Abstract
Mammalian cells are capable of delivering multiple types of membrane capsules extracellularly. The limiting membrane of late endosomes can fuse with the plasma membrane, leading to the extracellular release of multivesicular bodies (MVBs), initially contained within the endosomes, as exosomes. Budding viruses exploit the TSG101 protein and endosomal sorting complex required for transport (ESCRT) machinery used for MVB formation to mediate the egress of viral particles from host cells. Here we report the discovery of a virus-independent cellular process that generates microvesicles that are distinct from exosomes and which, like budding viruses, are produced by direct plasma membrane budding. Such budding is driven by a specific interaction of TSG101 with a tetrapeptide PSAP motif of an accessory protein, arrestin domain-containing protein 1 (ARRDC1), which we show is localized to the plasma membrane through its arrestin domain. This interaction results in relocation of TSG101 from endosomes to the plasma membrane and mediates the release of microvesicles that contain TSG101, ARRDC1, and other cellular proteins. Unlike exosomes, which are derived from MVBs, ARRDC1-mediated microvesicles (ARMMs) lack known late endosomal markers. ARMMs formation requires VPS4 ATPase and is enhanced by the E3 ligase WWP2, which interacts with and ubiquitinates ARRDC1. ARRDC1 protein discharged into ARMMs was observed in co-cultured cells, suggesting a role for ARMMs in intercellular communication. Our findings reveal an intrinsic cellular mechanism that results in direct budding of microvesicles from the plasma membrane, providing a formal paradigm for the evolutionary recruitment of ESCRT proteins in the release of budding viruses.
Keywords: Gag, receptor, ubiquitin, vesicle
The plasma membrane is a dynamic structure that can bud inward or outward to generate membrane-encapsulated vesicles (1, 2). Inward budding leads to the formation of endosomes, which can mediate the sorting and degradation of receptors and other plasma membrane proteins by the highly conserved endosomal sorting complex required for transport (ESCRT) machinery that recognizes and guides ubiquitinated receptors from plasma membranes to late endosomes and then to lysosomes, where the receptors are degraded (for reviews, see refs. 1, 3, and 4). The protein encoded by tumor susceptibility gene 101 (TSG101), an essential ESCRT component, is central to endosomal sorting (5, 6). Using a unique ubiquitin enzyme variant (UEV) domain that binds to ubiquitin and recognizes the tetrapeptide protein motif PS/TAP (7–9), TSG101 interacts with the early endosomal protein HRS (6, 10, 11) and the late endosome-associated protein ALIX (12) to mediate the trafficking of ubiquitinated receptors from early endosomes to late endosomes; there, additional ESCRT proteins and an ATPase (VPS4) drive the formation and delivery of endosomal cargo to endosomal membrane-derived vesicles known as “multivesicular bodies” (MVBs) (3, 13). Fusion of MVBs with lysosomes leads to degradation of the cargo, whereas trafficking and fusion of MVB-containing late endosomes with the plasma membrane deliver small vesicles termed “exosomes” into the extracellular space (2, 14, 15).
Outward budding of plasma membranes occurs during egress of budding viruses from cells. The best-characterized example of viral budding, which hijacks the ESCRT machinery, involves HIV (16–18). HIV budding is driven by the viral Gag protein, which localizes to the plasma membrane of host cells. In the late domain of HIV Gag is a critical PTAP motif that interacts with the UEV domain of TSG101 (19, 20), enabling Gag to recruit TSG101 and other ESCRT proteins, which normally reside on endosomal membranes, to the cell surface (12, 21, 22). By such recruitment, Gag can initiate plasma membrane budding. Disruption of the Gag–TSG101 interaction either through mutation of the PTAP motif in Gag or overexpression of mutant TSG101 can block the release of HIV viral particles (23–25). Many other viruses also exploit TSG101 or other ESCRT components to aid their budding at the cell surface (26, 27).
Although viral budding and MVB formation are topologically similar, in that both processes use the ESCRT machinery and involve the budding of membranes away from the cytosol, the two events in fact are functionally and structurally distinct. MVBs are formed by invagination of late endosome limiting membranes into the lumen of the organelle, whereas budding of viruses normally occurs at the cell surface and results in release of viral particles from the cytoplasm extracellularly (i.e., evagination). Thus, the analogy between MVB formation and viral budding (18, 26) is incomplete. Here we report an intrinsic cellular process, termed “direct plasma membrane budding” (DPMB), that underlies MVB-independent vesicle formation at the cell surface. We show that DPMB, which involves the recruitment of TSG101 to the cell surface by arrestin domain-containing protein 1 (ARRDC1) and is mechanistically distinct from MVB formation and from MVB-related exosome formation, leads directly to evagination of the plasma membrane and the release of a previously undescribed type of microvesicle into the extracellular space.
Results
ARRDC1 Recruits TSG101 to the Plasma Membrane.
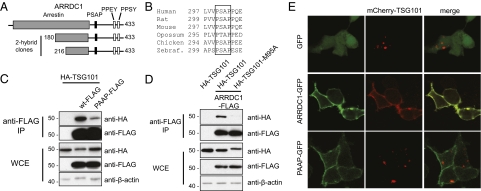
Using a yeast two-hybrid screen and the UEV domain of TSG101 as the bait, we identified two independent clonal isolates that correspond to the C-terminal fragments of ARRDC1 (Fig. 1A). This finding is consistent with a previous report of coimmunoprecipitation of ARRDC1 with TSG101 (28). ARRDC1 contains a highly conserved PSAP motif (Fig. 1B), and mutational alteration of this motif to PAAP markedly reduced the interaction between ARRDC1 and TSG101 (Fig. 1C). Additionally a mutation of amino acid residue methionine 95 of TSG101, which is known to be required for PS/TAP binding (7, 8), abolished the ARRDC1–TSG101 interaction (Fig. 1D). Together, these results demonstrate that ARRDC1 interacts with TSG101 through specific UEV/PSAP binding.
