Abstract
Machado-Joseph disease (MJD), also known as Spinocerebellar ataxia type 3 (SCA3), is the most common inherited spinocerebellar ataxia and one of many polyglutamine neurodegenerative diseases. In MJD, a CAG repeat expansion encodes an abnormally long polyglutamine (polyQ) tract in the disease protein, ATXN3. Here we review MJD, focusing primarily on the function and dysfunction of ATXN3 and on advances toward potential therapies. ATXN3 is a deubiquitinating enzyme (DUB) whose highly specialized properties suggest that it participates in ubiquitin-dependent proteostasis. By virtue of its interactions with VCP, various ubiquitin ligases and other ubiquitin-linked proteins, ATXN3 may help regulate the stability or activity of many proteins in diverse cellular pathways implicated in proteotoxic stress response, aging, and cell differentiation. Expansion of the polyQ tract in ATXN3 is thought to promote an altered conformation in the protein, leading to changes in interactions with native partners and to the formation of insoluble aggregates. The development of a wide range of cellular and animal models of MJD has been crucial to the emerging understanding of ATXN3 dysfunction upon polyQ expansion. Despite many advances, however, the principal molecular mechanisms by which mutant ATXN3 elicits neurotoxicity remain elusive. In a chronic degenerative disease like MJD, it is conceivable that mutant ATXN3 triggers multiple, interconnected pathogenic cascades that precipitate cellular dysfunction and eventual cell death. A better understanding of these complex molecular mechanisms will be important as scientists and clinicians begin to focus on developing effective therapies for this incurable, fatal disorder.
Keywords: polyglutamine disease, deubiquitinating enzyme, protein quality control, spinocerebellar ataxia, neurodegeneration, ataxin-3
1. Introduction
Many hereditary neurodegenerative diseases manifest later in life and are characterized by the progressive and selective loss of neuronal cell bodies, axons, dendrites and/ or synapses. For decades scientists have sought to clinically define specific neurodegenerative diseases and their genetic causes in order to achieve a molecular diagnosis, offer presymptomatic and prenatal testing to affected families, generate cellular and animal models toward understanding pathogenic mechanisms and facilitate the development of potential therapies. Studies over the past 20 years have established that an unusual type of mutation, dynamic repeat expansions, cause many inherited neurodegenerative diseases.
Among the dynamic repeat expansion diseases, the polyglutamine (polyQ) disorders caused by CAG repeat expansions represent the most common class, although each polyglutamine disease is relatively rare. In all polyQ diseases the CAG repeat expansion is translated into an abnormally long stretch of glutamine residues in the corresponding disease protein. Spinal Bulbar Muscular Atrophy (SBMA) was the first discovered polyQ disease, identified 20 years ago (La Spada et al., 1991). Since then nine additional polyQ diseases have been identified: the Spinocerebellar Ataxias (SCA) types 1, 2, 3 (also known as Machado-Joseph disease), 6, 7 and 17, Dentatorubral-Pallidoluysian Atrophy (DRPLA), Huntington disease (HD), and, most recently, Huntington Disease-like 2 (HDL2). All polyQ diseases are dominantly inherited disorders except SBMA, which is X-linked. The current review focuses on MJD/SCA3 and its disease protein, ataxin-3 (ATXN3).
Development of rational, targeted therapies for these diseases will be facilitated by knowing the pathogenic mechanism of the disease-causing mutation. As a class, polyQ diseases share certain features that suggest a general toxic mechanism triggered by expanded polyQ, which might be targetable in class-wide therapeutics. All ten polyglutamine diseases are characterized by selective neurodegeneration in the central nervous system (CNS) despite widespread expression of the disease proteins. Indeed there is little correlation between the expression pattern of polyQ proteins and the sites of CNS pathology. The disease proteins are widely expressed throughout the CNS with two notable exceptions: the CACNA1A calcium channel subunit in SCA6, which is mainly expressed in affected cerebellar Purkinje cells, and the androgen receptor in SBMA, which is primarily expressed in vulnerable motor neurons. Another shared feature of polyQ disease proteins is their propensity to misfold, oligomerize, and form intracellular aggregates and inclusions that constitute a pathological disease hallmark. The misfolding and aggregation of polyQ disease proteins have been targets of some proposed therapeutic strategies (Bauer and Nukina, 2009; Di Prospero and Fischbeck, 2005; Matos et al., 2011; Williams and Paulson, 2008).
Despite these shared features, however, each polyQ disease is a distinctive disorder with characteristic symptomatology and pathology occurring in specific brain regions. PolyQ disease proteins differ in size, cellular localization and biological function, suggesting that the toxic effect of a given polyQ expansion depends on the specific protein context and that the particular details of pathogenesis may be unique to each disease.
Here we review Machado-Joseph disease (MJD), also known as Spinocerebellar Ataxia Type 3 (SCA3), focusing primarily on the molecular properties of the disease protein, ATXN3, both in normal and pathogenic contexts, and on recent progress toward therapeutic development for this fatal disorder.
2. MJD
2.1. Clinical features
The discovery of MJD (OMIM#109150) illustrates the difficulty of defining a disease as a single entity when variable symptoms themselves represent a hallmark of the disease. MJD was first described in Northern American families of Azorean ancestry. Between 1972 and 1977 the disease was identified in four families, reported as four distinct entities named “Machado disease” (Nakano et al., 1972), “nigrospino-dentatal degeneration” (Woods and Schaumburg, 1972), “Joseph disease” (Rosenberg et al., 1976), and “Azorean disease of the nervous system” (Romanul et al., 1977). In 1975, Coutinho and Andrade studied 15 families from the Azorean Islands and proposed that the above mentioned diseases were simply variations of the same clinical disorder (Coutinho and Andrade, 1978). They defined it as “Machado-Joseph disease,” a single disorder characterized by an unusually high degree of clinical variability.
Most frequently in affected individuals, a slowly progressive “ataxia-plus” syndrome appears, typically beginning between the ages of 20 and 50 years (Coutinho and Andrade, 1978; Paulson, 1998 Oct 10 [updated 2011 March 17]). Cerebellar ataxia, progressive external ophthalmoplegia, dysarthria, dysphagia, pyramidal signs, dystonia, rigidity, and distal muscle atrophies are common features of MJD. The highly variable clinical presentation led to a description of four distinct clinical subtypes of MJD (Coutinho and Andrade, 1978; Lima and Coutinho, 1980; Paulson, 2007; Riess et al., 2008; Rosenberg, 1992). Type 1 begins early in life, often before age 20, may progress more quickly and is characterized by prominent pyramidal signs (rigidity and spasticity) and extrapyramidal features (bradykinesia and dystonia) as well as ataxia. Type 2, the most common type, has an intermediate age-at-onset (20–50 years) with cerebellar ataxia, progressive external ophthalmoplegia and pyramidal signs. Type 3 has a later onset (40–75 years) and is characterized by peripheral signs such as motor neuronopathy and muscle atrophy together with ataxia. Type 4, the rarest presentation, is characterized by parkinsonism associated with other core clinical features. More recently, a type 5 MJD was proposed for rare cases presenting pure spastic paraplegia (Landau et al., 2000; Sakai and Kawakami, 1996; Wang et al., 2009). Other common features not confined to a specific subtype are weight loss and restless legs syndrome (Paulson, 2007; Riess et al., 2008). Less commonly, mild cognitive and behavioral problems can be observed (Burk et al., 2003; Kawai et al., 2004).
Since the original clinical description of MJD, many affected families have been identified worldwide, both of Portuguese and nonPortuguese ancestry (Eto et al., 1990; Healton et al., 1980; Lima and Coutinho, 1980; Livingstone and Sequeiros, 1984; Sakai et al., 1983; Sequeiros and Suite, 1986; Takiyama et al., 1993; Taniguchi and Konigsmark, 1971). MJD is currently thought to be the most common dominantly inherited ataxia in the world, comprising 15–45% of dominantly inherited ataxia in different countries and ethnic populations (Margolis, 2002; Paulson, 2007; Schols et al., 2004).
2.2. The disease brain
Despite the fact that MJD is classified as a form of spincocerebellar ataxia, brain imaging and neuropathological studies indicate that the range of CNS involvement extends well beyond the brainstem and cerebellum.
Enlargement of the fourth ventricle is the most consistent feature observed by magnetic resonance imaging (MRI) in MJD. Neuroimaging studies have revealed atrophy of the pons, cerebellar vermis and hemispheres, basal ganglia (globus pallidus, caudate and putamen), midbrain and medulla oblongata (Etchebehere et al., 2001; Klockgether et al., 1998; Murata et al., 1998; Taniwaki et al., 1997; Yoshizawa et al., 2003). The atrophy in the cerebellum and brainstem is progressive and dependent on the length of the CAG repeat and the age of the patients (Abe et al., 1998; Eichler et al., 2011; Onodera et al., 1998). However, different brain regions present different rates of atrophy progression. While atrophy of the cerebellum and pontine base seem to correlate with patient’s age, atrophy of the midbrain and pontine tegmentum show no significant progression (Horimoto et al., 2008).
More recent quantitative imaging studies using large cohorts of patients show that MJD is a more widespread disorder throughout the CNS involving the cerebellar hemispheres and vermis, the thalamus, and the frontal, parietal, temporal, occipital and limbic lobes (D'Abreu et al., 2011; D'Abreu et al., 2010; De Oliveira et al., 2010; Etchebehere et al., 2001). Furthermore, magnetic resonance spectroscopy analysis of deep white matter has shown metabolic abnormalities suggestive of axonal dysfunction in MJD patients (D'Abreu et al., 2009). Glucose utilization deficits in cerebellum, brainstem and cerebral cortex can be observed in MJD carriers even before clinical signs of disease (Soong and Liu, 1998). Likewise, decreased binding for dopamine transporter in these regions and in the striatum is observed in symptomatic MJD patients (Taniwaki et al., 1997; Wullner et al., 2005). Brain abnormalities detected by advanced and quantitative neuroimaging techniques may offer effective biomarkers to monitor interventional trials in MJD patients.
Brains of MJD patients with advanced disease weigh significantly less than brains from individuals without neurological or psychiatric disease (Iwabuchi et al., 1999). Macroscopically, MJD brains show depigmentation of the substantia nigra and atrophy of the cerebellum, pons, medulla oblongata, as well as multiple cranial nerve nuclei (III to XII) (Rub et al., 2008).
Neurodegeneration in MJD was initially described in the olivopontocerebellar regions (Coutinho and Sequeiros, 1981; Ross, 1995). Recent pathoanatomical studies, however, revealed more extensive damage affecting areas of the cerebellothalamocortical motor loop, the basal ganglia-thalamocortical motor loop, and several other systems: visual, auditory, somatosensory, vestibular, oculomotor, ingestionrelated brainstem, precerebellar brainstem, cholinergic and dopaminergic midbrain, and pontine noradrenergic systems (Rub et al., 2008). Retained integrity of the cortical and subcortical regions of the limbic system and mild degeneration of the white matter of cerebellum, brainstem and spinal cord are also characteristic of MJD (Riess et al., 2008; Rub et al., 2008).
The disease protein, ATXN3, was initially reported to accumulate in neuronal nuclear inclusions (NNIs) in vulnerable regions of the MJD brain (Paulson et al., 1997b; Schmidt et al., 1998). The NNIs stain positively for ubiquitin (Ub) and contain other proteins such as Ub-like proteins, heat-shock proteins (HSPs), proteasome subunits, transcription factors, and other polyQ proteins (Chai et al., 1999a; Chai et al., 1999b; Chai et al., 2001; Mori et al., 2005; Paulson et al., 1997b; Schmidt et al., 1998; Takahashi et al., 2001). Recent studies using other pathoanatomical techniques have established the occurrence of NNIs in both affected and unaffected brain regions (Rub et al., 2008; Rüb et al., 2006; Rub et al., 2007; Yamada et al., 2004). In fact, no clear-cut correlation exists between the distribution of NNIs and the pattern of neurodegeneration, suggesting that NNIs do not play a direct role in determining the survival or death of an affected neuron (Rüb et al., 2006).
In addition to NNIs, neuronal cytoplasmic inclusions (NCIs) immunopositive for expanded polyQ ATXN3 have been described in MJD brains, displaying a similar distribution pattern as NNIs (Hayashi et al., 2003; Yamada et al., 2008; Yamada et al., 2004). NCIs are mainly Ub-negative, consist of fine granules about 1.5 µm in diameter, and are proposed to correspond to electron-dense minute structures scattered in pale, small primitive lysosomes (Yamada et al., 2002). ATXN3 aggregates are also observed in axons; these widespread axonal inclusions contain Ub and p62 and are found in fiber tracts known to degenerate in MJD (Seidel et al., 2010).
In summary, in the MJD brain different types of ATXN3 aggregates accumulate in specific cellular compartments. Defining the molecular composition of each type of aggregate structure may lead to a better understanding both of their possible role in disease pathogenesis and of their correlation to neurodegeneration and other aspects of disease.
3. The disease gene, ATXN3
3.1. Genetics of MJD
From its initial description in 1972, MJD was recognized to be a dominantly inherited genetic disorder. Eleven years later, the MJD disease gene was mapped to chromosome 14q32.1 (Takiyama et al., 1993). That same year, the presence of clinical features of ataxia apparently distinct from MJD in some French families that did not map to the SCA1 or SCA2 loci led researchers to propose the existence of a novel, dominantly inherited ataxia which they named Spinocerebellar Ataxia Type 3 (SCA3) (Gispert et al., 1993). Once the gene defect in MJD was discovered, however, it became clear that SCA3 and MJD are in fact the same disease.
The MJD1 gene was cloned in 1994 (Kawaguchi et al., 1994). Now designated ATXN3, the disease gene was found to have a polyglutamine-encoding CAG repeat that was expanded in affected individuals (Kawaguchi et al., 1994). MJD thus joined SBMA, SCA1 and HD as diseases caused by CAG/polyQ expansions.
The molecular diagnosis of MJD based on the ATXN3 CAG repeat expansion rapidly led to confirmation of the disease in families of many different ethnic origins (Gaspar et al., 2001; Higgins et al., 1996; Lindblad et al., 1996; Maciel et al., 1995). SCA3 was discovered to be the same disease as MJD (Durr et al., 1996; Haberhausen et al., 1995; Matilla et al., 1995), explaining why the disease is designated both as MJD and SCA3. Presymptomatic and prenatal testing soon became available to MJD families through genetic counseling programs (Lima et al., 2001; Maciel et al., 2001; Rolim et al., 2006; Sequeiros et al., 1998).
The expanded repeat in ATXN3 is nearly a pure CAG tract, interrupted by a single lysine codon near the beginning of the repeat (CAG)2CAAAAG(CAG)n (Kawaguchi et al., 1994). This trinucleotide repeat ranges from 12 to 44 triplets in healthy individuals and from ~60 to 87 in MJD patients (Lima et al., 2005; Maciel et al., 2001). Rare alleles of intermediate repeat length fall between the clearly normal and mutant ranges and are not associated with classical clinical features of disease (Gu et al., 2004; Maciel et al., 2001; Padiath et al., 2005; Paulson, 2007; Takiyama et al., 1997; van Alfen et al., 2001; van Schaik et al., 1997). Cases of homozygosity are extremely rare in MJD; the few described homozygous patients appear to show a more severe form of disease suggesting a gene dosage effect (Carvalho et al., 2008; Fukutake et al., 2002; Lang et al., 1994; Lerer et al., 1996; Sobue et al., 1996; Takiyama et al., 1995).
