantitatively in their defense elicitation ability, none induce responses as strongly as flg22Pto. flg22 is often considered the archetypical MAMP (23), nevertheless, there are numerous biological reasons why our candidates may not achieve this standard. Most importantly, the extremely strong elicitation activity of the PtoDC3000 flg22 peptide in Arabidopsis Col-0 is nontypical. As discussed previously, substantial variation in elicitation activity has been observed for both flagellin and EF-Tu epitopes, with few responses as strong as that observed for PtoDC3000 flg22 in Arabidopsis Col-0 (12, 18). Additionally, our synthesized peptide may not precisely correspond to the specific epitope or be properly folded. The peptides are also unmodified, and it has been well established that some MAMPs only elicit a strong defense response after posttranslational modification (24–26). Finally, we may have tested the candidates in the wrong host or under the incorrect environmental conditions. Consequently, we cannot state how many MTI elicitors are expressed by any one microbe or are circulating within microbial populations, but it is clear that there is the potential for tremendous diversity. Although each new elicitor provides opportunities for detailed characterization of the MTI response, given the nature of this screen, it was not feasible to systematically refine the analysis for any particular candidate.
Our results reveal a diversity of MTI elicitors that vary widely in their defense induction ability. It seems reasonable to hypothesize that different hosts have evolved specificity to different elicitors, and that elicitor variants are constantly turning over in microbial populations in response to host recognition. Consequently, genetic variation in these core bacterial proteins may turn out to play a critical role in governing host specificity.
This study has illustrated how patterns of natural selection in bacterial proteins can be used for identifying inducers of innate immunity. This method can be easily extended to identify elicitors of innate immunity in any pathosystem that has population genomic data. The identification of elicitors will provide important insight into the molecular and evolutionary mechanisms underlying host–pathogen interactions and coevolution. Because peptides can be synthesized in heterologous and in vitro systems, readily stored and transported, easily applied, and confer rapid, broad spectrum—although transient—resistance, these innate immunity elicitors may prove to be a ready source of new antimicrobial agents for both medical and agricultural systems.
Materials and Methods
Peptides.
All peptides were designed based on the sequence of PtoDC3000 and synthesized and purified by Sheldon Biotechnology Centre, McGill University (www.mcgill.ca/sheldon) and Bio Basic Inc (www.biobasic.com).
Plant Growth Conditions.
A. thaliana (Col-0) plants were grown in Enconair growth rooms at ≈20 °C under a 12-h photoperiod (100–150 μmol/m2s).
Callose Deposition Assays.
The putative elicitor peptides were diluted in water to a concentration of 10 μM (adjusted for the purity of each individual peptide) and pressure infiltrated into the abaxial side of 4- to 5-wk-old A. thaliana leaves. Leaves were excised after 24 h, cleared with lactophenol, stained with aniline blue, and examined under epifluorescent microscopy for the formation of callose deposits. A minimum of four randomly selected regions within each leaf was photographed from at least four leaves per plant and four plants per treatment. Each image was analyzed with ImageJ (29) by quantifying the proportion of the area occupied by callose, excluding deposits occurring in the vasculature. The entire experiment was replicated three times with similar results.
Virulence Suppression Assays.
A. thaliana plants were inoculated with the peptides at 10 μM (adjusted for the purity of each individual peptide) then infiltrated with PtoDC3000 or PmaM6 24 h later (OD600 0.0002–0.0005). Bacterial growth was monitored subsequent to bacterial infiltration on day 0 and 24 h later by grinding surface sterilized leaf discs in 10 mM MgSO4, serially diluting the homogenized tissue, plating on solid selective media, and counting the colony forming units. A minimum of four leaves per plant and four plants per treatment was used, and the entire experiment was replicated three times with similar results.
Statistical Analyses.
Significance was assessed for virulence suppression and callose deposition assays by using the Student t test to compare treatment effects with the mock (water) treatment. Fisher's combined probability metaanalysis was used with a FDR of 5% and 0.05% to assess the overall trend in the virulence suppression and callose deposition data, respectively.
Supplementary Material
Acknowledgments
We thank D. Desveaux, A. Lee, J. D. Lewis, H. Maughan, H. E. O'Brien, J. Stavrinides, P. W. Wang, and M. Wilton for their thoughtful input to this study as well as important contributions from the rest of the D.S.G. and Desveaux laboratories. This work was supported by Natural Sciences and Engineering Research Council of Canada awards (to D.S.G. and H.C.M.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 4029.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1113893109/-/DCSupplemental.