Fig. 1.
ARRDC1 interacts with and recruits TSG101 to the cell membrane via a conserved PSAP motif. (A) Schematic representation of the ARRDC1 protein, indicating relevant structural domains: the N-terminal arrestin domain (gray box), the C-terminal PSAP motif (filled box), and PPXY motifs (open boxes). Also depicted are the ARRDC1 fragments identified in the yeast two-hybrid screen using the UEV domain of TSG101 as the bait. (B) Alignment of fragments of ARRDC1 orthologs showing conservation of the PSAP motif. (C) Western blot analysis of the interaction between FLAG-tagged wild-type (wt) ARRDC1 or the PSAP mutant (PAAP) and HA-tagged TSG101. Anti-FLAG immunoprecipitates (IP) and whole-cell extracts (WCE) from HEK293T cells transfected with the indicated constructs were examined by immunoblotting using the indicated antibodies. (D) Western blot analysis of the interaction between HA-tagged wild-type or a mutant (M95A) TSG101 and ARRDC1, as in C. (E) Localization of mCherry-TSG101 fusion protein in HEK293T cells cotransfected with GFP, ARRDC1-GFP, or its PSAP mutant (PAAP-GFP). Cells were fixed and examined by confocal microscopy. More than 20 cells expressing both ARRDC1-GFP and mcherry-TSG101 were examined, and all showed a similar localization pattern. Representative images are shown in the figure.
To understand better the biological consequences of the ARRDC1–TSG101 interaction, we investigated the cellular locations of individually expressed or coexpressed proteins that were differentially labeled (ARRDC1-GFP and mCherry-TSG101). We discovered that ARRDC1-GFP localized almost exclusively to the plasma membrane (Fig. 1E), a finding that was reproduced using FLAG-tagged ARRDC1 in HEK293T cells (Fig. S1A) or ARRDC1-GFP in multiple cell lines tested (Fig. S1B). Consistent with our previous study (6), adventitiously expressed TSG101 was observed in the cytoplasm in multiple punctate foci, as is characteristic of endosome-associated localization (Fig. 1E). However, when coexpressed with ARRDC1, TSG101 was almost completely redistributed to the plasma membrane (Fig. 1E). Importantly, coexpression of TSG101 with the PSAP mutant of ARRDC1 did not result in redistribution of TSG101 to the cell membrane, indicating that recruitment of TSG101 to this location is dependent on the PSAP motif in ARRDC1.
ARRDC1 Drives the Release of Extracellular Microvesicles.
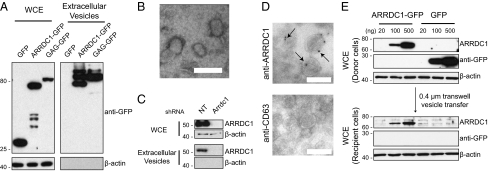
The dramatic relocalization of TSG101 from endosomes to plasma membrane by ARRDC1 is reminiscent of TSG101 recruitment by the HIV Gag protein during HIV budding (21, 22, 29). Given the striking parallel between ARRDC1 and Gag in recruitment of TSG101 from the cytosol to the plasma membrane, we hypothesized that ARRDC1 may function like Gag to mediate the direct release of plasma membrane-derived vesicles. To test this hypothesis, we expressed GFP, ARRDC1-GFP, or Gag-GFP in HEK293T cells and 24 h later harvested the conditioned medium and collected extracellular vesicles that were released by the cultured cells. Western blot analysis showed that Gag-GFP and ARRDC1-GFP proteins were robustly present in extracellular vesicles, but the control GFP protein was not (Fig. 2A). Consistent with this finding, ARRDC1 was included among the many proteins identified by mass spectrometry in microvesicles derived from human urine (30), bladder (31), and colon cancer cells (32). Electron microscopic examination of the released extracellular vesicles from ARRDC1-transfected cells indicated that the vesicles are <100 nm in diameter with a mean diameter of ∼45 nm (Fig. 2B).
Fig. 2.
ARRDC1 expression potentiates release of vesicles that are devoid of exosomal markers and can fuse with recipient cells. (A) Western blot analysis, using the indicated antibodies, of whole-cell extracts and of the corresponding extracellular vesicles produced by HEK293T cells transfected with constructs expressing GFP, wild-type ARRDC1-GFP, or HIV GAG-GFP. (B) Electron microscopy of extracellular vesicles derived from HEK293T cells transfected with an ARRDC1 expression construct. The average diameters of ∼ 100 individual extracellular microvesicles were measured using the ImageJ software (NIH), and utilized to calculate the mean diameter. (Scale bar, 100 nm.) (C) Western blot analysis of whole-cell extracts and extracellular vesicles from HEK293T cells transfected with nontargeting shRNA or ARRDC1-targeting shRNA lentiviruses. (D) Anti-ARRDC1 and anti-CD63 immunogold labeling of collagen-embedded extracellular vesicle cryosections. Arrows indicate immunogold-positive labeling. (Scale bars, 100 nm.) (E) Transfer of ARRDC1-containing vesicles from donor to recipient cells. Cells transfected by increasing amounts of constructs expressing ARRDC1-GFP or GFP were washed and seeded in 0.4-μm transwells atop untransfected cells. Western blot analysis of the corresponding lysates from transfected donor cells and untransfected recipient cells was carried out using the indicated antibodies.