As in other polyQ diseases, the CAG repeat size in MJD inversely correlates with age of disease onset and directly correlates with disease severity (Durr et al., 1996; Jardim et al., 2001; Maciel et al., 1995; Schols et al., 1996). Because intergenerational instability of the CAG repeat occurs in MJD families, the repeat may be of different lengths in progenitors and offspring. Paternal mutant alleles are slightly more unstable than maternal ones, and thus are more prone to expand or contract when transmitted to the next generation (Igarashi et al., 1996; Maciel et al., 1995; Maruyama et al., 1995). This dynamic feature of the disease mutation explains the phenomenon of anticipation observed in some MJD families, in which affected offspring tend to manifest disease earlier than an affected parent (Coutinho and Sequeiros, 1981; Sequeiros and Coutinho, 1993). CAG repeat instability can also occur in different cells from the same tissue, a phenomenon known as somatic mosaicism. In MJD, somatic mosaicism may occur in the brain, but larger repeats are not preferentially associated with affected brain regions (Lopes-Cendes et al., 1996; Maciel et al., 1997).
The existence of several single nucleotide polymorphisms (SNPs) and single tandem repeats (STRs) neighboring the CAG tract have allowed for a better understanding of both the mechanisms of repeat instability and the origin of the MJD mutation. Specific SNPs neighboring the repeat were shown to have both cis and trans effects on CAG repeat instability (Igarashi et al., 1996; Maciel et al., 1999; Martins et al., 2008). Evolution of the CAG repeat in the ATXN3 gene appears to have been driven by a multistep mutational mechanism (Martins et al., 2006). Interestingly, two mutational events may explain the fact that MJD is spread worldwide: the mutation probably originated in Asia, later spreading throughout Europe, with a founder effect explaining the high prevalence in Portugal and the second mutational event perhaps explained by Portuguese emigration (Gaspar et al., 2001; Martins et al., 2007).
Although CAG repeat length is strongly correlated with several clinical aspects of disease, other genetic and/or environmental factors likely contribute to disease presentation. For example, DNA methylation in the promoter region of the ATXN3 gene was recently proposed to have a small, positive effect on the age at onset of MJD patients, indicating that epigenetic factors might contribute to clinical variability in MJD (Emmel et al., 2011).
3.2. Genomic structure and transcripts
The ATXN3 gene spans ~48 Kb and comprises 11 exons with the CAG repeat residing in exon 10 (Ichikawa et al., 2001). Four different transcripts of approximately 1.4, 1.8, 4.5, and 7.5 Kb are ubiquitously expressed in human brain and in non-nervous tissues (Ichikawa et al., 2001; Schmitt et al., 1997). These multiple transcripts may result from alternative splicing in exons 2, 10, and 11 in combination with different polyadenylation signals (Goto et al., 1997; Ichikawa et al., 2001; Kawaguchi et al., 1994). A recent study proposed the existence of two novel exons 6a and 9a, located downstream of the corresponding exons, and 50 potential new alternative splice variants of the ATXN3 gene (Bettencourt et al., 2010). The biological relevance of these numerous variants, however, remains unclear.
The regulation of ATXN3 gene expression is still poorly understood. The 5’-flanking region is a TATA-less promoter, comprising GC-rich regions, a CCAAT box, multiple putative SP1 binding sites, and a core promoter region within ~300 bp of the start codon (Schmitt et al., 2003). The ATXN3 3’ untranslated region (UTR) remains unstudied but the existence of transcripts carrying different 3’UTRs suggests additional gene regulation at this level (Ichikawa et al., 2001). The field would benefit from greater clarification of the mechanisms regulating ATXN3 gene expression as they could represent potential therapeutic targets.
4. The ATXN3 product, ATXN3
An evolutionarily conserved protein, ATXN3 has a long list of orthologs in a wide range of species (Costa et al., 2004; Linhartova et al., 1999; Rodrigues et al., 2007; Schmitt et al., 1997). Normal (i.e. nonexpanded) human ATXN3 has a molecular weight of approximately 42 KDa, varying slightly in size depending on the length of the polymorphic glutamine repeat. Defining the function, localization, stability and physiological role of wild-type ATXN3 is critically important if scientists want to understand how polyQ expansion in this protein causes its dysfunction and triggers a toxic mechanism.
4.1. Structure and function as a deubiquitinating enzyme
ATXN3 is a deubiquitinating enzyme (DUB) that binds Ub and polyUb chains, and is itself regulated by ubiquitination (Burnett et al., 2003; Chai et al., 2004; Donaldson et al., 2003; Nicastro et al., 2009; Scheel et al., 2003; Todi et al., 2009). Current data support the view that ATXN3 functions, at least in part, to edit polyUb chains added by Ub ligases to target proteins (Kuhlbrodt et al., 2011; Scaglione et al., 2011). Ubiquitination of proteins occurs through sequential reactions involving the Ub activating enzyme (E1), Ub conjugating enzymes (E2), and Ub ligases (E3). Several types of polyUb chains can be formed by a Ub ligase depending on the specific lysine linkages made between Ub molecules. Different linkage types confer divergent functions on polyUb chains. For example, K48-linked polyUb chains typically target proteins for proteasomal degradation whereas K63-linked chains play diverse roles in subcellular localization, vesicular trafficking, DNA repair, and translation (Glickman and Ciechanover, 2002; Woelk et al., 2007).
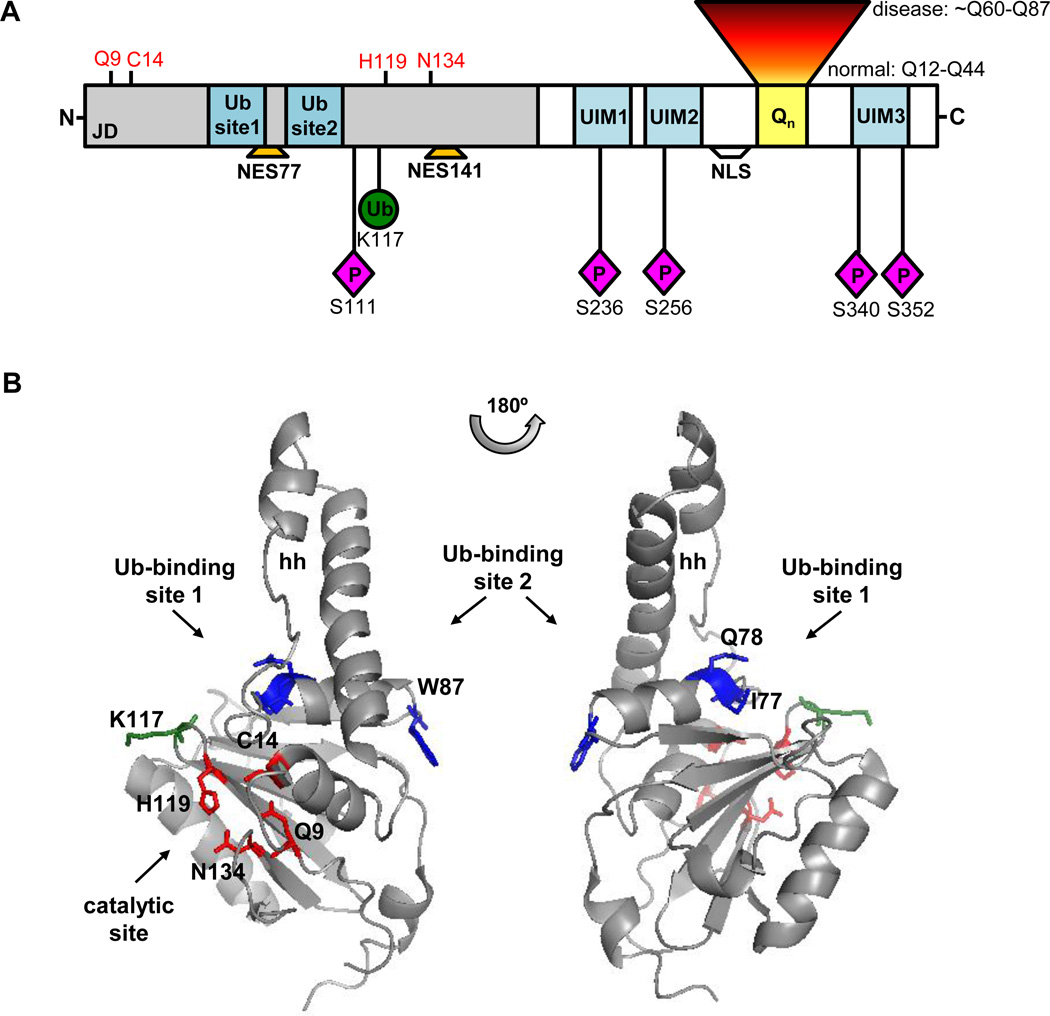
ATXN3 contains a structured globular N-terminus of 198 amino acids – the catalytic Josephin domain (JD) - followed by an unstructured, flexible C-terminus containing the polyQ stretch and two or three ubiquitin interacting motifs (UIM), depending on the protein isoform (Goto et al., 1997; Masino et al., 2003) (Figure 1).
Figure 1.
Functional domains and posttranslational modifications of ATXN3. A) ATXN3 contains a N-terminal DUB catalytic domain (Josephin domain, JD) centered on residues Q9, C14, H119, and N134 (red), two Ub-binding sites, and two nuclear export signals (NES77 and NES141). The C-terminal region of ATXN3 contains 2 or 3 UIMs (depending on the isoform), the polyQ tract (Qn), and the NLS. ATXN3 is mono-ubiquitinated primarily at residue K117 and phosphorylated at residues S111, S236, S256, S340, and S352. B) Solution structure of JD (PDB: 1YZB) highlighting the catalytic residues (red) and critical residues of Ub-binding sites (blue). Ub-binding site 1 (I77Q78) resides near the catalytic pocket while Ub-binding site 2 (W87) resides on the opposite side, separated from site 1 by a helical hairpin (hh). The main ATXN3 mono-ubiquitination site, K117 (green), also localizes near the catalytic cleft.
While several potential ATXN3 isoforms may be translated, only two have been studied in detail. Both are full length proteins that contain the polyQ tract and UIMs1 and 2 but differ in their C-termini, either lacking or containing UIM3 (designated 2UIM ATXN3 and 3UIM ATXN3, respectively). Studies using antibodies that recognize either both isoforms or only 3UIM ATXN3 indicate that 3UIM ATXN3 is the predominant isoform expressed in brain (Harris et al., 2010; Schmidt et al., 1998; Trottier et al., 1998).
The JD adopts a semi-elongated L-structure composed of a globular catalytic subdomain and a helical hairpin (Mao et al., 2005; Nicastro et al., 2006; Nicastro et al., 2005). The ubiquitin protease activity resides in the catalytic subdomain, which comprises the cleavage pocket (Q9, C14, H119, N134) typical of papain-like cysteine proteases and two binding sites for Ub (Mao et al., 2005; Nicastro et al., 2006; Nicastro et al., 2009). ATXN3 Ub-protease activity requires the active site cysteine 14 (Berke and Paulson, 2003; Burnett et al., 2003; Chai et al., 2004). PolyUb chains with at least four Ub units, but not shorter chains, are cleaved in vitro by ATXN3. ATXN3 also shows preference for K63-linked and K48/K63-mixed linkage polyUb chains over K48-linked chains in vitro (Burnett et al., 2003; Chai et al., 2004; Winborn et al., 2008). Although favoring the cleavage of long polyUb chains, ATXN3 can also deubiquitinate specific monoubiquitinated substrates once in functional protein complexes (Scaglione et al., 2011).
Ub binding to the JD occurs through an induced-fit mechanism mediated by the helical hairpin (Nicastro et al., 2009). Lying close to the active cleft, Ub-binding site 1 is essential for cleavage of all Ub chains whereas site 2, residing on the opposite side and overlapping with the interaction surface of the HHR23B Ub-like (Ubl) domain, may confer polyUb-linkage preference since mutating this site reduces cleavage of K48-linked and K63/K48-mixed, but not K63-linked, polyUb chains (Nicastro et al., 2010). Intriguingly, only a K48-linkage di-Ub molecule appears capable of simultaneously occupying both Ub sites on the isolated JD (Nicastro et al., 2010) (Figure 1).
Specific recognition and positioning of Ub chains for proteolytic cleavage by ATXN3 requires cooperation between its Ub-binding sites in the JD and the UIMs in the C-terminus. The UIMs are essential for higher affinity polyUb chain binding but dispensable for cleavage (Winborn et al., 2008). Though capable of binding K48 or K63-linkage polyUb chains, the UIMs likely position chains in a way that promotes K63-linked chain cleavage and inhibits K48-linked chain cleavage in vitro.
A fraction of the cellular pool of ATXN3 is itself ubiquitinated. Although ATXN3 can be mono- and oligo-ubiquitinated, the major ATXN3-Ub species is monoubiquitinated (Berke et al., 2005; Todi et al., 2009). This posttranslational modification enhances ATXN3 DUB activity toward ubiquitinated substrates and free polyUb chains (Todi et al., 2010; Todi et al., 2009). Among the several lysine residues that can be ubiquitinated on ATXN3, residue K117 near the catalytic pocket is predominantly modified (Todi et al., 2010) (Figure 1). Ubiquitination at K117 increases ATXN3 activity independent of other cofactors/interactors, other types of posttranslational modification, or the known Ub binding domains (Todi et al., 2010).
How does ATXN3 function as a DUB? Most likely, C-terminal UIMs and JD Ub binding site 1 cooperate to position the polyUb chains for Ub isopeptide-bond cleavage at the catalytic site. Although ATXN3 shows preference for cleaving long polyUb chains containing at least four units, it can also deubiquitinate select monoubiquitinated substrates. The properties described for ATXN3 in vitro may represent an overly simplistic view of how ATXN3 actually functions in cells when interacting with its multiple known partners and potential substrates.
4.2. Localization
ATXN3 is widely expressed throughout peripheral and neuronal tissues in many different cell types (Paulson et al., 1997a; Schmidt et al., 1998; Trottier et al., 1998; Wang et al., 1997). A similar expression pattern is observed for ATXN3 orthologs in other species (Costa et al., 2004; Rodrigues et al., 2007; Schmitt et al., 1997). Intracellular localization of ATXN3 is regulated at different levels, and its primary subcellular site of action remains uncertain. ATXN3 has been reported in the cytoplasm, nucleus and even in mitochondria (Paulson et al., 1997a; Pozzi et al., 2008; Tait et al., 1998; Trottier et al., 1998).
ATXN3 is highly mobile in the cytoplasm and nucleus with its diffusion limited by the rate of transport across the nuclear membrane (Chai et al., 2002). Nucleocytoplasmic shuttling of ATXN3 is mediated, in part, by a weak nuclear localization signal (NLS), 282RKRR285, and two nuclear export signals (NES), NES77 and NES141 (CRM1/exportin-dependent) (Figure 1) (Antony et al., 2009; Macedo-Ribeiro et al., 2009; Reina et al., 2010; Tait et al., 1998). Interestingly, NES77 overlaps with Ub-binding site 1, but whether Ub binding actually modulates transport into or out of the nucleus remains unknown.
Under basal conditions nuclear import of ATXN3 appears to be mainly controlled by casein kinase 2 (CK2)-mediated phosphorylation of residues S236 in UIM1 and S340/S352 in UIM3 (Mueller et al., 2009). Posttranslational modification by ubiquitination does not affect subcellular localization of ATXN3 in cell lines, but enzymatically active ATXN3 has been reported to localize to the nucleus more often than catalytically inactive ATXN3 (Todi et al., 2007; Todi et al., 2010).
Intracellular localization of ATXN3 is also regulated by specific proteotoxic stressors. Heat shock or oxidative stress leads ATXN3 to accumulate in the nucleus (Reina et al., 2010). Whether CK2-mediated phosphorylation contributes to ATXN3 nuclear translocation under these stress conditions is currently uncertain (Mueller et al., 2009; Reina et al., 2010). Phosphorylation of S111 in the JD seems to be required for nuclear localization of ATXN3 upon heat-shock (Reina et al., 2010).