References
- 1.Janeway CA, Jr, Medzhitov R. Innate immune recognition. Annu Rev Immunol. 2002;20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- 2.Nürnberger T, Brunner F, Kemmerling B, Piater L. Innate immunity in plants and animals: Striking similarities and obvious differences. Immunol Rev. 2004;198:249–266. doi: 10.1111/j.0105-2896.2004.0119.x. [DOI] [PubMed] [Google Scholar]
- 3.Medzhitov R. Recognition of microorganisms and activation of the immune response. Nature. 2007;449:819–826. doi: 10.1038/nature06246. [DOI] [PubMed] [Google Scholar]
- 4.Chisholm ST, Coaker G, Day B, Staskawicz BJ. Host-microbe interactions: Shaping the evolution of the plant immune response. Cell. 2006;124:803–814. doi: 10.1016/j.cell.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 5.Schwessinger B, Zipfel C. News from the frontline: Recent insights into PAMP-triggered immunity in plants. Curr Opin Plant Biol. 2008;11:389–395. doi: 10.1016/j.pbi.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 6.Li J, Brader G, Palva ET. Kunitz trypsin inhibitor: An antagonist of cell death triggered by phytopathogens and fumonisin b1 in Arabidopsis. Mol Plant. 2008;1:482–495. doi: 10.1093/mp/ssn013. [DOI] [PubMed] [Google Scholar]
- 7.Rippa S, Eid M, Formaggio F, Toniolo C, Béven L. Hypersensitive-like response to the pore-former peptaibol alamethicin in Arabidopsis thaliana. ChemBioChem. 2010;11:2042–2049. doi: 10.1002/cbic.201000262. [DOI] [PubMed] [Google Scholar]
- 8.Boller T, Felix G. A renaissance of elicitors: Perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol. 2009;60:379–406. doi: 10.1146/annurev.arplant.57.032905.105346. [DOI] [PubMed] [Google Scholar]
- 9.Dodds PN, Rathjen JP. Plant immunity: Towards an integrated view of plant-pathogen interactions. Nat Rev Genet. 2010;11:539–548. doi: 10.1038/nrg2812. [DOI] [PubMed] [Google Scholar]
- 10.Jones JDG, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 11.Kunze G, et al. The N terminus of bacterial elongation factor Tu elicits innate immunity in Arabidopsis plants. Plant Cell. 2004;16:3496–3507. doi: 10.1105/tpc.104.026765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zipfel C, et al. Bacterial disease resistance in Arabidopsis through flagellin perception. Nature. 2004;428:764–767. doi: 10.1038/nature02485. [DOI] [PubMed] [Google Scholar]
- 13.Ingle RA, Carstens M, Denby KJ. PAMP recognition and the plant-pathogen arms race. Bioessays. 2006;28:880–889. doi: 10.1002/bies.20457. [DOI] [PubMed] [Google Scholar]
- 14.Bent AF, Mackey D. Elicitors, effectors, and R genes: The new paradigm and a lifetime supply of questions. Annu Rev Phytopathol. 2007;45:399–436. doi: 10.1146/annurev.phyto.45.062806.094427. [DOI] [PubMed] [Google Scholar]
- 15.Urwin R, Holmes EC, Fox AJ, Derrick JP, Maiden MC. Phylogenetic evidence for frequent positive selection and recombination in the meningococcal surface antigen PorB. Mol Biol Evol. 2002;19:1686–1694. doi: 10.1093/oxfordjournals.molbev.a003991. [DOI] [PubMed] [Google Scholar]
- 16.Ross HA, Rodrigo AG. Immune-mediated positive selection drives human immunodeficiency virus type 1 molecular variation and predicts disease duration. J Virol. 2002;76:11715–11720. doi: 10.1128/JVI.76.22.11715-11720.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frank SA. Immunology and Evolution of Infectious Disease. Princeton, NJ: Princeton Univ Press; 2002. [PubMed] [Google Scholar]
- 18.Sun WX, Dunning FM, Pfund C, Weingarten R, Bent AF. Within-species flagellin polymorphism in Xanthomonas campestris pv campestris and its impact on elicitation of Arabidopsis FLAGELLIN SENSING2-dependent defenses. Plant Cell. 2006;18:764–779. doi: 10.1105/tpc.105.037648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Felix G, Duran JD, Volko S, Boller T. Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J. 1999;18:265–276. doi: 10.1046/j.1365-313x.1999.00265.x. [DOI] [PubMed] [Google Scholar]
- 20.Andersen-Nissen E, et al. Evasion of Toll-like receptor 5 by flagellated bacteria. Proc Natl Acad Sci USA. 2005;102:9247–9252. doi: 10.1073/pnas.0502040102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cai RM, et al. The plant pathogen Pseudomonas syringae pv. tomato is genetically monomorphic and under strong selection to evade tomato immunity. PLoS Path. 2011;7:e1002130. doi: 10.1371/journal.ppat.1002130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gómez-Gómez L, Felix G, Boller T. A single locus determines sensitivity to bacterial flagellin in Arabidopsis thaliana. Plant J. 1999;18:277–284. doi: 10.1046/j.1365-313x.1999.00451.x. [DOI] [PubMed] [Google Scholar]
- 23.Wall DP, Fraser HB, Hirsh AE. Detecting putative orthologs. Bioinformatics. 2003;19:1710–1711. doi: 10.1093/bioinformatics/btg213. [DOI] [PubMed] [Google Scholar]
- 24.Yang ZH, Nielsen R, Goldman N, Pedersen AMK. Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics. 2000;155:431–449. doi: 10.1093/genetics/155.1.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 26.Yang ZH, Wong WSW, Nielsen R. Bayes empirical bayes inference of amino acid sites under positive selection. Mol Biol Evol. 2005;22:1107–1118. doi: 10.1093/molbev/msi097. [DOI] [PubMed] [Google Scholar]
- 27.Ham JH, Kim MG, Lee SY, Mackey D. Layered basal defenses underlie non-host resistance of Arabidopsis to Pseudomonas syringae pv. phaseolicola. Plant J. 2007;51:604–616. doi: 10.1111/j.1365-313X.2007.03165.x. [DOI] [PubMed] [Google Scholar]
- 28.Kim MG, Mackey D. Measuring cell-wall-based defenses and their effect on bacterial growth. In: Ewbank J, Vivier E, editors. Arabidopsis. Methods in Molecular Biology: Innate Immunity. Vol 415. Totowa, NJ: Humana; 2007. [Google Scholar]
- 29.Sheffield JB. ImageJ, a useful tool for biological image processing and analysis. Microsc Microanal. 2007;13:200–201. [Google Scholar]
- 30.Matzinger P. The danger model: A renewed sense of self. Science. 2002;296:301–305. doi: 10.1126/science.1071059. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.