We next determined whether native, endogenously produced ARRDC1 is released into microvesicles. As shown in Fig. 2C, extracellular vesicles collected from HEK293T cells contained ARRDC1 protein, indicating that ARRDC1 release into extracellular microvesicles is a process intrinsic to these cells. Additionally, depletion of ARRDC1 by shRNA resulted in loss of the extracellular ARRDC1 immunopositive signal (Fig. 2C), indicating that the released ARRDC1 protein is of cellular origin and is not derived from the FBS supplementing the culture medium. To demonstrate further the presence of ARRDC1 in the microvesicles we collected, we prepared collagen-embedded sections of extracellular vesicle pellets collected from HEK293T cells and stained the vesicles with ARRDC1 antibody. ARRDC1 indeed was detected in the microvesicles (Fig. 2D, Upper). In contrast, these microvesicles stained negative for the exosomal marker LAMP3/CD63 (Fig. 2D, Lower). We refer to these extracellular microvesicles as “ARRDC1-mediated microvesicles” (ARMMs).
Using a co-culture system, we observed that ARRDC1 protein contained within ARMMs can be transferred between cells. ARMMs donor cells transfected with control GFP or ARRDC1-GFP were seeded atop untransfected recipient cells on a 0.4-μm porous membrane in a transwell (Materials and Methods). After 30 h, cell lysates were collected from both donor and recipient cells and were analyzed for ARRDC1-GFP or GFP. As shown in Fig. 2E, ARRDC1-GFP but not discrete GFP was detected in the recipient cells, indicating transfer of ARRDC1, which served in these experiments as a surrogate marker for ARMMs, to the recipient cells. Furthermore, the amount of ARRDC1 transferred to recipient cells was proportional to the abundance of adventitiously expressed ARRDC1 in the donor cells, providing further evidence of the functional role of ARRDC1 in ARMMs formation and indicating ARMMs-mediated protein transfer.
Release of ARMMs Requires TSG101 and VPS4 ATPase.
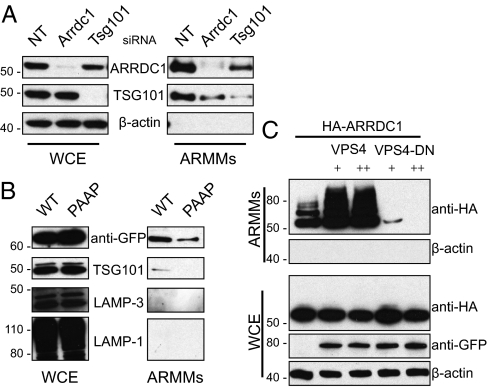
TSG101 is essential for MVB formation (33) and HIV budding (19). We determined whether the release of ARMMs also requires TSG101. As shown in Fig. 3A, knockdown of TSG101 expression reduced ARRDC1 in ARMMs by ∼two thirds compared with control. TSG101 also was released into ARMMs, as occurs for viral particles produced by Gag-mediated budding (34). Moreover, when ARRDC1 was reduced, TSG101 abundance in ARMMs was also reduced by ∼70%. These data indicate a mutual dependency of ARRDC1 and TSG101 for inclusion in ARMMs. We next determined whether ARMMs release specifically requires the ARRDC1–TSG101 interaction. As shown in Fig. 3B, PSAP-mutant ARRDC1 exhibited greatly decreased ARMMs release. Moreover, TSG101 was undetectable in culture medium fractions containing vesicles produced by cells expressing the ARRDC1 PSAP mutant (Fig. 3B). Collectively, these findings indicate that ARMMs release requires both TSG101 and its interaction with ARRDC1. Consistent with our observations using immunogold staining (Fig. 2D), known exosomal markers LAMP3/CD63 and LAMP1 were not detected in the ARRDC1-containing vesicles by Western blot analysis (Fig. 3B), indicating the non-exosomal nature of ARMMs.
Fig. 3.
The ARRDC1–TSG101 interaction and the budding-related protein VPS4a are required for release of ARMMs. (A) ARRDC1 and TSG101 are required for ARMMs release. Western blot analysis of ARMMs and whole-cell extracts from HEK293T cells subjected to transfection by indicated siRNAs. The results shown are representative of three independent replication experiments. (B) PSAP-mutant ARRDC1 is released less efficiently into ARMMs. Cells were transfected with wild-type or PSAP-mutant (PAAP) ARRDC1-GFP, and the corresponding whole-cell extracts and ARMMs were analyzed by Western blotting probed by the indicated antibodies. (C) ARMMs release requires functional VPS4 ATPase activity. HEK293T cells were cotransfected with constructs expressing HA-ARRDC1 and 0.5 or 1 μg of VPS4a (VPS4) or VPS4a E228Q (VPS4-DN) GFP fusion proteins. Whole-cell extracts and ARMMs were analyzed by Western blotting using the indicated antibodies.
The VPS4 ATPase catalyzes the final “pinch-off” of the membranes in the formation of both MVBs and viral buds (35, 36) and is another essential component of the endosomal-sorting pathway. We found that this ATPase also is necessary for ARMMs production. As shown in Fig. 3C, although expression of wild-type VPS4 enhanced ARMMs release, expression of a catalytically inactive mutant of the ATPase (E228Q) almost completely blocked such release. The inhibitory effect is greater than was observed for TSG101 knockdown and may be attributable to the more potent effect by the VPS4 dominant-negative overexpression on the function of the ESCRT pathway. Similarly greater effects of VPS4 in comparison with TSG101 have been observed for HIV Gag budding (19). Together, our data argue strongly that the ESCRT pathway is essential for the release of ARMMs, as it is for MVB formation and viral budding.