In summary, the subcellular localization of ATXN3 is highly regulated, depending on interactions between internal localization signals, posttranslational modifications, protein-protein interactions and specific cellular conditions (Antony et al., 2009; Macedo-Ribeiro et al., 2009; Mueller et al., 2009; Reina et al., 2010; Trottier et al., 1998). Understanding the events that modulate intracellular trafficking of ATXN3 may help to elucidate disease pathogenesis, as mutant ATXN3 tends to accumulate in the nucleus and the nucleus is a preferential site for polyQ-induced toxicity.
4.3. Stability
Knowledge of specific cellular events that modulate ATXN3 stability and function is important to understand the cellular dysfunction caused by polyQ expansion. The stability of ATXN3 can be affected by its solubility or propensity to aggregate, by its susceptibility to proteolytic cleavage, and by signals that alter its rate of degradation.
4.3.1. Aggregation
ATXN3 has an intrinsic propensity to aggregate in vitro under native conditions. As is true for other polyQ disease proteins, the fibrillization of ATXN3 is modulated by flanking domains in the protein – in the case of ATXN3, the N-terminal JD influences aggregation. Nonpathogenic (i.e. nonexpanded) ATXN3 is able to undergo a single step aggregation event via JD self-association into dimers. These dimers then aggregate into spheroidal oligomers that in turn assemble into elongated “beads-on-strings” fibrils that are SDS-soluble and Thioflavin T (ThT)-positive (Ellisdon et al., 2007; Ellisdon et al., 2006; Gales et al., 2005; Masino et al., 2011b).
In vitro fibrils formed by nonpathogenic ATXN3 are structurally similar to the ones formed by other amyloidogenic proteins (Masino et al., 2011b). Importantly, ATXN3 enzymatic activity is lost in the fibrils, probably because the native α-helical structure of the JD is converted into a β-sheet-enriched conformation as aggregation proceeds (Masino et al., 2011b). To slow down ATXN3 aggregation, strategies have been pursued to increase JD thermodynamic stability and inhibit self-interaction (Masino et al., 2011a; Robertson et al., 2010; Saunders et al., 2011). Interaction of Ub or alpha B-crystallin (αβ-c) with JD potently inhibits JD self-association in vitro (Masino et al., 2011a; Robertson et al., 2010). Interestingly, in cells the 2UIM ATXN3 isoform carrying a hydrophobic C-terminal is more prone to aggregate than the 3UIM isoform (Harris et al., 2010).
These results suggest that protein-protein interactions involving the JD and C-terminus of ATXN3 may help prevent ATXN3 aggregation in vivo and thereby preserve its enzymatic function. Such interactions might explain why ATXN3 aggregates are not seen in normal individuals despite the propensity of nonpathogenic ATXN3 to aggregate in vitro. Because aggregation of mutant ATXN3 is a pathological hallmark of MJD, it will be even more important to understand the fibrillization pathway of mutant ATXN3 and identify potential altered intermolecular interactions that modulate the formation of insoluble aggregates.
4.3.2. Proteolysis
Caspases and calpains have been reported to cleave ATXN3 at specific sites, both in cell lines and in animal models (Berke et al., 2004; Colomer Gould et al., 2007; Goti et al., 2004; Haacke et al., 2006; Haacke et al., 2007; Jung et al., 2009; Mauri et al., 2006; Wellington et al., 1998). Caspase-1 and caspase-3 cleave ATXN3 in vitro (Wellington et al., 1998) but in cell lines undergoing apoptosis ATXN3 is cleaved mainly by caspase-1, resulting in the release of a polyQ-containing fragment (Berke et al., 2004). Apoptotic cleavage of ATXN3 is abolished by mutating all nine potential caspase recognition sites in ATXN3 and is markedly reduced by mutating a cluster of aspartate residues within UIM2 (D241/244/248) (Berke et al., 2004). Evidence in Drosophila shows that proteolytic processing of ATXN3 is conserved across species and may be caspase-dependent (Jung et al., 2009).
The role of calpains in ATXN3 proteolysis is unsettled, given the opposing results in the literature (Berke et al., 2004; Haacke et al., 2007; Jung et al., 2009; Wellington et al., 1998). Some studies suggest that ATXN3 proteolysis is not affected by calpain inhibition (Berke et al., 2004; Jung et al., 2009; Wellington et al., 1998), however another study in vitro and in cells showed that calcium-dependent calpains cleave ATXN3 in regions around amino acids 60, 200, 260 and 318 (Haacke et al., 2007). Divergent results regarding calpain-mediated proteolysis of ATXN3 are probably explained by the use of different systems in which protein-protein interactions essential for this process may be affected by cellular environment and overexpression of ATXN3, among other factors.
Though details of ATXN3 cleavage in vivo remain uncertain, perhaps multiple proteolytic events can occur as in other polyQ disease proteins, carried out by different proteases acting sequentially or concomitantly. The specific cellular conditions under which normal and mutant ATXN3 are cleaved, and the functional properties of the protein fragments generated, remain to be determined. Further defining the proteolytic processing of ATXN3 is important because a leading hypothesis regarding polyQ-mediated toxicity is the generation of aggregation-prone, “toxic” protein fragments. Thus, implicated proteases could prove to be druggable targets.
4.3.3. Degradation
A relatively long-lived protein, ATXN3 is degraded at least partly by the proteasome (Berke et al., 2005; Jana et al., 2005; Matsumoto et al., 2004). In transfected cells, 3UIM ATXN3, which might also be degraded by macroautophagy, is more stable than 2UIM ATXN3 (Harris et al., 2010) perhaps because additional phosphorylation within UIM3 stabilizes the protein (Mueller et al., 2009). Several E3 ligases and proteasome shuttle proteins are reportedly involved in ATXN3 polyubiquitination and shuttling to the proteasome for degradation, including the E3/shuttle complexes E4B/VCP, CHIP/Hsp70, and E6-AP/Hsp70 (Jana et al., 2005; Matsumoto et al., 2004; Mishra et al., 2008). Gp78, an ER-associated E3, also promotes ATXN3 polyubiquitination and proteasomal degradation (Ying et al., 2009). Interestingly, ATXN3 turnover may also be controlled by its own catalytic activity, though this has not been confirmed in cells expressing physiological levels of the protein (Todi et al., 2007). Once ATXN3 reaches the proteasome its degradation seems to be enhanced by direct binding to the 19S proteasome (Wang et al., 2007). Because ATXN3 is a DUB and participates in protein quality control pathways, its interactions with E3/shuttle complexes might even regulate their function while also affecting ATXN3 turnover.
Wild-type ATXN3 can be degraded by the proteasome, but whether other protein quality control pathways are involved in its turnover remains unknown. Further clarification of ATXN3 degradation pathways might suggest therapeutic strategies to facilitate clearance of the mutant protein in the disease state.
4.4. Normal cellular and physiological role of ATXN3
Identification of ATXN3-interacting proteins and studies in various model systems have shed light on the biological roles of ATXN3. Because knockout models of ATXN3 orthologs in mouse and C. elegans do not display an obvious phenotype, ATXN3 is a nonessential protein (Rodrigues et al., 2007; Schmitt et al., 2007). Conceivably, three other JD-like containing proteins may exert similar functions to ATXN3 and compensate for its absence in knockout models (Todi and Paulson, 2011). Given the diversity of ATXN3’s identified protein partners (at least 46) (Table 1) and putative interactors (at least 93) (Araujo et al., 2011; Sowa et al., 2009), ATXN3 likely participates in many cellular pathways.
Table 1.
Known protein interactors of ATXN3.
| Protein name | Description | Reference |
|---|---|---|
| ACCN2 | amiloride-sensitive cation channel 2, neuronal | Shen et al., 2006 |
| ARHGDIA | Rho GDP dissociation inhibitor (GDI) alpha | Shen et al., 2006 |
| ARHGAP19 | Rho GTPase activating protein 19 | Lim et al., 2006 |
| ATXN3 | ataxin-3 | Todi et al., 2007 |
| CBP | CREB binding protein | Chai et al., 2002 |
| CHIP | STIP1 homology and U-box containing protein 1 | Jana et al., 2005 |
| CK2 beta | casein kinase II subunit beta | Tao et al., 2008 |
| Derlin-1 | Der1-like domain family, member 1 | Wang et al., 2006 |
| Dynein | dynein, cytoplasmic 2, heavy chain 1 | Burnett et al., 2005 |
| E6-AP | ubiquitin ligase E3A | Mishra et al., 2010 |
| EWSR1 | Ewing sarcoma breakpoint region 1 | Lim et al., 2006 |
| FOXO4 | forkhead box other 4 | Araujo et al., 2011 |
| Gp78 | autocrine motility factor receptor (AMFR) | Ying et al., 2009 |
| GSK 3β | glycogen synthase kinase 3β | Fei et al., 2007 |
| H3 | histone H3 | Li et al., 2002 |
| H4 | histone H4 | Li et al., 2002 |
| HAP1 | huntingtin-associated protein 1 | Takeshita et al., 2011 |
| HDAC3 | histone deacetylase 3 | Evert et al., 2006 |
| HDAC6 | histone deacetylase 6 | Burnett et al., 2005 |
| HHR23A | RAD23 homolog A (S. cerevisiae) | Wang et al., 2000 |
| HHR23B | RAD23 homolog B (S. cerevisiae) | Wang et al., 2000 |
| Itga5 | α5 integrin subunit (Mus musculus) | do Carmo Costa et al., 2010 |
| KCTD10 | BTB/POZ-domain containing protein KCTD10 | Sowa et al., 2010 |
| Lin-10 | lin-10 | Lim et al., 2006 |
| NCOR1 | nuclear receptor co-repressor 1 | Evert et al., 2006 |
| NEDD8 | neural precursor cell expressed, developmentally down-regulated 8 | Ferro et al., 2007 |
| OTUB2 | ubiquitin thioesterase OTUB2 | Sowa et al., 2009 |
| PARK2 | parkin | Durcan et al., 2010 |
| PCAF | p300/CREBBP-associated factor | Li et al., 2002 |
| PLIC-1 | protein linking IAP to the cytoskeleton | Heir et al., 2006 |
| PML | promyelocytic leukemia antigen | Chai et al., 2001 |
| PRKCABP | protein kinase C-alpha-binding protein | Lim et al., 2006 |
| p300 | E1A binding protein p300 | Li et al., 2002 |
| p45 | proteasome (prosome, macropain) 26S subunit, ATPase, 5 | Wang et al., 2007 |
| SELS | selenoprotein S | Wang et al., 2006 |
| SUMO1 | SMT3 suppressor of mif two 3 homolog 1 (S. cerevisiae) | Shen et al., 2006 |
| SYVN1 | synovial apoptosis inhibitor 1, synoviolin | Wang et al., 2006 |
| TBP | TATA box binding protein | Araujo et al., 2011 |
| TEX11 | testis expressed 11 | Lim et al., 2006 |
| TUBB | tubulin, beta | Zhong et al., 2006 |
| UBB | ubiquitin B | Ferro et al., 2007 |
| UBE4B | ubiquitination factor E4B (UFD2 homolog, yeast) | Matsumoto et al., 2004 |
| UBXN-5 | UBX-containing protein in Nematode family member (ubxn-5, C. elegans) | Rodrigues et al., 2009 |
| USP13 | ubiquitin carboxyl-terminal hydrolase 13 | Sowa et al., 2009 |
| VCP | valosin-containing protein | Kobayashi et al., 2002 |
| WT1 | Wilms tumor 1 | Araujo et al., 2011 |
Multiple lines of evidence implicate ATXN3 in cellular protein quality control, particularly the ubiquitin proteasome system (UPS) responsible for degradation of short-lived and misfolded proteins. ATXN3 may regulate the ubiquitination status of many proteins since total ubiquitinated protein levels are increased in Atxn3 knockout mouse brain (Scaglione et al., 2011; Schmitt et al., 2007). Overexpression of ATXN3 in polyQ-neurodegeneration models in Drosophila suppresses toxicity and cell death, implying that ATXN3 is a neuroprotective protein; its neuroprotective action, moreover, depends both on its DUB activity and proper functioning of the proteasome (Warrick JM et al., 2005). ATXN3 also appears to be involved in the cellular response to heat stress (Reina et al., 2010; Rodrigues et al., 2011).
ATXN3 associates with several E3 ligases, cleaves and edits long polyUb, and participates in substrate delivery to the proteasome by interacting with shuttle proteins (Durcan et al., 2010; Kuhlbrodt et al., 2011; Scaglione et al., 2011; Wang et al., 2006; Zhong and Pittman, 2006). Table 1 lists the E3 ligases and proteasome shuttle factors that interact with ATXN3. ATXN3 interacts via an arginine/lysine motif (aa 277–291) with the AAA ATPase Valosin-Containing Protein or ATPase p97 (VCP/p97) (Boeddrich et al., 2006; Doss-Pepe et al., 2003), which functions coordinately with ubiquitinating complexes to target proteins for proteasomal degradation. The VCP/ATXN3 complex might serve to transfer polyubiquitinated substrates, after editing by ATXN3, directly to the proteasome or to other proteasomal shuttling factors like Ubiquilin/PLIC1 and the Rad23 homologues HHRB23A/B (Doss-Pepe et al., 2003; Heir et al., 2006; Kuhlbrodt et al., 2011; Wang et al., 2000).
The VCP/ATXN3 functional complex also regulates endoplasmic reticulum-associated degradation (ERAD) (Wang et al., 2006; Zhong and Pittman, 2006). Through interaction with ER membrane components including ER-specific E3 ligases, VCP and ATXN3 control the dislocation and degradation of misfolded proteins from the ER (Wang et al., 2006; Zhong and Pittman, 2006). As a scaffold protein, VCP interacts with many adaptors that modulate its activity. SARKS, an ubiquitin regulatory X (UBX) domain-containing protein and VCP adaptor, is able to inhibit VCP/ATXN3-mediated ERAD (LaLonde and Bretscher, 2011). The synergistic cooperation of VCP and ATXN3 in proteostasis is also important in aging as double knockouts in C. elegans have a longer lifespan than wild-type worms (Kuhlbrodt et al., 2011). In coordination with VCP and possibly the E3 ligase E4B, ATXN3 also seems to regulate the ubiquitination status and subsequent degradation of components of the insulin/ insulin-like growth factor 1 (IGF1) signaling pathway implicated in lifespan regulation (Kuhlbrodt et al., 2011).
C-terminus of Hsc70 interacting protein (CHIP) is an ATXN3-interacting E3 (Jana et al., 2005) that has been linked to many neurodegenerative diseases. Important insights into the functional interaction between ATXN3 and CHIP have recently been elucidated (Scaglione et al., 2011). ATXN3 is recruited to the ubiquitination complex by monoubiquitinated CHIP, where it then limits the length of polyUb chains formed on substrates and terminates the ubiquitination cycle by removing monoubiquitin from CHIP (Scaglione et al., 2011). Based on these findings, we speculate that ATXN3 could regulate the activity of E3s in a manner similar to the way deneddylases modulate E3 activity in SCF complexes (Scaglione et al., 2011). In fact, ATXN3 also displays deneddylase activity in vitro, cleaving isopeptide bonds between a substrate and the Ubl protein neural precursor cell expressed developmentally downregulated 8, or NEDD8 (Ferro et al., 2007). ATXN3-mediated cleavage of monoubiquitinated or neddylated E3s may prove to regulate several ubiquitinating E3 complexes.
Parkin, an E3 implicated in Parkinson disease, also interacts functionally with ATXN3. Parkin undergoes autoubiquitination in vitro, forming mainly K27 and K29-linked polyUb chains which are linked to lysosomal and autophagic degradation. Normal ATXN3 interacts with and deubiquitinates parkin but does not affect its stability (Durcan et al., 2010). It is possible that ATXN3 controls the abundance and edits the architecture of the Ub chains linked to parkin, thereby targeting this protein for specific cellular pathways (Durcan et al., 2010).
When the ubiquitin-proteasome system is compromised or overwhelmed, accumulating proteins become concentrated in perinuclear inclusions called aggresomes. ATXN3 helps regulate the formation of aggresomes in a manner that requires an active DUB site and the UIMS (Burnett and Pittman, 2005). Additional interactions of ATXN3 with other components implicated in aggresome organization, such as dynein, histone deacetylase 6 (HDAC6), protein linking IAP to the cytoskeleton (PLIC1) and microtubules, support the importance of ATXN3 to this cellular process (Burnett and Pittman, 2005; Heir et al., 2006; Mazzucchelli et al., 2009; Rodrigues et al., 2010).
Indeed, ATXN3 is important to cytoskeletal organization and to the formation of focal adhesions (Rodrigues et al., 2010). In the specific case of myoblast differentiation into muscle fibers (myogenesis), ATXN3 is critical to the initial differentiation steps to organize the cytoskeleton and to regulate the levels of integrin subunits and other proteins involved in integrin-mediated signaling (do Carmo Costa et al., 2010). Because ATXN3 interacts with and stabilizes α5 integrin in a DUB activity-dependent manner, it most likely regulates the degradation of this protein through its role in ubiquitin-dependent proteostasis (do Carmo Costa et al., 2010). This basal function of ATXN3 may be common to many cell types.
ATXN3’s ability to bind DNA and interact with transcription regulators points toward a role for ATXN3 in transcriptional regulation, most likely as a transcriptional corepressor (Li et al., 2002). Through interaction with cAMP-response element binding (CREB)-binding protein (CBP), p300, and p300/CREBBP associated factor (PCAF), ATXN3 inhibits CREB-mediated transcription (Chai et al., 2001; Li et al., 2002). ATXN3 also regulates histone acetylation, inhibiting p300-mediated histone acetylation and promoting histone deacetylation by interaction with histone deacetylase 3 (HDAC3) and nuclear receptor co-repressor 1 (NCOR1) (Evert et al., 2006; Li et al., 2002). The specific biochemical role of ATXN3 in these reactions, however, is unknown.
Although ATXN3 has been shown to bind a consensus site in DNA (GAGGAA) through a putative basic leucine zipper motif (bZIP) located in its C-terminus (223–270 aa) (Evert et al., 2006), it is still unclear whether ATXN3 functions as a classical repressor. The UPS modulates transcription by regulating chromatin and controlling levels of various transcriptional machinery components, and thus the involvement of ATXN3 in transcriptional regulation might be coupled to its DUB activity. A potential mechanism of action of ATXN3 in transcriptional regulation is to target chromatin by directly binding DNA or histones, which might then favor the ability of ATXN3 to inhibit histone acetylation, recruit corepressors like HDAC3 or NCOR1, and, through its DUB activity, stabilize repressor complexes that enhance histone deacetylation (Evert et al., 2006).
Recently, a specific activity of ATXN3 in transcriptional modulation was suggested by its potential physiological role in response to oxidative stress. ATXN3 interacts with and stabilizes the forkhead box O (FOXO) transcription factor FOXO4, and upon oxidative stress they both translocate to the nucleus and activate manganese superoxide dismutase (SOD2) transcription which in turn protects cells from oxidative damage (Araujo et al., 2011).
In summary, the unique properties of ATXN3 as a DUB suggest that it helps regulate the stability or activity of many proteins in diverse cellular pathways implicated in a wide range of physiological events. It is conceivable that the diverse physiological roles of ATXN3 result not only from its DUB activity, but also from its ubiquitin binding capacity and potential protease activity against other Ub-like proteins such as NEDD8 and SUMO.
5. Mutant ATXN3 and disease pathogenesis
Expansion of the polyQ track likely induces a conformational change in ATXN3 that affects many properties of the protein: stability and degradation, subcellular localization, molecular interactions with other proteins, and propensity to aggregate. These altered properties result in loss and/or gain of function, leading to cellular dysfunction and selective neuronal cell death.
5.1. Dysfunction of mutant ATXN3
Mutant (expanded) ATXN3 still binds K48- and K63-linked polyUb chains, gets activated by ubiquitination, and retains DUB catalytic activity in vitro against K48 and K63 chains similarly to normal ATXN3 (Burnett et al., 2003; Todi et al., 2007; Todi et al., 2009; Winborn et al., 2008), but expanded ATXN3 does appear to have an enhanced capacity to deubiquitinate K27- and K29-linked Ub chains (Durcan et al., 2010). Ubiquitination of expanded ATXN3 in cells still occurs at many of the same sites as in normal ATXN3 (Todi et al., 2010). The neuroprotective features of ATXN3 are also at least partly preserved in expanded ATXN3 (Warrick et al., 2005).
Much like normal ATXN3, soluble expanded ATXN3 diffuses rapidly in cytoplasm and nucleoplasm, but unlike normal ATXN3 is highly prone to aggregate, primarily in the nucleus (Chai et al., 2002). The nuclear compartment appears to be the primary site of cellular toxicity in polyQ diseases (Perez et al., 1998). Translocation of both normal and pathogenic ATXN3 from the cytoplasm to the nucleus is mediated by CK2 phosphorylation under basal conditions, and increases in heat-shock and oxidative stress (Mueller et al., 2009; Reina et al., 2010).
Aggregates formed by mutant ATXN3 differ from aggregates generated by normal ATXN3. In contrast to normal ATXN3, mutant ATXN3 undergoes a two-stage aggregation process in vitro (Ellisdon et al., 2006). The first stage (formation of SDS-soluble fibrils) is similar to the one occurring for normal ATXN3 but occurs at a faster rate, and the second stage (generation of SDS-insoluble aggregates) occurs through the polyQ segment (Ellisdon et al., 2006). Glutamine side-chain hydrogen bonding in the polyQ track, possibly adopting a β-helical turn or hairpin conformation, contributes to the irreversible aggregation of expanded ATXN3 (Natalello et al., 2011; Perutz et al., 2002; Sikorski and Atkins, 2005). Functional interactions of wild-type ATXN3 with other molecules have been shown to reduce its aggregation propensity and increase solubility (see section 4.3.1). In the case of mutant ATXN3 it is possible that these modifier interactions are lessened, leading to a faster rate of aggregation.
As in other polyQ diseases, the “toxic fragment hypothesis” may apply to MJD. Aggregation of mutant ATXN3 is thought to be enhanced by proteolysis that generates C-terminal fragments containing expanded polyQ tract, which then can act as seeds for aggregation (Goti et al., 2004; Ikeda et al., 1996; Paulson et al., 1997b; Teixeira-Castro et al., 2011). With the exception of one study reporting less proteolysis of expanded ATXN3 than the normal protein (Pozzi et al., 2008), pathogenic and normal ATXN3 are thought to undergo the same types of caspase- or calcium-dependent calpain cleavage (Berke et al., 2004; Haacke et al., 2007; Jung et al., 2009). The stability of generated fragments containing the polyQ track, however, probably differs for normal and expanded ATXN3. For example, ATXN3 C-terminal fragments are not found in brains from healthy humans but are present in brain homogenates from MJD patients and MJD transgenic mice (Goti et al., 2004). These fragments, however, have not been detected in human MJD brains in all studies (e.g., Berke et al., 2004), possibly reflecting the use of different antibodies. C-terminal ATXN3 fragments cleaved proximal to amino acid 190 and containing the UIMs, expanded polyQ and NLS, are more abundant in the nuclear fraction from affected brain regions of MJD patients (Colomer Gould et al., 2007; Goti et al., 2004). Nuclear accumulation of these fragments may be due less to the presence of a weak NLS than to the absence of NES signals and, consequently, escape from chaperone-mediated clearance by the UPS in the cytoplasm (Antony et al., 2009; Breuer et al., 2010). The generation of C-terminal polyQ-containing fragments not only favors aggregation but also eliminates the putative protective action of the JD which is cleaved away by proteolysis (Warrick et al., 2005).
At least two E3s, parkin and the mitochondrial ubiquitin ligase MITOL, are reportedly involved in the proteasomal degradation of mutant ATXN3 C-terminal fragments (Sugiura et al., 2010; Tsai et al., 2003). Though, the mode of action of MITOL with full-length ATXN3 is unknown, parkin seems not to be involved in degradation of full-length wild-type or mutant ATXN3 (Durcan et al., 2010). Like normal ATXN3, expanded ATXN3 is polyubiquitinated and degraded by the UPS (Jana et al., 2005; Matsumoto et al., 2004; Mishra et al., 2008; Wang et al., 2009; Ying et al., 2009) but may have a longer half-life (Matsumoto et al., 2004). While it is still unknown if wild-type ATXN3 is appreciably degraded by macroautophagy, some evidence suggests that mutant ATXN3 can be degraded by this protein quality control pathway (Berger et al., 2006). Furthermore, pathogenic ATXN3 induces autophagy in a Drosophila model (Bilen and Bonini, 2007).
Increased stability of expanded ATXN3 might be explained by enhanced binding to VCP, thereby delaying its release from the VCP/E4B complex for subsequent proteasomal degradation (Boeddrich et al., 2006; Matsumoto et al., 2004). The stronger interaction between expanded ATXN3 and VCP may also impair ERAD and other cellular processes that depend on this functional interaction (Zhong and Pittman, 2006).
Expanded ATXN3 binds with higher affinity to CHIP and may target CHIP for degradation (Scaglione et al., 2011). Though normal and expanded ATXN3 bind similarly to polyubiquitinated parkin, pathogenic ATXN3 shows increased DUB activity towards polyUb-parkin, promoting its degradation via autophagy (Durcan et al., 2010). Consistent with these findings, CHIP and parkin levels are decreased in brains of transgenic mice expressing expanded ATXN3 (Durcan et al., 2010; Scaglione et al., 2011).
Mutant ATXN3 displays altered DNA binding which reduces its ability to form deacetylase complexes and repress transcription of target genes (Evert et al., 2006; Li et al., 2002). Another example of ATXN3-mediated transcriptional regulation in which expanded ATXN3 is impaired is its ability to promote FOXO4-mediated SOD2 transcription during oxidative stress (Araujo et al., 2011).
In summary, mutant ATXN3 appears to be dysfunctional at several levels: (i) it forms insoluble aggregates; (ii) it is susceptible to proteolysis resulting in the generation of expanded polyQ-containing ATXN3 fragments that favor aggregation, primarily in the nucleus; (iii) and it interacts abnormally with at least some of its native partners, impeding its own degradation and leading in some cases to a gain-offunction and in other cases to partial loss of normal ATXN3 function. Examples of gain-of-function of mutant ATXN3 are insoluble aggregate formation and induction of parkin degradation. Conversely, ATXN3’s reduced ability to form deacetylating repressor complexes at target genes and to function as an activator of FOXO4-mediated SOD2 transcription exemplify partial loss of function of expanded ATXN3. As has been well established for SCA1 (Lim et al., 2008; Zoghbi and Orr, 2009), it will be interesting to determine whether ATXN3 dysfunction induced by polyQ expansion includes a partial loss of wild-type ATXN3 function.
5.2. Animal models of ATXN3 overexpression
Many animal models overexpressing specific forms of ATXN3 are available to study the molecular and phenotypic aspects of MJD (Table 2). Models recapitulating aspects of disease exist in mice, rats, flies and worms.
Table 2.
Animal models of ATXN3 overexpression.
| Species | Gene promoter |
Expressed protein (isoform) | PolyQ size |
Line | Motor dysfunction |
Neuropathology | Reference | |
|---|---|---|---|---|---|---|---|---|
| Aggregates | Cell death | |||||||
| M. musculus | L7 | HA-ATXN3(2UIM) | Q79 | MJDQ79 | − | − | − | Ikeda et al., 1996 |
| HA-[aa290-Ct]ATXN3(2UIM) | Q35 | Q35C | − | − | − | |||
| Q79 | Q79C | +++ | ND | ++ | ||||
| ATXN3 | all ATXN3 isoforms | Q15 | MJD15.4 | − | − | − | Cemal et al., 2002 | |
| Q84 | MJD84.2 | +++ | +++ | ++ | ||||
| Prnp | ATXN3(2UIM) | Q20 | Q20-A | − | − | − | Goti et al., 2004 | |
| Q71 | Q71-B | +++ | +++ | − | ||||
| Q71-C | +++ | ++ | + | |||||
| Prnp | ATXN3delta[aa190–220](2UIM) | Q20 | deltaQ20 | − | − | − | Colomer Gould et al., 2007 | |
| Q71 | deltaQ71 | +++ | ND | ND | ||||
| Prnp | ATXN3(3UIM) | Q15 | 15.1 | − | − | − | Bichelmeier et al., 2007 | |
| Q70 | 70.61 | +++ | ++ | + | ||||
| Q148 | 148.19 | +++ | ++ | ND | ||||
| ATXN3-3NLS(3UIM) | Q148 | 148.NLS | +++ | +++ | + | |||
| ATXN3-NES(3UIM) | Q148 | 148.NES | + | + | − | |||
| Prnp | HA-ATXN3(2UIM) | Q22 | Ataxin-3-Q22 | − | − | − | Chou et al., 2008 | |
| Q79 | Ataxin-3-Q79 | ++ | + | + | ||||
| L7 | HA-[aa287-Ct]ATXN3(2UIM) | Q69 | polyQ | +++ | +++ | ND | Torashima et al., 2008 | |
| TetOff-Prnp | ATXN3(3UIM) | Q77 | Prp/MJD77 | ++ | ++ | + | Boy et al., 2009 | |
| Htt | ATXN3(3UIM) | Q148 | HDpromMJD148 | + | + | + | Boy et al., 2009 | |
| CMV | ATXN3(3UIM) | Q83 | CMVMJD83 | − | − | − | Silva-Fernandes et al., 2010 | |
| Q94 | CMVMJD94 | ++ | + | ++ | ||||
| R. norvegicus | PGK (lentivirus) | Myc-ATXN3(2UIM) | Q27 | − | ND | − | − | Alves et al., 2008 |
| Q72 | − | + | ++ | ++ | ||||
| D. melanogaster | elav-GAL4 or gmr-GAL4 | HA-[aa281-Ct]ATXN3(2UIM) | Q27 | UAS-MJDtr-Q27 | ND | − | − | Warrick et al., 1998 |
| Q78 | UAS-MJDtr-Q78 | +++ | ++ | +++ | ||||
| elav-GAL4 or gmr-GAL4 | Myc-ATXN3(2UIM) | Q27 | UAS-SCA3-Q27 | − | − | − | Warrick et al., 1998 | |
| Q84 | UAS-SCA3-Q84 | +++ | ++ | ++ | ||||
| HA-ATXN3(2UIM) | Q78 | UAS-MJDtr-Q78 | +++ | ++ | ++ | |||
| Myc-ATXN3UIM*(2UIM) | Q27 | UAS-SCA3-Q27-UIM* | ND | ND | ND | |||
| Q80 | UAS-SCA3-Q80-UIM* | ND | ND | ++ | ||||
| Myc-ATXN3C14A(2UIM) | Q27 | UAS-SCA3-Q27-C14A | ND | ND | ND | |||
| Q80 | UAS-SCA3-Q88-C14A | ND | ND | +++ | ||||
| Myc-[Nt-aa287]ATXN3(2UIM) | − | UAS-SCA3-delta | ND | ND | ND | |||
| HA-[aa281-Ct]ATXN3-CAA/G(2UIM) | Q78 | UAS-SCA3tr-Q78 | + | ++ | + | Li et al., 2008 | ||
| elav-GAL4 | Myc-ATXN3-FLAG(2UIM) | Q84 | UAS-Myc-Atx3Q84-FLAG(WT) | ND | ++ | +++ | Jung et al., 2009 | |
| Myc-ATXN3(6M)-FLAG(2UIM) | Q84 | UAS-Myc-Atx3Q84-FLAG(6M) | ND | ++ | ++ | |||
| C. elegans | unc-119 | [Nt-aa344]ATXN3-GFP(2UIM) | Q17 | MJD1-17Q-GFP | ND | − | ND | Khan et al., 2006 |
| Q91 | MJD1-91Q-GFP | ND | − | ND | ||||
| Q130 | MJD1-130Q-GFP | + | + | ND | ||||
| [aa283–344]ATXN3-GFP(2UIM) | Q19 | 19Q-GFP | − | − | ND | |||
| Q33 | 33Q-GFP | ND | − | ND | ||||
| Q63 | 63Q-GFP | ND | + | ND | ||||
| Q127 | 127Q-GFP | ++ | ++ | ND | ||||
| F25B3.3 | ATXN3-YFP(3UIM) | Q14 | AT3q14 | − | − | ND | Teixeira-Castro et al., 2011 | |
| Q75 | AT3q75 | − | − | ND | ||||
| Q130 | AT3q130 | ++ | + | ND | ||||
| [aa257-Ct]ATXN3-YFP(3UIM) | Q14 | 257cAT3q14 | − | − | ND | |||
| Q75 | 257cAT3q75 | + | + | ND | ||||
| Q80 | 257cAT3q80 | ++ | ++ | ND | ||||
| Q128 | 257cAT3q128 | +++ | +++ | ND | ||||
absent;
mild;
intermediate;
severe;
ND, Not Determined; Ct, C-terminus; Nt, N-terminus; UIM*. S236/256A;(WT), wild type; (6M), D171/208/217/223/225/228N
Most in vivo MJD models are stable transgenic lines that show highly reproducible features over time. Known for their ease of maintenance and genetic manipulation, several invertebrate models of ATXN3 overexpression in Drosophila and C. elegans have provided important insights regarding the pathogenic mechanisms involving aggregation, proteolysis and toxicity of expanded ATXN3, as well as the apparent neuroprotective role of wild-type ATXN3 (Jung et al., 2009; Khan et al., 2006; Teixeira-Castro et al., 2011; Warrick et al., 2005; Warrick et al., 1998). These models have also proved to be important tools for screening potential therapeutic molecules and genetic modifiers of disease.