Arrestin Domain-Mediated ARRDC1 Localization at the Plasma Membrane Is Required for ARMMs Release.
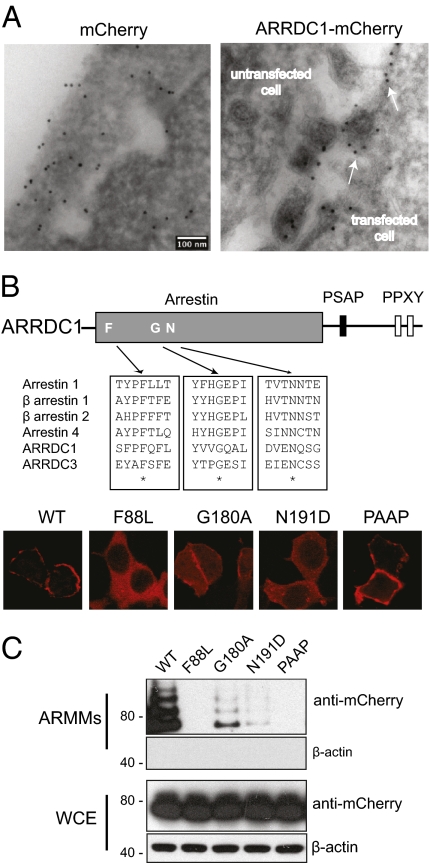
To establish further that ARMMs are derived from the plasma membrane, we examined frozen sections of ARRDC1-expressing cells by electron microscopy. Cells were transfected with constructs expressing mCherry or ARRDC1-mCherry and treated with immunogold-labeled anti-mCherry antibody. While mCherry displayed ubiquitous localization, ARRDC1-mCherry was limited to the cell membrane, and staining was observed specifically in budding vesicles emanating from the cell surface and also in ARMMs secreted into the extracellular space (Fig. 4A). This result supports the notion that ARMMs originate from and are formed at the cell surface.
Fig. 4.
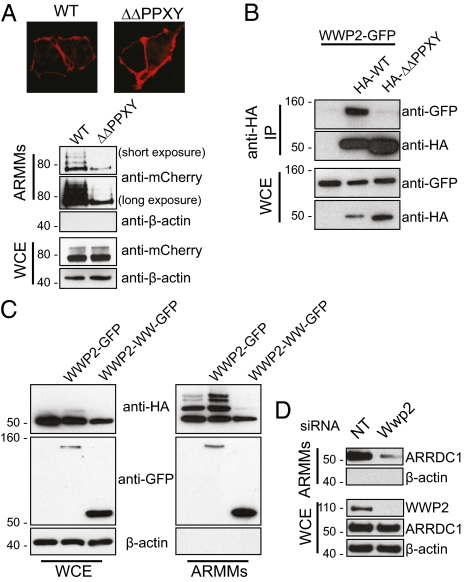
Arrestin domain-mediated association of ARRDC1 with the plasma membrane is essential for cell surface-derived budding of ARMMs. (A) ARMMs bud at the cell surface. Cryosections of HEK293T cells transfected with constructs expressing mCherry or ARRDC1-mCherry were immunogold labeled with anti-mCherry. White arrows (Right) indicate presumptive sites of outward budding at the cell membrane adjacent to immunogold-positive ARMMs captured from the intercellular space. An untransfected cell is indicated in the same field and shows no staining. (Scale bar, 100 nm.) (B) Disruption of conserved arrestin residues alters localization of ARRDC1. Conserved arrestin-domain residues among arrestins 1–4 and the known membrane-associated ARRDC1 and ARRDC3 were identified by ClustalW alignment and were mutated. The corresponding mCherry fusion mutants were expressed in HEK293T cells and showed altered localization compared with wild-type or PAAP ARRDC1. (C) HEK293T cells were transfected with constructs expressing the indicated ARRDC1 mutants fused to mCherry. The corresponding ARMMs and whole-cell extracts were analyzed by Western blotting using the indicated antibodies.
ARRDC1 contains an N-terminal domain that is homologous to arrestins (37). The arrestin proteins and particularly the β-arrestins have been shown to associate with receptors at the cell membrane to regulate their signaling (38). We hypothesized that the arrestin domain of ARRDC1 mediates its localization to the plasma membrane. To find highly conserved residues that might be important for the localization of ARRDC1 to the plasma membrane, we aligned amino acid sequences of human arrestin paralogs with ARRDC1 and with another ARRDC protein, ARRDC3, which also is known to localize to plasma membrane (Fig. 4B, Upper) (39, 40). Several conserved residues (F88, G180, and N191) in the arrestin domain of ARRDC1 were identified. We mutated these conserved residues and determined the cellular location of the corresponding ARRDC1-mCherry fusion proteins. The F88L mutation completely abolished ARRDC1 association with the plasma membrane, leaving the mutant protein evenly distributed in the cytosol, whereas the G180A and N191D mutants retained partial plasma membrane localization (Fig. 3B). Consistent with data shown in Fig. 1E, the plasma membrane localization of ARRDC1 protein was not perturbed by mutation of PSAP to PAAP. We then examined the effects of arrestin-domain mutants on ARMMs release. Unlike PAAP-mutant ARRDC1, which localizes to the plasma membrane but is not released, arrestin-domain mutants of ARRDC1 proteins interfered with ARMMs release in a manner that correlated with the extent of the disruption of plasma membrane association by the mutations (Fig. 4C). The F88L mutant completely blocked ARMMs release; the other two mutants had partial inhibitory effects (Fig. 4C). These results demonstrate that the arrestin domain mediates ARRDC1 localization to the plasma membrane and is required for the release of ARMMs.