Albeit more laborious to generate and less genetically manipulable than invertebrate models, rodent models share important molecular, anatomical and physiological similarities with humans. Numerous transgenic mouse models of MJD have been generated with the aim of mimicking general clinical and molecular aspects of the human disease or of studying a specific hypothesis regarding disease pathogenesis (Table 2).
With the exception of a mouse model generated by genomic integration of the entire human ATXN3 gene housed in a yeast artificial chromosome (YAC) (Cemal et al., 2002), the transgene employed in most mouse models has been a complementary DNA (cDNA) encoding a particular isoform of ATXN3 driven behind a foreign promoter (Table 2). Resulting from DNA random integration, these models differ from each other partly with respect to the copy number of integrated transgenes. Severity of MJD-like symptoms and pathology in the mouse model is generally proportional to the expression levels of mutant ATXN3 in MJD transgenic mice, with homozygous mice displaying a more pronounced phenotype than heterozygous mice, similar to the case in MJD patients. Table 2 describes the major pathological and phenotypic presentations for each mouse model.
Different mouse models expressing full-length ATXN3 isoforms are currently available. Selective expression of mutant ATXN3 in Purkinje cells did not have an apparent deleterious effect in the first generated MJD mouse model, which might have been expected as these cells are not primarily affected in MJD patients (Ikeda et al., 1996). In contrast, three models expressing a pathogenic ATXN3 isoform in the CNS under the direction of the prion protein (Prnp) promoter present relatively early onset motor dysfunction with NNI, although there is little neurodegeneration (Bichelmeier et al., 2007; Chou et al., 2008; Goti D et al., 2004; Goti et al., 2004). Interestingly, the use of the rat huntingtin (Htt) promoter to direct expression of mutant ATXN3 in brain more closely recapitulates the human disease, presenting late onset symptoms, intranuclear inclusions and significant neurodegeneration (Boy et al., 2010). Aiming to reproduce the ubiquitous pattern of ATXN3 expression, an additional model was generated in which the cytomegalovirus (CMV) promoter directs expression of an expanded 3UIM ATXN3 isoform (Silva-Fernandes et al., 2010). These transgenic mice show relatively early onset, non-progressive, mild motor incoordination together with ATXN3-positive cytoplasmic puncta and neurodegeneration in several brain regions affected in MJD patients (Silva-Fernandes et al., 2010).
Other models expressing an ATXN3 fragment or modified versions of ATXN3 have been generated to study specific aspects of disease pathogenesis. Models generated to study the potential toxicity of an ATXN3 C-terminal fragment showed that its expression in Purkinje cells resulted in the formation of intranuclear inclusions and a severe neurodegenerative phenotype in mice (Ikeda H et al., 1996; Torashima et al., 2008). Transgenic mice expressing ATXN3 lacking the protein segment between amino acid residues 190 and 220 revealed that the potential major proteolytic cleavage site on ATXN3 is N-terminal of amino acid residue 190 (Colomer Gould et al., 2007). And mice expressing ATXN3 with additional NLS or NES signals established that mutant ATXN3 is more toxic in the nucleus (Bichelmeier et al., 2007).
While some of the above models recapitulate aspects of the human disease, they all overexpress a single isoform of ATXN3 under the control of an exogenous promoter. In this respect, the YAC MJD transgenic model more closely mirrors what happens in human MJD because all elements of the ATXN3 gene are present, including the 5’ and 3’ regulatory regions (Cemal et al., 2002). As a result, all potential ATXN3 protein isoforms can be expressed. The YAC MJD84.2 transgenic mice also reproduce certain MJD-like features includingearly onset motor incoordination, cytoplasmic and nuclear ATXN3 aggregates, and neurodegeneration in later stages (Cemal et al., 2002; Chen et al., 2008; Shakkottai et al., 2011).
Some properties associated with the expanded CAG repeat in MJD are also replicated in the YAC MJD mice and two cDNA models, HDpromMJD148 and CMVMJD94 (Boy et al., 2010; Cemal et al., 2002; Silva-Fernandes et al., 2010). Similar to MJD patients, in one or more of these models the CAG repeat results in: (i) intergenerational repeat instability, tending to expand upon paternal transmission and contract upon maternal transmission (Boy et al., 2010; Cemal et al., 2002; Silva-Fernandes et al., 2010); (ii) somatic mosaicism, although expansions in repeat length are not preferentially associated with affected brain regions (Silva-Fernandes et al., 2010); and (iii) a direct correlation between repeat length and disease severity (Silva-Fernandes et al., 2010).
A conditional mouse model using the Tet-off system was generated to test whether disease symptoms, once manifest, can be reversed by switching off expression of mutant ATXN3 (Boy et al., 2009). Though expanded ATXN3 is mainly expressed in glial cells in this model, the mice do present phenotypic and pathological MJD-like features that are indeed rescued once the transgene is turned off (Boy et al., 2009). While this result indicates that reducing levels of pathogenic ATXN3 could be a promising strategy to treat SCA3, it will be important to confirm the reversibility of disease features in an equivalent inducible model expressing ATXN3 in neurons.
Finally, the injection of lentivirus expressing human ATXN3 into rat brains reproduces several aspects of MJD neuropathology and provides a very useful tool to study disease pathogenesis in specific brain regions (Alves et al., 2008a; Alves et al., 2008b), although there are some inherent limitations to acute overexpression models.
The importance of having many different animal models of MJD cannot be overestated, as collectively they advance our understanding of many different aspects of MJD and disease pathogenesis. Currently, some models are useful to study the mechanisms of disease progression, others display early MJD-like signs and thus allow for testing therapeutic approaches, and still others serve to test specific hypotheses about pathomechanisms. While the higher severity typically observed in homozygous models probably reflects increased levels of mutant ATXN3, we need to keep in mind the possibility that silencing an endogenous gene at the site of transgene integration could confound analysis. Despite their unquestioned value, the models described above are all overexpression models. Hence, the observed physiological signs could result, in part, from cellular overload of an exogenous protein that is unrelated to the intrinsic pathogenicity of mutant ATXN3. The generation of a knock-in mouse model expressing murine ATXN3 with a polyQ expansion under the control of its endogenous regulatory regions would be a welcome addition to the collection of MJD models, as it should represent a genetically and physiologically accurate model of the human disease.
5.3. Pathogenic mechanisms
While the precise pathogenic mechanism triggered by CAG repeat expansion in the ATXN3 gene in MJD patients remains unknown, numerous in vitro and in vivo studies have begun to shed light on the problem. Although a hyperexpanded CAG repeat RNA transcript is toxic and causes degeneration in a Drosophila model of MJD (Li et al., 2008), most evidence suggests that the key toxic species is instead the mutant ATXN3 protein with its polyQ expansion. Expanded ATXN3 is thought to undergo conformational changes and acquire toxic properties, either as a monomer or as part of oligomeric/ aggregate species, resulting in altered molecular interactions. The fact that the majority of suppressors of MJD toxicity affect protein misfolding and protein quality control pathways (Table 3) supports the view that altered conformation and protein misfolding are central to the disease process.
Table 3.
Genetic modifiers of mutant ATXN3 toxicity.
| Protein name | Function | Effect on toxicity | MJD model | Reference |
|---|---|---|---|---|
| ATXN3 | DUB | suppressor | D. melanogaster | Warrick et al., 2005 |
| none | R. norvegicus | Alves et al., 2010 | ||
| none | M. musculus | Hubener et al., 2010 | ||
| USP47 | DUB | suppressor | D. melanogaster | Bilen et al., 2007 |
| CHIP | Ub E3 ligase | suppressor | M. musculus | Williams et al., 2009 |
| E4B | Ub E3 ligase | suppressor | HEK293T cells | Matsumoto et al., 2004 |
| Ubiquitin | post-translational tag | suppressor | D. melanogaster | Bilen et al., 2007 |
| VCP | AAA ATPase | suppressor | D. melanogaster | Boeddrich et al., 2006 |
| HSP70 | chaperone | suppressor | D. melanogaster | Warrick et al., 1999 |
| D. melanogaster | Bilen et al., 2007 | |||
| HSP40 | chaperone | suppressor | D. melanogaster | Bilen et al., 2007 |
| αB-Crystallin | chaperone | suppressor | D. melanogaster | Bilen et al., 2007 |
| HSPB8 | chaperone | suppressor | HEK293T cells | Carra et al., 2010 |
| D. melanogaster | ||||
| TPR2 | chaperone | suppressor | D. melanogaster | Bilen et al., 2007 |
| CRAG | Guanosine triphosphatase | suppressor | M. musculus | Torashima et al., 2008 |
| Beclin-1 | protein binding | suppressor | neuronal cells | Nascimento-Ferreira et al., 2011 |
| R. norvegicus | ||||
| Bantam | microRNA | suppressor | D. melanogaster | Bilen et al., 2006 |
| MBNL1 | nucleic acid binding | suppressor | D. melanogaster | Li et al., 2008 |
| IGF2BP2 | mRNA binding | suppressor | D. melanogaster | Bilen et al., 2007 |
| PABP | poly(A) RNA binding | suppressor | D. melanogaster | Lessing et al., 2008 |
| UBXD2 | UBX protein | suppressor | D. melanogaster | Bilen et al., 2007 |
| UBXD8 | UBX protein | suppressor | D. melanogaster | Bilen et al., 2007 |
| Exportin1 | protein transporter | suppressor | D. melanogaster | Bilen et al., 2007 |
| HSF1 | transcription factor | suppressor | C. elegans | Teixeira-Castro et al., 2011 |
| DAF16 | transcription factor | suppressor | C. elegans | Teixeira-Castro et al., 2011 |
| NFAT5 | transcription factor | suppressor | D. melanogaster | Bilen et al., 2007 |
| ATF2 | transcription factor | suppressor | D. melanogaster | Bilen et al., 2007 |
| SIN3A | transcription regulator | suppressor | D. melanogaster | Bilen et al., 2007 |
| TRIM71 | transcription regulator | suppressor | D. melanogaster | Bilen et al., 2007 |
| Dbr | zinc ion binding | suppressor | D. melanogaster | Bilen et al., 2007 |
| ACOX1 | palmitoyl-CoA oxidase | suppressor | D. melanogaster | Bilen et al., 2007 |
| Sup35 | yeast prion domain | suppressor | D. melanogaster | Li et al., 2007 |
| UL97 | HCMV kinase | supressor | HeLa cells | Tower et al., 2011 |
| FBXL11 | transcription regulator | enhancer | D. melanogaster | Bilen et al., 2007 |
| ATXN2 | RNA and protein binding | enhancer | D. melanogaster | Lessing et al., 2008 |
How does mutant ATXN3 cause cellular dysfunction and cell death? As discussed below, several hypotheses have been put forth as the potential toxic mechanism triggered by misfolded mutant ATXN3 and its altered protein interactions: (i) formation of aggregates; (ii) failure of cellular protein homeostasis; (iii) impairment of axonal transport; (iv) transcriptional dysregulation; (v) mitochondrial dysfunction and oxidative stress; and (vi) abnormal neuronal signaling. These hypotheses are not mutually exclusive.
Neuronal inclusions formed by mutant ATXN3 – both NNIs and NCIs (described in section 2.2) - represent pathological hallmarks of MJD. Whether intraneuronal inclusions formed by polyQ disease proteins are directly toxic, however, has been hotly debated. When first discovered, inclusions in human MJD brain were speculated to mediate neurodegeneration as their abundance correlated with CAG repeat size and disease severity. Moreover, the inclusions also contain several key proteins including ubiquitin, proteasomal components, chaperones, transcription factors, and wild-type ATXN3 suggesting that various cellular pathways might be depleted of crucial components (Chai et al., 1999a; Chai et al., 1999b; Mori et al., 2005; Paulson et al., 1997b; Schmidt et al., 1998; Takahashi et al., 2001). Neuronal inclusions, however, do not correlate directly with degeneration and are currently viewed instead as biomarkers of cellular failure to clear mutant ATXN3.
As direct toxicity of large inclusions appears doubtful, what is the toxic species in MJD? Because misfolded β-rich polyQ protein monomers and oligomers are toxic to cells (Nagai et al., 2007) and a conformational change from α to β structure is observed in ATXN3 during its aggregation, β-rich ATXN3 monomers and oligomers may exist and be toxic to cells in MJD (Masino et al., 2011b). Efforts to detect and identify such species in brains of MJD patients and animal models of disease should help elucidate the aggregation process of ATXN3 and might support a strategy of targeting ATXN3 misfolding and early oligomerization as potential therapy for MJD.