Ubiquitination of ARRDC1 by HECT Domain Ubiquitin Ligase WWP2 Facilitates ARMMs Release.
ARRDC1 contains two highly conserved PPXY motifs near its C terminus. Although cellular localization of PPXY-mutant ARRDC1 proteins was not perturbed, expression of these proteins markedly inhibited ARMMs release (Fig. 5A). In addition, we observed a corresponding decrease in ARRDC band laddering (Fig. 5A). Such laddering is suggestive of ubiquitination, and to learn whether ARRDC1 in ARMMs is in fact ubiquitinated, we cotransfected HEK293T cells with constructs expressing HA-tagged ubiquitin and ARRDC1-GFP, and collected ARMMs for immunoprecipitation by anti-HA antibody. As shown in Fig. S2, total lysates of ARMMs collected from ARRDC1-expressing cells also showed increased ubiquitination as compared with a GFP control (Fig. S2), supporting the notion that the observed laddering of ARMMs-associated ARRDC1 protein results from ubiquitination and additionally suggesting that ubiquitination facilitates ARMMs release.
Fig. 5.
WWP2 interacts with and ubiquitinates ARRDC1 to enhance ARMMs release. (A) Deletion of PPXY motifs reduced ARRDC1 ubiquitination and ARMMs release. HEK293T cells were transfected with wild-type or PPXY-deleted (ΔΔPPXY) ARRDC1 fused to mCherry and visualized by confocal microscopy. The corresponding cell lysates and ARMMs were analyzed by Western blotting as indicated. (B) PPXY motifs in ARRDC1 interact with WWP2. HEK293T cells were cotransfected with GFP-tagged WWP2 and HA-tagged wild-type or ΔΔPPXY ARRDC1. Anti-HA immunoprecipitates and whole-cell extracts were examined by Western blotting as indicated. (C) WWP2 expression potentiates ARRDC1 ubiquitination in ARMMs. HEK293T cells were cotransfected with constructs expressing HA-ARRDC1 and WWP2-GFP or the WW domain of WWP2 fused to GFP. Whole-cell extracts and ARMMs from each transfected group were analyzed by immunoblotting as indicated. (D) WWP2 knockdown diminishes ARRDC1 in ARMMs. HEK293T was subjected to transfection with nontargeting (NT) or WWP2-targeting siRNAs. Corresponding whole-cell extracts and ARMMs were analyzed by Western blotting.
To identify possible candidates for the ubiquitin E3 ligase that mediates ARRDC1 ubiquitination and which may have a role in ARMMs release, anti-ARRDC1 rabbit antibody was used to immunoprecipitate endogenous ARRDC1 and associated proteins from HEK293T cell lysates, and immunoprecipitates were analyzed by mass spectrometry. Among the proteins identified in ARRDC1 immunoprecipitates was WWP2, which is a member of the NEDD4 E3 ligase family (41, 42). We confirmed the interaction between ARRDC1 and WWP2 by coimmunoprecipitation (Fig. 5B). WWP2 contains WW domains that are known to interact with PPXY motifs (41, 42). We found that mutations in the PPXY domains in ARRDC1 almost completely inhibited its interaction with WWP2 (Fig. 5B). Additionally, as shown in Fig. 5C, ubiquitination of ARRDC1 in ARMMs was enhanced by WWP2 but was not observed in cells expressing the fragment containing only the WW domains, indicating a dominant-negative effect of the WW domain expression on ARRDC1 ubiquitination. Interestingly, both WWP2 and the WW domain fragment also were detected in extracellular vesicle fractions, consistent with the observed WW domain-mediated interaction between ARRDC1 and WWP2. Furthermore, a specific role for WWP2 in ARMMs release was demonstrated, because siRNA-mediated WWP2 knockdown markedly reduced the amount of ARRDC1 released in ARMMs (Fig. 5D). The data obtained in this series of experiments indicate that WWP2, through interaction with PPXY motifs in ARRDC1, ubiquitinates ARRDC1 and enhances ARMMs release.
Discussion
Mammalian cells are capable of secreting into the extracellular milieu a variety of microvesicles, among which are exosomes, which are derived from MVBs of late endosomes (2, 14, 43). The results we report reveal a type of microvesicle that is generated by DPMB and is distinct from exosomes. Although TSG101 is located at the surface of late endosomes during the formation of MVBs, TSG101 is recruited by ARRDC1 to the surface of cells for production of ARMMs. Consistent with the plasma membrane origin of ARMMs, our data show that ARMMs lack late endosomal markers such as CD63 and LAMP1 (Figs. 2D and 3B).
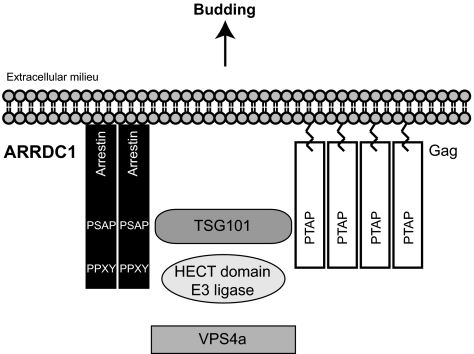
Our findings suggest a direct analogy between ARMMs formation and budding mediated by Gag and other viral proteins (Fig. 6). It has been well established that many viruses, including HIV, use cellular machinery for their egress (16–18). HIV Gag uses PTAP motifs to recruit TSG101 and other endosomal pathway proteins to bud from the cell surface. It has been suggested that Gag protein functionally mimics the host protein Hrs, which interacts with TSG101 through a PSAP motif (11). However, although Hrs and Gag interact similarly with TSG101, Hrs is an early endosome-associated protein and does not localize to the plasma membrane where viral budding occurs (6, 44). In contrast, ARRDC1 is directed to the plasma membrane by its arrestin-like domain and multimerizes there (Fig. S3), as does Gag (45). Moreover, both Gag and ARRDC1, when adventitiously expressed, are sufficient to induce vesicle formation. Additionally, our results demonstrate that, like HIV Gag budding, ARMMs release requires both TSG101 and VPS4. These data strongly suggest that HIV Gag and likely other viral proteins mimic ARRDC1 to mediate the release of viral particles from host cells. Whether ARMMs production also requires ESCRT-III and ALIX which are involved in Gag-mediated HIV budding (46), is not known at present. Recently, the shedding of ESCRT-dependent microvesicles that may prove to be ARMMs has been found to be increased in Caenorhabditis elegans embryos by loss of the evolutionarily conserved P4-ATPase, TAT-5, and consequent exposure of phosphatidylethanolamine at the cell surface (47).
Fig. 6.
A model for ARMMs formation and Gag-mediated viral budding. Both ARRDC1 and Gag interact with TSG101, are ubiquitinated, and require VPS4 ATPase to produce budding vesicles.
ARRDC1 belongs to a family of ARRDC proteins that share a common domain structure: an arrestin domain at the N termini and two PPXY motifs at the C termini (37). ARRDC1 contains a highly conserved PSAP motif that is not found in other ARRDC proteins. The PSAP motif interacts directly with TSG101 and, in cells that adventitiously overexpress ARRDC1, redirects TSG101 from endosomes to the cell surface. Potentially, such ARRDC1-mediated relocalization of TSG101 may alter endosomal trafficking and sorting and, consequently, signal transduction by receptors subjected to endosomal sorting mechanisms. Because the arrestin domain of ARRDC1 mediates localization of this protein to the plasma membrane and because β-arrestins are known to bind to phosphorylated receptor proteins (38), we suggest that ARRDC1 may prove to be a multifaceted regulator of receptor-mediated signaling.
Our data also indicate a role for ARRDC1 ubiquitination in ARMMs release. We showed that ARRDC1 PPXY motifs interact with the ubiquitin E3 ligase WWP2 (Fig. 5B) and that WWP2 mediates the ubiquitination of ARRDC1, in particular the ARRDC1 species that are incorporated into ARMMs. A recent study has shown that another E3 ligase, WWP1, also can interact with and ubiquitinate ARRDC1 (28). It remains unclear whether WWP1 also has a role in ARMMs production. Interestingly, our previous study showed that the ARRDC1 relative ARRDC3 also interacts with members of the NEDD4 E3 ligase family (40). Through interaction with NEDD4 E3 ligases, ARRDC3 mediates ubiquitination of β2-adrenergic receptors. Whether ARRDC1 exerts a similar ubiquitination adaptor function has not been determined.
Extracellular microvesicles have the potential to function as mediators of cell–cell communication (2). Indeed, several studies have identified functional proteins and RNA molecules in exosomal microvesicles and have shown that macromolecules transferred by exosomes can produce functional effects in recipient cells (48, 49). ARMMs may function similarly in cellular communication, and, consistent with this notion, we found that ARMMs can transfer ARRDC1 between cells. We speculate that ARMMs, like exosomes, may contain RNAs or additional proteins involved in cell–cell communication, and, because ARRDC1 is able to localize to the plasma membrane, we also hypothesize that some of these proteins may be plasma membrane receptors. Identification of the potential signaling proteins or RNAs in ARMMs will facilitate elucidation of the biological role of these microvesicles.
Materials and Methods
Additional information about materials and methods can be found in SI Materials and Methods.
Isolation and Transfer of ARMMs.
Culture medium atop nascent or transfected cells was collected and precleared of cellular debris by two consecutive centrifugations (500 × g and 2,000 × g). The medium then was passed through a 0.2-μm filter (Sarstedt) and subjected to ultracentrifugation using the Beckman LTA110 rotor in an Optima LTX centrifuge (Beckman) or using the SW41 Ti rotor in a L8-M centrifuge (Beckman) at 120,000 × g for 2 h, depending on the volume of centrifugate. Medium then was aspirated, and pellets were washed twice with ice-cold PBS. ARMMs then were resuspended either in 1.3× lithium dodecyl sulfate (Invitrogen) supplemented with β-mercaptoethanol or in PBS. To calibrate ARMMs production with cell number, protein concentrations of the corresponding cytosolic lysates were measured using the Protein 660 nm assay (Pierce). For ARMMs transfer assays, transfected cells were washed thoroughly and seeded atop a 0.4-μm transwell membrane (Costar) for 16 h; then the transwell was transferred to a plate containing untransfected HEK293T cells. ARMMs transfer was allowed to proceed for 30 h before harvesting. Alternatively, purified ARMMs resuspended in PBS were added to culture medium containing untransfected HEK293T cells and were incubated for 24–30 h before harvesting.
Immunogold Labeling and Electron Microscopy.
For immunogold staining of budding vesicles or vesicles in intercellular spaces, cells were washed, gently detached with 0.5 mM EDTA, and layered on a cushion of 8% (wt/vol) paraformaldehyde (PFA) in 0.1 M sodium phosphate buffer. Cell pellets were collected by centrifugation at 2,000 × g for 3 min, and pellets were replenished with 4% (wt/vol) PFA fixative for 2 h at room temperature before freezing and processing for visualization.