Cells have different protein quality control systems to clear misfolded proteins and maintain cellular homeostasis. Indeed, molecular chaperones, the UPS and autophagy have all been implicated in the refolding and clearance of mutant ATXN3 (Berger et al., 2006; Chai et al., 1999a; Chai et al., 1999b). A study in a mouse model revealed a failure of some proteostasis systems in later stages of disease but before appreciable cell death (Chou et al., 2008). At earlier stages of disease, pathogenic ATXN3 induces cellular stress pathways, resulting in increased expression of certain molecular chaperones that are known to suppress expanded ATXN3 toxicity by facilitating its folding and decreasing aggregates (Bilen and Bonini, 2007; Chai et al., 1999a; Chou et al., 2008; Huen and Chan, 2005; Warrick et al., 1999). Consistent with this, heat-shock factor 1 (HSF1) was recently shown to have an early protective role by decreasing protein aggregation in C. elegans models of MJD (Teixeira-Castro et al., 2011). In later stages of disease in MJD transgenic mice, however, there is downregulation of HSP70 and HSP40 which would impair the ability of neurons to handle the cellular stress caused by mutant ATXN3 (Chou et al., 2008; Huen and Chan, 2005). This depletion of chaperones was recently extended to the small heat-shock protein HSP27 in cellular models of MJD (Chang et al., 2009). Pathogenic ATXN3 may have an equivalent effect on autophagy as disease progresses, because this clearance pathway is induced in MJD Drosophila larvae but is depleted in brains of MJD patients and transgenic mouse models (Bilen and Bonini, 2007; Nascimento-Ferreira et al., 2011). It will be important to determine whether this decrease in authophagic function happens before neuronal loss. Interestingly, proteasomal protein degradation does not seem to be compromised in cellular and Drosophila models of MJD (Berke et al., 2005; Bilen and Bonini, 2007; Chai et al., 1999b). Thus, the decrease of some cellular protein clearance pathways correlates with the accumulation of mutant ATXN3 with aging, implying an impairment of cellular protein homeostasis. This impairment of proteostasis (at least of the chaperone machinery) supports the view that neuronal dysfunction precedes cell loss in MJD.
Possibly as a byproduct of a failure in cellular homeostasis, ATXN3 aggregates are found not only in the soma but also in fiber tracts known to undergo neurodegeneration in MJD patients (Seidel et al., 2010). In C.elegans and Drosophila models of MJD, mutant ATXN3 aggregates induce swelling and aberrant branching of neuronal processes, which impairs synaptic transmission (Gunawardena and Goldstein, 2005; Khan et al., 2006). ATXN3 interacts with cytoskeletal components and is important for cytoskeletal organization, thus it might normally play a role in cytoskeletal transport that becomes dysregulated in mutant ATXN3 (Burnett and Pittman, 2005; do Carmo Costa et al., 2010; Rodrigues et al., 2010). Insights from these studies suggest that axonal dysfunction may play a role in MJD pathogenesis and may explain why MJD patients often have motor neuropathy or neuronopathy.
Abnormal interactions of mutant ATXN3 with its native protein partners, or novel interactions with new partners, may be a recurring theme underlying many of the potential pathogenic mechanisms in MJD. Numerous interactors of wild-type ATXN3 have been idenitified, but little is known about their relative interaction with mutant ATXN3. For example, expanded ATXN3 interacts differently with two native partners, CHIP and parkin, which may contribute tothe depletion of these neuroprotective proteins in models of MJD (Durcan et al., 2010; Scaglione et al., 2011). Because ATXN3 is implicated in ubiquitin and protein quality control pathways, aberrant interactions of mutant ATXN3 could, in principle, lead to altered stability of a numerous substrate proteins including critically important, cell-specific factors. This could help explain selective neurodegeneration in MJD. Comparative analysis of the behavior of wild-type versus mutant ATXN3 in specific protein complexes will likely be crucial to a full understanding of cellular dysfunction in MJD.
Aberrant interactions of mutant ATXN3 also could influence its activity as a putative transcriptional regulator. Upon polyQ expansion, the action of ATXN3 as a transcriptional corepressor or activator seems to be compromised (Araujo et al., 2011; Evert et al., 2006). Altered interactions with transcription factors and co-activators and the recruitment of certain transcription factors to polyQ protein aggregates are common themes in the polyQ diseases, suggesting that transcriptional dysregulation contributes to disease pathogenesis in MJD and other polyQ disorders. Transcriptome analyses in a neuronal cell model of MJD and cerebella from transgenic MJD mice revealed altered transcription of many genes (Chou et al., 2008; Evert et al., 2001; Evert et al., 2003). Mutant ATXN3 leads to downregulation of genes involved in glutamatergic neurotransmission, intracellular calcium signaling/mobilization or MAP kinase pathways, GABAA/B receptor subunits, HSPs, and transcription factors regulating neuronal survival and differentiation (Chou et al., 2008). Conversely, upregulated genes include ones that are involved in neuronal cell death and inflammation (Chou et al., 2008; Evert et al., 2001; Evert et al., 2003). As posttranslational modifications including ubiquitination are known to regulate transcription, an alteration in the DUB activity of mutant ATXN3 could contribute to cell-specific dysregulation of target genes.
One recently described example is the reduced ability of expanded ATXN3 to activate FOXO4-mediated SOD2 transcription thereby leading to increased cellular susceptibility to oxidative stress (Araujo et al., 2011). Interestingly, levels of HSP27, which also has anti-oxidant features, are decreased in MJD suggesting that, as in other polyQ diseases, the cellular defenses to reactive oxygen species (ROS) may be depleted in this disease (Chang et al., 2009). In fact, some cellular models of MJD show decreased antioxidant enzyme activity and increased mitochondrial-mediated cell death via apoptosis (Tsai et al., 2004; Yu et al., 2009). However, question of apoptosis-mediated cell death in MJD is controversial as it isnot activated in several other cellular and mouse MJD models (Evert et al., 1999; Silva-Fernandes et al., 2010). Nonetheless, the view that mitochondrial damage and increased oxidative stress contribute to disease has gained strength recently as a potential pathomechanism in MJD.
The upregulation of inflammatory genes in cells expressing expanded ATXN3 was confirmed by detection of neuroinflammatory markers in the pons of MJD patients, suggesting that glia contribute to MJD pathogenesis (Evert et al., 2001; Evert et al., 2003). Neuroinflammation, however, may not be activated until later stages of disease as it was not detected in early stages of disease in a transgenic mouse model of MJD (Silva-Fernandes et al., 2010). Although there is evidence that glial cells expressing expanded ATXN3 can alter Purkinje cell function in another MJD transgenic mouse model (Boy et al., 2009), the precise role of glia in the pathogenic mechanism of MJD is largely unexplored. Systematic temporal and functional analysis of the several types of glial cells in brains from MJD patients and MJD mouse models will help to understand their specific role during the development of this disease.
Consistent with the downregulation of genes involved in intracellular calcium signaling/mobilization and MAP kinase pathways (Chou et al., 2008), neurons expressing mutant ATXN3 display a decrease in intracellular Ca2+ through an abnormal association with the type 1 inositol 1,4,5-triphosphate receptor (InsP3R1) (Chen et al., 2008). Indeed, as in other polyQ diseases, the electrophysiological properties of neurons are altered in MJD. In addition to impaired calcium signaling (Chen et al., 2008), potassium channel dysfunction and depolarized resting membrane potential were observed in neuronal cells expressing mutant ATXN3 (Jeub et al., 2006). Consistent with this finding, in a mouse model of MJD we recently identified robust early changes in cerebellar Purkinje neuron firing that are associated with altered kinetics of voltage-activated potassium (Kv) channels (Shakkottai et al., 2011). Importantly, the silencing of repetitively firing Purkinje cells in MJD transgenic mice occurs at an early stage of disease when mice display motor incoordination but have not yet manifested any neuronal degeneration (Shakkottai et al., 2011). In parallel studies in transfected cells, mutant ATXN3 affected slightly the kinetics of Kv3 and Kv1 channels, suggesting that early Purkinje cell dysfunction in MJD mice may reflect a direct effect of ATXN3 on channel activity (Shakkottai et al., 2011). Clearly, further studies are needed to explore potential electrophysiological changes in other affected neuronal populations in MJD, but early evidence suggests that altered neuronal physiology contributes to MJD pathogenesis and likely underlies some motor symptoms before neuronal cell death has begun to occur (Shakkottai et al., 2011). Understanding the role of altered neuronal physiology in MJD and other polyQ diseases may reveal new, potentially shared therapeutic targets for these incurable neurodegenerative diseases.
In summary, mutant ATXN3 misfolding and its altered molecular interactions with numerous proteins may well trigger a series of interrelated pathogenic cascades in MJD. In a slowly progressive chronic disease extending over decades and involving various types of cells within the brain, it may be unrealistic to expect a single mechanism to emerge as the cause. Even though much has already been revealed about MJD pathogenesis, much more needs to be known before we will clearly understand the molecular events driving this process.
6. Development of MJD therapeutics
MJD and the other polyQ diseases are currently untreatable. Despite the absence of preventive treatment, many symptoms of disease can be treated using pharmacological and nonpharmacological measures. Details about symptomatic therapy and clinical trials in MJD can be found in several recent reviews (Bettencourt and Lima, 2011; Paulson, 1998 Oct 10 [updated 2011 March 17]).
Seeking a preventive therapy, MJD researchers have explored pharmacological and genetic approaches, both of which have shown some promise. As described above, efforts are also being made to generate animal models of disease with robust phenotypes that will facilitate therapeutic testing and further advance our understanding of the pathogenic mechanisms in MJD (see section 5). Silencing strategies that target the causative RNA or disease protein have the advantage of hitting proximally in pathogenic cascade and thus not requiring a detailed understanding of downstream mechanisms, which remain unclear in MJD. On the other hand, approaches targeting crucial events in disease pathogenesis may be more potent but depend on accurate knowledge of the central pathogenic pathways and epiphenomena elicited by a specific mutation. Both of these strategies have been explored in the development of potential therapies for MJD.
The fact that phenotypic and pathological features of disease could be reversed In a conditional mouse model of MJD by switching off the transgene indicates that strategies targeting RNA or protein clearance might be useful as future preventive therapies for patients even after symptoms and signs of disease have begun (Boy et al., 2009). Current nucleotide-based gene suppression strategies (e.g. RNAi and antisense molecules) can target specific genes with high specificity, mediating specific suppression of alleles differing by as little as one or a few base pairs. Hence, a gene silencing strategy in MJD is within the realm of possibility.
Silencing ATXN3 transcripts by RNA interference (RNAi) has been shown to mitigate degeneration in a rat model of MJD (Alves et al., 2008a; Alves et al., 2010). This rescue occurred in a nonallele-specific manner as intracerebral viral-injections of short hairpin RNAs (shRNAs) specific or nonspecific for the mutant ATXN3 transcript showed similar effects (Alves et al., 2008a; Alves et al., 2010). Thus, silencing of wild type ATXN3 along with mutant ATXN3 appears not to be deleterious, at least in this model, which concurs with the lack of neurodegeneration in Atxn3 knockout mice (Schmitt et al., 2007). Other types of RNA-targeting molecules such as peptide nucleic acid (PNA) and locked nucleic acid (LNA) antisense oligomers also have been shown to be effective in reducing ATXN3 levels in fibroblasts from MJD patients and constitute promising molecules to test in vivo (Hu et al., 2011; Hu et al., 2009).
Targeting mutant ATXN3 clearance is also a promising approach toward therapy. Pharmacological induction of autophagy using a rapamycin ester (temsirolimus) increases mutant ATXN3 degradation, reduces the number of aggregates, and improves the motor phenotype in a mouse model of MJD (Menzies et al., 2010). As temsirolimus has been designed for long-term use in patients, it represents a potential therapy for MJD. Additional evidence supporting autophagy induction as a viable therapeutic target is the fact that lentiviral-mediated overexpression of the autophagy protein beclin-1 increases clearance of mutant ATXN3 and decreases aggregates in a MJD rat model (Nascimento-Ferreira et al., 2011). Thus, protein quality control systems involved in mutant ATXN3 degradation represent promising therapeutic targets in MJD.
Though much remains to be learned about the pathogenic mechanisms of MJD, several key molecular events in pathogenesis including aggregate formation, transcriptional dysregulation and abnormal neuronal signaling, have been targeted in the development of potential therapies. Additionally, several genetic modifiers of mutant ATXN3 toxicity have been identified which themselves represent potential targets (Table 3).
Mutant ATXN3 aggregation has been successfully decreased by treatment of Drosophila and C. elegans models of MJD with 17-(allylamino)-17-demethoxygeldanamycin (17-AAG), an HSP90 inhibitor (Fujikake et al., 2008; Teixeira-Castro et al., 2011). 17-AAG also improved locomotor activity in MJD transgenic C.elegans (Teixeira-Castro et al., 2011). Y-27632, an inhibitor of Rho kinases, also decreased soluble and insoluble expanded ATXN3 in cells (Bauer et al., 2009).
Mutant ATXN3-mediated transcriptional downregulation and H3/H4 histone hypoacetylation has been significantly reversed by treating MJD transgenic mice with sodium butyrate, a HDAC inhibitor (Chou et al., 2010) with a corresponding improvement in motor performance (Chou et al., 2010). Recently, treatment of MJD transgenic C.elegans with valproic acid, another HDAC inhibitor, also led to improved locomotor activity accompanied by a decrease in aggregates (Teixeira-Castro et al., 2011). Therefore, compounds that promote histone acetylation over deacetylation may hold promise as preventive therapy in MJD.
Another therapeutic strategy is to use pharmacological approaches to correct abnormal neuronal signaling. The use of dantrolene to stabilize intracellular Ca2+ signaling in MJD transgenic mice resulted in improved motor phenotype and reduced neuronal loss (Chen et al., 2008). And finally, short term administration of an activator of calcium-activated potassium channels, SKA-31, partially corrected Purkinje neuronal firing and improved motor function in MJD transgenic mice (Shakkottai et al., 2011).
In conclusion, there have been several promising developments in the search for therapies for MJD. Agents targeting different molecular events thought to contribute to disease pathogenesis have shown some success in animal models of MJD, though none has yet been advanced to the point of clinical trials in MJD patients. It may be necessary to develop safer, and more efficacious, second generation molecules with reduced toxicity or adverse effects and enhanced delivery to the CNS. Pre-clinical trials of promising molecules using gene delivery or pharmacological approaches in mammalian models of MJD will be crucial for the eventual, successful generation of an effective treatment for MJD.
7. Final remarks
This year marks the 20th anniversary of an historical moment in human genetics: the discovery that heritable human diseases can be caused by dynamic repeat mutations. In 1991, expanded CCG and CAG repeat sequences were reported as the molecular basis of Fragile X syndrome and SBMA, respectively (Kremer et al.1991; LaSpada et al. 1991). Soon afterward, expansion of polyQ-encoding CAG repeats was revealed to be the cause of the largest class of dynamic repeat disorders, the polyQ neurodegenerative diseases, of which MJD is one of at least ten. In the 17 years since the discovery of the MJD disease gene, many advances have taken place both with respect to the disease itself and to the underlying pathomechanism. ATXN3, the MJD protein, is now known to be a highly specialized DUB that participates in several ubiquitin-related cellular pathways. This knowledge of ATXN3 function, together with the development of a wide range of cellular and animal models of MJD, has greatly advanced our understanding of disease pathogenesis in MJD. This progress notwithstanding, much more needs to be learned before we have a firm grasp of the molecular mechanisms driving neuronal dysfunction and neuronal cell death in MJD. As is true for all polyQ diseases, clarifying the normal functions of ATXN3 and defining the ways it is dysfunctional in the disease state are critical steps toward a full understanding of disease mechanisms. Several potential pathogenic pathways triggered by expanded polyQ proteins are shared among the polyQ diseases. Thus, there is the potential for class-wide therapeutic strategies to disease prevention; indeed, several therapeutic agents along these lines have shown some success in animal models of MJD. At the same time, the search is on for disease-specific strategies that target proximal steps in the pathogenic cascade (e.g. drugs or nucleotide-based approaches to reduce levels of the ATXN3 transcript or disease protein) or target pathways implicated specifically in MJD further downstream in the cascade (e.g. pharmacological agents that alter ion channel dysfunction, as recently described in MJD). To date, no potential preventive strategies have been tested in MJD patients. But it is our hope that the remarkable, recent advances in MJD research will open the door to such clinical trials soon.
Article Highlights.