Supplementary Material
Acknowledgments
We thank Maria Ericsson (Harvard Medical School electron microscopy facility) for her expert help in electron microscopy. This work was supported by startup funding from the Harvard School of Public Health, by a Biomedical Research grant from the American Lung Association, and by National Institute of Environmental Health Sciences Superfund Research Program Grant P42ES016454 (to Q.L.). R.H. is supported by a doctoral research award from the Canadian Institutes of Health Research. S.N.C. received support from a K.-T. Li Professorship from the Friends of Stanford University Foundation in Taiwan.
Footnotes
The authors declare no conflict of interest.
See Commentary on page 4025.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1200448109/-/DCSupplemental.
References
- 1.Hurley JH, Boura E, Carlson LA, Różycki B. Membrane budding. Cell. 2010;143:875–887. doi: 10.1016/j.cell.2010.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Théry C, Ostrowski M, Segura E. Membrane vesicles as conveyors of immune responses. Nat Rev Immunol. 2009;9:581–593. doi: 10.1038/nri2567. [DOI] [PubMed] [Google Scholar]
- 3.Henne WM, Buchkovich NJ, Emr SD. The ESCRT pathway. Dev Cell. 2011;21:77–91. doi: 10.1016/j.devcel.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 4.Katzmann DJ, Odorizzi G, Emr SD. Receptor downregulation and multivesicular-body sorting. Nat Rev Mol Cell Biol. 2002;3:893–905. doi: 10.1038/nrm973. [DOI] [PubMed] [Google Scholar]
- 5.Babst M, Odorizzi G, Estepa EJ, Emr SD. Mammalian tumor susceptibility gene 101 (TSG101) and the yeast homologue, Vps23p, both function in late endosomal trafficking. Traffic. 2000;1:248–258. doi: 10.1034/j.1600-0854.2000.010307.x. [DOI] [PubMed] [Google Scholar]
- 6.Lu Q, Hope LW, Brasch M, Reinhard C, Cohen SN. TSG101 interaction with HRS mediates endosomal trafficking and receptor down-regulation. Proc Natl Acad Sci USA. 2003;100:7626–7631. doi: 10.1073/pnas.0932599100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pornillos O, Alam SL, Davis DR, Sundquist WI. Structure of the Tsg101 UEV domain in complex with the PTAP motif of the HIV-1 p6 protein. Nat Struct Biol. 2002;9:812–817. doi: 10.1038/nsb856. [DOI] [PubMed] [Google Scholar]
- 8.Pornillos O, et al. Structure and functional interactions of the Tsg101 UEV domain. EMBO J. 2002;21:2397–2406. doi: 10.1093/emboj/21.10.2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sundquist WI, et al. Ubiquitin recognition by the human TSG101 protein. Mol Cell. 2004;13:783–789. doi: 10.1016/s1097-2765(04)00129-7. [DOI] [PubMed] [Google Scholar]
- 10.Bache KG, Brech A, Mehlum A, Stenmark H. Hrs regulates multivesicular body formation via ESCRT recruitment to endosomes. J Cell Biol. 2003;162:435–442. doi: 10.1083/jcb.200302131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pornillos O, et al. HIV Gag mimics the Tsg101-recruiting activity of the human Hrs protein. J Cell Biol. 2003;162:425–434. doi: 10.1083/jcb.200302138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.von Schwedler UK, et al. The protein network of HIV budding. Cell. 2003;114:701–713. doi: 10.1016/s0092-8674(03)00714-1. [DOI] [PubMed] [Google Scholar]
- 13.Hurley JH, Stenmark H. Molecular mechanisms of ubiquitin-dependent membrane traffic. Annu Rev Biophys. 2011;40:119–142. doi: 10.1146/annurev-biophys-042910-155404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schorey JS, Bhatnagar S. Exosome function: From tumor immunology to pathogen biology. Traffic. 2008;9:871–881. doi: 10.1111/j.1600-0854.2008.00734.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Théry C, Zitvogel L, Amigorena S. Exosomes: Composition, biogenesis and function. Nat Rev Immunol. 2002;2:569–579. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- 16.Bieniasz PD. The cell biology of HIV-1 virion genesis. Cell Host Microbe. 2009;5:550–558. doi: 10.1016/j.chom.2009.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Demirov DG, Freed EO. Retrovirus budding. Virus Res. 2004;106:87–102. doi: 10.1016/j.virusres.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 18.Morita E, Sundquist WI. Retrovirus budding. Annu Rev Cell Dev Biol. 2004;20:395–425. doi: 10.1146/annurev.cellbio.20.010403.102350. [DOI] [PubMed] [Google Scholar]
- 19.Garrus JE, et al. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell. 2001;107:55–65. doi: 10.1016/s0092-8674(01)00506-2. [DOI] [PubMed] [Google Scholar]
- 20.VerPlank L, et al. Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55(Gag) Proc Natl Acad Sci USA. 2001;98:7724–7729. doi: 10.1073/pnas.131059198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin-Serrano J, Zang T, Bieniasz PD. HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat Med. 2001;7:1313–1319. doi: 10.1038/nm1201-1313. [DOI] [PubMed] [Google Scholar]
- 22.Martin-Serrano J, Zang T, Bieniasz PD. Role of ESCRT-I in retroviral budding. J Virol. 2003;77:4794–4804. doi: 10.1128/JVI.77.8.4794-4804.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Demirov DG, Ono A, Orenstein JM, Freed EO. Overexpression of the N-terminal domain of TSG101 inhibits HIV-1 budding by blocking late domain function. Proc Natl Acad Sci USA. 2002;99:955–960. doi: 10.1073/pnas.032511899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Göttlinger HG, Dorfman T, Sodroski JG, Haseltine WA. Effect of mutations affecting the p6 gag protein on human immunodeficiency virus particle release. Proc Natl Acad Sci USA. 1991;88:3195–3199. doi: 10.1073/pnas.88.8.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang M, Orenstein JM, Martin MA, Freed EO. p6Gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J Virol. 1995;69:6810–6818. doi: 10.1128/jvi.69.11.6810-6818.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Freed EO, Mouland AJ. The cell biology of HIV-1 and other retroviruses. Retrovirology. 2006;3:77. doi: 10.1186/1742-4690-3-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martin-Serrano J, Neil SJ. Host factors involved in retroviral budding and release. Nat Rev Microbiol. 2011;9:519–531. doi: 10.1038/nrmicro2596. [DOI] [PubMed] [Google Scholar]
- 28.Rauch S, Martin-Serrano J. Multiple interactions between the ESCRT machinery and arrestin-related proteins: Implications for PPXY-dependent budding. J Virol. 2011;85:3546–3556. doi: 10.1128/JVI.02045-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ono A, Freed EO. Cell-type-dependent targeting of human immunodeficiency virus type 1 assembly to the plasma membrane and the multivesicular body. J Virol. 2004;78:1552–1563. doi: 10.1128/JVI.78.3.1552-1563.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pisitkun T, Shen RF, Knepper MA. Identification and proteomic profiling of exosomes in human urine. Proc Natl Acad Sci USA. 2004;101:13368–13373. doi: 10.1073/pnas.0403453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Welton JL, et al. Proteomics analysis of bladder cancer exosomes. Mol Cell Proteomics. 2010;9:1324–1338. doi: 10.1074/mcp.M000063-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mathivanan S, et al. Proteomics analysis of A33 immunoaffinity-purified exosomes released from the human colon tumor cell line LIM1215 reveals a tissue-specific protein signature. Mol Cell Proteomics. 2010;9:197–208. doi: 10.1074/mcp.M900152-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Razi M, Futter CE. Distinct roles for Tsg101 and Hrs in multivesicular body formation and inward vesiculation. Mol Biol Cell. 2006;17:3469–3483. doi: 10.1091/mbc.E05-11-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hammarstedt M, Garoff H. Passive and active inclusion of host proteins in human immunodeficiency virus type 1 gag particles during budding at the plasma membrane. J Virol. 2004;78:5686–5697. doi: 10.1128/JVI.78.11.5686-5697.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Babst M. A protein's final ESCRT. Traffic. 2005;6:2–9. doi: 10.1111/j.1600-0854.2004.00246.x. [DOI] [PubMed] [Google Scholar]
- 36.Scott A, et al. Structural and mechanistic studies of VPS4 proteins. EMBO J. 2005;24:3658–3669. doi: 10.1038/sj.emboj.7600818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alvarez CE. On the origins of arrestin and rhodopsin. BMC Evol Biol. 2008;8:222. doi: 10.1186/1471-2148-8-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lefkowitz RJ, Shenoy SK. Transduction of receptor signals by beta-arrestins. Science. 2005;308:512–517. doi: 10.1126/science.1109237. [DOI] [PubMed] [Google Scholar]
- 39.Draheim KM, et al. ARRDC3 suppresses breast cancer progression by negatively regulating integrin beta4. Oncogene. 2010;29:5032–5047. doi: 10.1038/onc.2010.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nabhan JF, Pan H, Lu Q. Arrestin domain-containing protein 3 recruits the NEDD4 E3 ligase to mediate ubiquitination of the beta2-adrenergic receptor. EMBO Rep. 2010;11:605–611. doi: 10.1038/embor.2010.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chantry A. WWP2 ubiquitin ligase and its isoforms: New biological insight and promising disease targets. Cell Cycle. 2011;10:2437–2439. doi: 10.4161/cc.10.15.16874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rotin D, Kumar S. Physiological functions of the HECT family of ubiquitin ligases. Nat Rev Mol Cell Biol. 2009;10:398–409. doi: 10.1038/nrm2690. [DOI] [PubMed] [Google Scholar]
- 43.Denzer K, Kleijmeer MJ, Heijnen HF, Stoorvogel W, Geuze HJ. Exosome: From internal vesicle of the multivesicular body to intercellular signaling device. J Cell Sci. 2000;113:3365–3374. doi: 10.1242/jcs.113.19.3365. [DOI] [PubMed] [Google Scholar]
- 44.Komada M, Soriano P. Hrs, a FYVE finger protein localized to early endosomes, is implicated in vesicular traffic and required for ventral folding morphogenesis. Genes Dev. 1999;13:1475–1485. doi: 10.1101/gad.13.11.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ono A, Demirov D, Freed EO. Relationship between human immunodeficiency virus type 1 Gag multimerization and membrane binding. J Virol. 2000;74:5142–5150. doi: 10.1128/jvi.74.11.5142-5150.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fujii K, Hurley JH, Freed EO. Beyond Tsg101: The role of Alix in ‘ESCRTing’ HIV-1. Nat Rev Microbiol. 2007;5:912–916. doi: 10.1038/nrmicro1790. [DOI] [PubMed] [Google Scholar]
- 47.Wehman AM, Poggioli C, Schweinsberg P, Grant BD, Nance J. The P4-ATPase TAT-5 Inhibits the Budding of Extracellular Vesicles in C. elegans Embryos. Curr Biol. 2011;21:1951–1959. doi: 10.1016/j.cub.2011.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Skog J, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10:1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Valadi H, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.