MJD is a neurodegenerative disease caused by polyQ expansion in ATXN3
ATXN3 is a deubiquitinating enzyme participating in ubiquitin-mediated proteostasis
PolyQ expansion in ATXN3 triggers cellular dysfunction and selective neuronal death
Improved understanding of disease mechanisms is suggesting routes to therapy
Acknowledgements
We would like to thank Drs. Sokol V. Todi and K. Mathew Scaglione for critical reading of the manuscript. MCC is the recipient of fellowships from Fundação para a Ciência e a Tecnologia (FCT) (SFRH/BPD/28560/2006) and National Ataxia Foundation (NAF Research Fellowship Award 2011). HLP is funded by NIH NS038712, the Mateus family fund, and the Ataxia Medical Research Foundation.
Abbreviations
- ATXN3
ataxin-3
- bZIP
basic leucine zipper motif
- CBP
CREB binding protein
- cDNA
complementary DNA
- CHIP
C-terminus of Hsc70 interacting protein
- CK2
casein kinase 2
- CMV
cytomegalovirus
- CNS
central nervous system
- CREB
cAMP-response element binding
- DRPLA
Dentatorubral pallidoluysian atrophy
- DUB
deubiquitinating enzyme
- ERAD
endoplasmic reticulum-associated degradation
- FOXO
forkhead box O
- HD
Huntington disease
- HDAC
histone deacetylase
- HDL2
Huntington Disease-like 2
- HSF1
heat-shock factor 1
- HSP
heat-shock protein
- Htt
huntingtin
- IGF1
insulin/ insulin-like growth factor 1
- JD
Josephin domain
- Kv
voltage-activated potassium
- LNA
locked nucleic acid
- MITOL
mitochondrial ubiquitin ligase
- MJD
Machado-Joseph disease
- MRI
magnetic resonance imaging
- NCIs
neuronal cytoplasmic inclusions
- NCOR1
nuclear receptor co-repressor 1
- NEDD8
neural precursor cell expressed developmentally downregulated 8
- NES
nuclear export signal
- NLS
nuclear localization signal
- NNIs
neuronal nuclear inclusions
- p97
ATPase p97
- PCAF
p300/CREBBP associated factor
- PLIC1
protein linking IAP to the cytoskeleton
- PNA
peptide nucleic acid
- polyQ
polyglutamine
- Prnp
prion protein
- RNAi
RNA interference
- ROS
reactive oxygen species
- SCA
Spinocerebellar ataxia
- SBMA
Spinal bulbar muscular atrophy
- shRNAs
short hairpin RNAs
- SNPs
single nucleotide polymorphisms
- SOD
superoxide dismutase
- STRs
single tandem repeats
- SUMO
small ubiquitin-like modifier
- Ub
ubiquitin
- Ubl
ubiquitin-like
- UIM
ubiquitin interacting motif
- UPS
ubiquitin proteasome system
- UTR
untranslated region
- VCP
valosin containing protein
- YAC
yeast artificial chromosome
- 17-AAG
17-allylamino-17-demethoxygeldanamycin
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Maria do Carmo Costa, Email: mariadoc@med.umich.edu.
Henry L. Paulson, Email: henryp@med.umich.edu.
References
- Abe Y, Tanaka F, Matsumoto M, Doyu M, Hirayama M, Kachi T, Sobue G. CAG repeat number correlates with the rate of brainstem and cerebellar atrophy in Machado-Joseph disease. Neurology. 1998;51:882–884. doi: 10.1212/wnl.51.3.882. [DOI] [PubMed] [Google Scholar]
- Alves S, Nascimento-Ferreira I, Auregan G, Hassig R, Dufour N, Brouillet E, Pedroso de Lima MC, Hantraye P, Pereira de Almeida L, Deglon N. Allele-specific RNA silencing of mutant ataxin-3 mediates neuroprotection in a rat model of Machado-Joseph disease. PLoS One. 2008a;3:e3341. doi: 10.1371/journal.pone.0003341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves S, Nascimento-Ferreira I, Dufour N, Hassig R, Auregan G, Nobrega C, Brouillet E, Hantraye P, Pedroso de Lima MC, Deglon N, de Almeida LP. Silencing ataxin-3 mitigates degeneration in a rat model of Machado-Joseph disease: no role for wild-type ataxin-3? Hum Mol Genet. 2010;19:2380–2394. doi: 10.1093/hmg/ddq111. [DOI] [PubMed] [Google Scholar]
- Alves S, Regulier E, Nascimento-Ferreira I, Hassig R, Dufour N, Koeppen A, Carvalho AL, Simoes S, de Lima MC, Brouillet E, Gould VC, Deglon N, de Almeida LP. Striatal and nigral pathology in a lentiviral rat model of Machado-Joseph disease. Hum Mol Genet. 2008b;17:2071–2083. doi: 10.1093/hmg/ddn106. [DOI] [PubMed] [Google Scholar]
- Antony PM, Mantele S, Mollenkopf P, Boy J, Kehlenbach RH, Riess O, Schmidt T. Identification and functional dissection of localization signals within ataxin-3. Neurobiol Dis. 2009;36:280–292. doi: 10.1016/j.nbd.2009.07.020. [DOI] [PubMed] [Google Scholar]
- Araujo J, Breuer P, Dieringer S, Krauss S, Dorn S, Zimmermann K, Pfeifer A, Klockgether T, Wuellner U, Evert BO. FOXO4-dependent upregulation of superoxide dismutase-2 in response to oxidative stress is impaired in spinocerebellar ataxia type 3. Hum Mol Genet. 2011;20:2928–2941. doi: 10.1093/hmg/ddr197. [DOI] [PubMed] [Google Scholar]
- Bauer PO, Nukina N. The pathogenic mechanisms of polyglutamine diseases and current therapeutic strategies. J Neurochem. 2009;110:1737–1765. doi: 10.1111/j.1471-4159.2009.06302.x. [DOI] [PubMed] [Google Scholar]
- Bauer PO, Wong HK, Oyama F, Goswami A, Okuno M, Kino Y, Miyazaki H, Nukina N. Inhibition of Rho kinases enhances the degradation of mutant huntingtin. J Biol Chem. 2009;284:13153–13164. doi: 10.1074/jbc.M809229200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger Z, Ravikumar B, Menzies FM, Oroz LG, Underwood BR, Pangalos MN, Schmitt I, Wullner U, Evert BO, O'Kane CJ, Rubinsztein DC. Rapamycin alleviates toxicity of different aggregate-prone proteins. Hum Mol Genet. 2006;15:433–442. doi: 10.1093/hmg/ddi458. [DOI] [PubMed] [Google Scholar]
- Berke SJ, Chai Y, Marrs GL, Wen H, Paulson HL. Defining the role of ubiquitin-interacting motifs in the polyglutamine disease protein, ataxin-3. J Biol Chem. 2005;280:32026–32034. doi: 10.1074/jbc.M506084200. [DOI] [PubMed] [Google Scholar]
- Berke SJ, Paulson HL. Protein aggregation and the ubiquitin proteasome pathway: gaining the UPPer hand on neurodegeneration. Curr Opin Genet Dev. 2003;13:253–261. doi: 10.1016/s0959-437x(03)00053-4. [DOI] [PubMed] [Google Scholar]
- Berke SJ, Schmied FA, Brunt ER, Ellerby LM, Paulson HL. Caspase-mediated proteolysis of the polyglutamine disease protein ataxin-3. J Neurochem. 2004;89:908–918. doi: 10.1111/j.1471-4159.2004.02369.x. [DOI] [PubMed] [Google Scholar]
- Bettencourt C, Lima M. Machado-Joseph Disease: from first descriptions to new perspectives. Orphanet J Rare Dis. 2011;6:35. doi: 10.1186/1750-1172-6-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettencourt C, Santos C, Montiel R, Costa MC, Cruz-Morales P, Santos LR, Simoes N, Kay T, Vasconcelos J, Maciel P, Lima M. Increased transcript diversity: novel splicing variants of Machado-Joseph disease gene (ATXN3) Neurogenetics. 2010;11:193–202. doi: 10.1007/s10048-009-0216-y. [DOI] [PubMed] [Google Scholar]
- Bichelmeier U, Schmidt T, Hubener J, Boy J, Ruttiger L, Habig K, Poths S, Bonin M, Knipper M, Schmidt WJ, Wilbertz J, Wolburg H, Laccone F, Riess O. Nuclear localization of ataxin-3 is required for the manifestation of symptoms in SCA3: in vivo evidence. J Neurosci. 2007;27:7418–7428. doi: 10.1523/JNEUROSCI.4540-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilen J, Bonini NM. Genome-wide screen for modifiers of ataxin-3 neurodegeneration in Drosophila. PLoS Genet. 2007;3:1950–1964. doi: 10.1371/journal.pgen.0030177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeddrich A, Gaumer S, Haacke A, Tzvetkov N, Albrecht M, Evert BO, Muller EC, Lurz R, Breuer P, Schugardt N, Plassmann S, Xu K, Warrick JM, Suopanki J, Wullner U, Frank R, Hartl UF, Bonini NM, Wanker EE. An arginine/lysine-rich motif is crucial for VCP/p97-mediated modulation of ataxin-3 fibrillogenesis. EMBO J. 2006;25:1547–1558. doi: 10.1038/sj.emboj.7601043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boy J, Schmidt T, Schumann U, Grasshoff U, Unser S, Holzmann C, Schmitt I, Karl T, Laccone F, Wolburg H, Ibrahim S, Riess O. A transgenic mouse model of spinocerebellar ataxia type 3 resembling late disease onset and gender-specific instability of CAG repeats. Neurobiol Dis. 2010;37:284–293. doi: 10.1016/j.nbd.2009.08.002. [DOI] [PubMed] [Google Scholar]
- Boy J, Schmidt T, Wolburg H, Mack A, Nuber S, Bottcher M, Schmitt I, Holzmann C, Zimmermann F, Servadio A, Riess O. Reversibility of symptoms in a conditional mouse model of spinocerebellar ataxia type 3. Hum Mol Genet. 2009;18:4282–4295. doi: 10.1093/hmg/ddp381. [DOI] [PubMed] [Google Scholar]
- Breuer P, Haacke A, Evert BO, Wullner U. Nuclear aggregation of polyglutamine-expanded ataxin-3: fragments escape the cytoplasmic quality control. J Biol Chem. 2010;285:6532–6537. doi: 10.1074/jbc.M109.036335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burk K, Globas C, Bosch S, Klockgether T, Zuhlke C, Daum I, Dichgans J. Cognitive deficits in spinocerebellar ataxia type 1, 2, and 3. J Neurol. 2003;250:207–211. doi: 10.1007/s00415-003-0976-5. [DOI] [PubMed] [Google Scholar]
- Burnett B, Li F, Pittman RN. The polyglutamine neurodegenerative protein ataxin-3 binds polyubiquitylated proteins and has ubiquitin protease activity. Hum Mol Genet. 2003;12:3195–3205. doi: 10.1093/hmg/ddg344. [DOI] [PubMed] [Google Scholar]
- Burnett BG, Pittman RN. The polyglutamine neurodegenerative protein ataxin 3 regulates aggresome formation. Proc Natl Acad Sci U S A. 2005;102:4330–4335. doi: 10.1073/pnas.0407252102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho DR, La Rocque-Ferreira A, Rizzo IM, Imamura EU, Speck-Martins CE. Homozygosity enhances severity in spinocerebellar ataxia type 3. Pediatr Neurol. 2008;38:296–299. doi: 10.1016/j.pediatrneurol.2007.12.006. [DOI] [PubMed] [Google Scholar]
- Cemal CK, Carroll CJ, Lawrence L, Lowrie MB, Ruddle P, Al-Mahdawi S, King RH, Pook MA, Huxley C, Chamberlain S. YAC transgenic mice carrying pathological alleles of the MJD1 locus exhibit a mild and slowly progressive cerebellar deficit. Hum Mol Genet. 2002;11:1075–1094. doi: 10.1093/hmg/11.9.1075. [DOI] [PubMed] [Google Scholar]
- Chai Y, Berke SS, Cohen RE, Paulson HL. Poly-ubiquitin binding by the polyglutamine disease protein ataxin-3 links its normal function to protein surveillance pathways. J Biol Chem. 2004;279:3605–3611. doi: 10.1074/jbc.M310939200. [DOI] [PubMed] [Google Scholar]
- Chai Y, Koppenhafer SL, Bonini NM, Paulson HL. Analysis of the role of heat shock protein (Hsp) molecular chaperones in polyglutamine disease. J Neurosci. 1999a;19:10338–10347. doi: 10.1523/JNEUROSCI.19-23-10338.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai Y, Koppenhafer SL, Shoesmith SJ, Perez MK, Paulson HL. Evidence for proteasome involvement in polyglutamine disease: localization to nuclear inclusions in SCA3/MJD and suppression of polyglutamine aggregation in vitro. Hum Mol Genet. 1999b;8:673–682. doi: 10.1093/hmg/8.4.673. [DOI] [PubMed] [Google Scholar]
- Chai Y, Shao J, Miller VM, Williams A, Paulson HL. Live-cell imaging reveals divergent intracellular dynamics of polyglutamine disease proteins and supports a sequestration model of pathogenesis. Proc Natl Acad Sci U S A. 2002;99:9310–9315. doi: 10.1073/pnas.152101299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai Y, Wu L, Griffin JD, Paulson HL. The role of protein composition in specifying nuclear inclusion formation in polyglutamine disease. J Biol Chem. 2001;276:44889–44897. doi: 10.1074/jbc.M106575200. [DOI] [PubMed] [Google Scholar]
- Chang WH, Tien CL, Chen TJ, Nukina N, Hsieh M. Decreased protein synthesis of Hsp27 associated with cellular toxicity in a cell model of Machado-Joseph disease. Neurosci Lett. 2009;454:152–156. doi: 10.1016/j.neulet.2009.03.004. [DOI] [PubMed] [Google Scholar]
- Chen X, Tang TS, Tu H, Nelson O, Pook M, Hammer R, Nukina N, Bezprozvanny I. Deranged calcium signaling and neurodegeneration in spinocerebellar ataxia type 3. J Neurosci. 2008;28:12713–12724. doi: 10.1523/JNEUROSCI.3909-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou AH, Chen SY, Yeh TH, Weng YH, Wang HL. HDAC inhibitor sodium butyrate reverses transcriptional downregulation and ameliorates ataxic symptoms in a transgenic mouse model of SCA3. Neurobiol Dis. 2010;41:481–488. doi: 10.1016/j.nbd.2010.10.019. [DOI] [PubMed] [Google Scholar]
- Chou AH, Yeh TH, Ouyang P, Chen YL, Chen SY, Wang HL. Polyglutamine-expanded ataxin-3 causes cerebellar dysfunction of SCA3 transgenic mice by inducing transcriptional dysregulation. Neurobiol Dis. 2008;31:89–101. doi: 10.1016/j.nbd.2008.03.011. [DOI] [PubMed] [Google Scholar]
- Colomer Gould VF, Goti D, Pearce D, Gonzalez GA, Gao H, Bermudez de Leon M, Jenkins NA, Copeland NG, Ross CA, Brown DR. A mutant ataxin-3 fragment results from processing at a site N-terminal to amino acid 190 in brain of Machado-Joseph disease-like transgenic mice. Neurobiol Dis. 2007;27:362–369. doi: 10.1016/j.nbd.2007.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa MC, Gomes-da-Silva J, Miranda CJ, Sequeiros J, Santos MM, Maciel P. Genomic structure, promoter activity, and developmental expression of the mouse homologue of the Machado-Joseph disease (MJD) gene. Genomics. 2004;84:361–373. doi: 10.1016/j.ygeno.2004.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutinho P, Andrade C. Autosomal dominant system degeneration in Portuguese families of the Azores Islands. A new genetic disorder involving cerebellar, pyramidal, extrapyramidal and spinal cord motor functions. Neurology. 1978;28:703–709. doi: 10.1212/wnl.28.7.703. [DOI] [PubMed] [Google Scholar]
- Coutinho P, Sequeiros J. Clinical, genetic and pathological aspects of Machado-Joseph disease. J Genet Hum. 1981;29:203–209. [PubMed] [Google Scholar]
- D'Abreu A, Franca MC, Jr, Yasuda CL, Campos BA, Lopes-Cendes I, Cendes F. Neocortical Atrophy in Machado-Joseph Disease: A Longitudinal Neuroimaging Study. J Neuroimaging. 2011 doi: 10.1111/j.1552-6569.2011.00614.x. [DOI] [PubMed] [Google Scholar]
- D'Abreu A, Franca M, Jr, Appenzeller S, Lopes-Cendes I, Cendes F. Axonal dysfunction in the deep white matter in Machado-Joseph disease. J Neuroimaging. 2009;19:9–12. doi: 10.1111/j.1552-6569.2008.00260.x. [DOI] [PubMed] [Google Scholar]
- D'Abreu A, Franca MC, Jr, Yasuda CL, Souza MS, Lopes-Cendes I, Cendes F. Thalamic volume and dystonia in Machado-Joseph disease. J Neuroimaging. 2010;21:e91–e93. doi: 10.1111/j.1552-6569.2010.00464.x. [DOI] [PubMed] [Google Scholar]
- De Oliveira MS, D'Abreu A, Franca MC, Jr, Lopes-Cendes I, Cendes F, Castellano G. MRI-Texture Analysis of Corpus Callosum, Thalamus, Putamen, and Caudate in Machado-Joseph Disease. J Neuroimaging. 2010 doi: 10.1111/j.1552-6569.2010.00553.x. [DOI] [PubMed] [Google Scholar]
- Di Prospero NA, Fischbeck KH. Therapeutics development for triplet repeat expansion diseases. Nat Rev Genet. 2005;6:756–765. doi: 10.1038/nrg1690. [DOI] [PubMed] [Google Scholar]
- do Carmo Costa M, Bajanca F, Rodrigues AJ, Tome RJ, Corthals G, Macedo-Ribeiro S, Paulson HL, Logarinho E, Maciel P. Ataxin-3 plays a role in mouse myogenic differentiation through regulation of integrin subunit levels. PLoS One. 2010;5:e11728. doi: 10.1371/journal.pone.0011728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson KM, Li W, Ching KA, Batalov S, Tsai CC, Joazeiro CA. Ubiquitin-mediated sequestration of normal cellular proteins into polyglutamine aggregates. Proc Natl Acad Sci U S A. 2003;100:8892–8897. doi: 10.1073/pnas.1530212100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doss-Pepe EW, Stenroos ES, Johnson WG, Madura K. Ataxin-3 interactions with rad23 and valosin-containing protein and its associations with ubiquitin chains and the proteasome are consistent with a role in ubiquitin-mediated proteolysis. Mol Cell Biol. 2003;23:6469–6483. doi: 10.1128/MCB.23.18.6469-6483.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durcan TM, Kontogiannea M, Thorarinsdottir T, Fallon L, Williams AJ, Djarmati A, Fantaneanu T, Paulson HL, Fon EA. The Machado-Joseph disease-associated mutant form of ataxin-3 regulates parkin ubiquitination and stability. Hum Mol Genet. 2010;20:141–154. doi: 10.1093/hmg/ddq471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durr A, Stevanin G, Cancel G, Duyckaerts C, Abbas N, Didierjean O, Chneiweiss H, Benomar A, Lyon-Caen O, Julien J, Serdaru M, Penet C, Agid Y, Brice A. Spinocerebellar ataxia 3 and Machado-Joseph disease: clinical, molecular, and neuropathological features. Ann Neurol. 1996;39:490–499. doi: 10.1002/ana.410390411. [DOI] [PubMed] [Google Scholar]
- Eichler L, Bellenberg B, Hahn HK, Koster O, Schols L, Lukas C. Quantitative assessment of brain stem and cerebellar atrophy in spinocerebellar ataxia types 3 and 6: impact on clinical status. AJNRAm J Neuroradiol. 2011;32:890–897. doi: 10.3174/ajnr.A2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellisdon AM, Pearce MC, Bottomley SP. Mechanisms of Ataxin-3 Misfolding and Fibril Formation: Kinetic Analysis of a Disease-associated Polyglutamine Protein. J Mol Biol. 2007 doi: 10.1016/j.jmb.2007.02.058. [DOI] [PubMed] [Google Scholar]
- Ellisdon AM, Thomas B, Bottomley SP. The two-stage pathway of ataxin-3 fibrillogenesis involves a polyglutamine-independent step. J Biol Chem. 2006;281:16888–16896. doi: 10.1074/jbc.M601470200. [DOI] [PubMed] [Google Scholar]
- Emmel VE, Alonso I, Jardim LB, Saraiva-Pereira ML, Sequeiros J. Does DNA methylation in the promoter region of the ATXN3 gene modify age at onset in MJD (SCA3) patients? Clin Genet. 2011;79:100–102. doi: 10.1111/j.1399-0004.2010.01508.x. [DOI] [PubMed] [Google Scholar]
- Etchebehere EC, Cendes F, Lopes-Cendes I, Pereira JA, Lima MC, Sansana CR, Silva CA, Camargo MF, Santos AO, Ramos CD, Camargo EE. Brain single-photon emission computed tomography and magnetic resonance imaging in Machado-Joseph disease. Arch Neurol. 2001;58:1257–1263. doi: 10.1001/archneur.58.8.1257. [DOI] [PubMed] [Google Scholar]
- Eto K, Sumi SM, Bird TD, McEvoy-Bush T, Boehnke M, Schellenberg G. Family with dominantly inherited ataxia, amyotrophy, and peripheral sensory loss. Spinopontine atrophy or Machado-Joseph Azorean disease in another non-Portuguese family? Arch Neurol. 1990;47:968–974. doi: 10.1001/archneur.1990.00530090038011. [DOI] [PubMed] [Google Scholar]
- Evert BO, Araujo J, Vieira-Saecker AM, de Vos RA, Harendza S, Klockgether T, Wullner U. Ataxin-3 represses transcription via chromatin binding, interaction with histone deacetylase 3, and histone deacetylation. J Neurosci. 2006;26:11474–11486. doi: 10.1523/JNEUROSCI.2053-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evert BO, Vogt IR, Kindermann C, Ozimek L, de Vos RA, Brunt ER, Schmitt I, Klockgether T, Wullner U. Inflammatory genes are upregulated in expanded ataxin-3-expressing cell lines and spinocerebellar ataxia type 3 brains. J Neurosci. 2001;21:5389–5396. doi: 10.1523/JNEUROSCI.21-15-05389.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evert BO, Vogt IR, Vieira-Saecker AM, Ozimek L, de Vos RA, Brunt ER, Klockgether T, Wullner U. Gene expression profiling in ataxin-3 expressing cell lines reveals distinct effects of normal and mutant ataxin-3. J Neuropathol Exp Neurol. 2003;62:1006–1018. doi: 10.1093/jnen/62.10.1006. [DOI] [PubMed] [Google Scholar]
- Evert BO, Wullner U, Schulz JB, Weller M, Groscurth P, Trottier Y, Brice A, Klockgether T. High level expression of expanded full-length ataxin-3 in vitro causes cell death and formation of intranuclear inclusions in neuronal cells. Hum Mol Genet. 1999;8:1169–1176. doi: 10.1093/hmg/8.7.1169. [DOI] [PubMed] [Google Scholar]
- Ferro A, Carvalho AL, Teixeira-Castro A, Almeida C, Tome RJ, Cortes L, Rodrigues AJ, Logarinho E, Sequeiros J, Macedo-Ribeiro S, Maciel P. NEDD8: a new ataxin-3 interactor. Biochim Biophys Acta. 2007;1773:1619–1627. doi: 10.1016/j.bbamcr.2007.07.012. [DOI] [PubMed] [Google Scholar]
- Fujikake N, Nagai Y, Popiel HA, Okamoto Y, Yamaguchi M, Toda T. Heat shock transcription factor 1-activating compounds suppress polyglutamine-induced neurodegeneration through induction of multiple molecular chaperones. J Biol Chem. 2008;283:26188–26197. doi: 10.1074/jbc.M710521200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukutake T, Shinotoh H, Nishino H, Ichikawa Y, Goto J, Kanazawa I, Hattori T. Homozygous Machado-Joseph disease presenting as REM sleep behaviour disorder and prominent psychiatric symptoms. Eue J Neurol. 2002;9:97–100. doi: 10.1046/j.1468-1331.2002.00335.x. [DOI] [PubMed] [Google Scholar]
- Gales L, Cortes L, Almeida C, Melo CV, Costa MC, Maciel P, Clarke DT, Damas AM, Macedo-Ribeiro S. Towards a structural understanding of the fibrillization pathway in Machado-Joseph's disease: trapping early oligomers of non-expanded ataxin-3. J Mol Biol. 2005;353:642–654. doi: 10.1016/j.jmb.2005.08.061. [DOI] [PubMed] [Google Scholar]
- Gaspar C, Lopes-Cendes I, Hayes S, Goto J, Arvidsson K, Dias A, Silveira I, Maciel P, Coutinho P, Lima M, Zhou YX, Soong BW, Watanabe M, Giunti P, Stevanin G, Riess O, Sasaki H, Hsieh M, Nicholson GA, Brunt E, Higgins JJ, Lauritzen M, Tranebjaerg L, Volpini V, Wood N, Ranum L, Tsuji S, Brice A, Sequeiros J, Rouleau GA. Ancestral origins of the Machado-Joseph disease mutation: a worldwide haplotype study. Am J Hum Genet. 2001;68:523–528. doi: 10.1086/318184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gispert S, Twells R, Orozco G, Brice A, Weber J, Heredero L, Scheufler K, Riley B, Allotey R, Nothers C, et al. Chromosomal assignment of the second locus for autosomal dominant cerebellar ataxia (SCA2) to chromosome 12q23–24.1. Nat Genet. 1993;4:295–299. doi: 10.1038/ng0793-295. [DOI] [PubMed] [Google Scholar]
- Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev. 2002;82:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- Goti D, Katzen SM, Mez J, Kurtis J, Kiluk J, Ben-Haïem L, Jenkins NA, Copeland NG, Kakizuka A, Sharp AH, Ross CA, Mouton PR, Colomer V. A mutant ataxin-3 putative-cleavage fragment in brains of Machado-Joseph disease patients and transgenic mice is cytotoxic above a critical concentration. The Journal of Neuroscience. 2004;24:10266–10279. doi: 10.1523/JNEUROSCI.2734-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goti D, Katzen SM, Mez J, Kurtis N, Kiluk J, Ben-Haiem L, Jenkins NA, Copeland NG, Kakizuka A, Sharp AH, Ross CA, Mouton PR, Colomer V. A mutant ataxin-3 putative-cleavage fragment in brains of Machado-Joseph disease patients and transgenic mice is cytotoxic above a critical concentration. J Neurosci. 2004;24:10266–10279. doi: 10.1523/JNEUROSCI.2734-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto J, Watanabe M, Ichikawa Y, Yee SB, Ihara N, Endo K, Igarashi S, Takiyama Y, Gaspar C, Maciel P, Tsuji S, Rouleau GA, Kanazawa I. Machado-Joseph disease gene products carrying different carboxyl termini. Neurosci Res. 1997;28:373–377. doi: 10.1016/s0168-0102(97)00056-4. [DOI] [PubMed] [Google Scholar]
- Gu W, Ma H, Wang K, Jin M, Zhou Y, Liu X, Wang G, Shen Y. The shortest expanded allele of the MJD1 gene in a Chinese MJD kindred with autonomic dysfunction. Eur Neurol. 2004;52:107–111. doi: 10.1159/000080221. [DOI] [PubMed] [Google Scholar]
- Gunawardena S, Goldstein LS. Polyglutamine diseases and transport problems: deadly traffic jams on neuronal highways. Arch Neurol. 2005;62:46–51. doi: 10.1001/archneur.62.1.46. [DOI] [PubMed] [Google Scholar]
- Haacke A, Broadley SA, Boteva R, Tzvetkov N, Hartl FU, Breuer P. Proteolytic cleavage of polyglutamine-expanded ataxin-3 is critical for aggregation and sequestration of non-expanded ataxin-3. Hum Mol Genet. 2006;15:555–568. doi: 10.1093/hmg/ddi472. [DOI] [PubMed] [Google Scholar]
- Haacke A, Hartl FU, Breuer P. Calpain Inhibition Is Sufficient to Suppress Aggregation of Polyglutamine-expanded Ataxin-3. J Biol Chem. 2007;282:18851–18856. doi: 10.1074/jbc.M611914200. [DOI] [PubMed] [Google Scholar]
- Haberhausen G, Damian MS, Leweke F, Muller U. Spinocerebellar ataxia, type 3 (SCA3) is genetically identical to Machado-Joseph disease (MJD) J Neurol Sci. 1995;132:71–75. doi: 10.1016/0022-510x(95)90927-i. [DOI] [PubMed] [Google Scholar]
- Harris GM, Dodelzon K, Gong L, Gonzalez-Alegre P, Paulson HL. Splice isoforms of the polyglutamine disease protein ataxin-3 exhibit similar enzymatic yet different aggregation properties. PLoS One. 2010;5:e13695. doi: 10.1371/journal.pone.0013695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M, Kobayashi K, Furuta H. Immunohistochemical study of neuronal intranuclear and cytoplasmic inclusions in Machado-Joseph disease. Psychiatry Clin Neurosci. 2003;57:205–213. doi: 10.1046/j.1440-1819.2003.01102.x. [DOI] [PubMed] [Google Scholar]
- Healton EB, Brust JC, Kerr DL, Resor S, Penn A. Presumably Azorean disease in a presumably non-Portuguese family. Neurology. 1980;30:1084–1089. doi: 10.1212/wnl.30.10.1084. [DOI] [PubMed] [Google Scholar]
- Heir R, Ablasou C, Dumontier E, Elliott M, Fagotto-Kaufmann C, Bedford FK. The UBL domain of PLIC-1 regulates aggresome formation. EMBO Rep. 2006;7:1252–1258. doi: 10.1038/sj.embor.7400823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins JJ, Nee LE, Vasconcelos O, Ide SE, Lavedan C, Goldfarb LG, Polymeropoulos MH. Mutations in American families with spinocerebellaRataxia (SCA) type 3: SCA3 is allelic to Machado-Joseph disease. Neurology. 1996;46:208–213. doi: 10.1212/wnl.46.1.208. [DOI] [PubMed] [Google Scholar]
- Horimoto Y, Matsumoto M, Yuasa H, Kojima A, Nokura K, Katada E, Yamamoto T, Yamamoto H, Mitake S. Brainstem in Machado-Joseph disease: atrophy or small size? Eur J Neurol. 2008;15:102–105. doi: 10.1111/j.1468-1331.2007.02008.x. [DOI] [PubMed] [Google Scholar]
- Hu J, Gagnon KT, Liu J, Watts JK, Syeda-Nawaz J, Bennett CF, Swayze EE, Randolph J, Chattopadhyaya J, Corey DR. Allele-selective inhibition of ataxin-3 (ATX3) expression by antisense oligomers and duplex RNAs. Biol Chem. 2011;392:315–325. doi: 10.1515/BC.2011.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Matsui M, Gagnon KT, Schwartz JC, Gabillet S, Arar K, Wu J, Bezprozvanny I, Corey DR. Allele-specific silencing of mutant huntingtin and ataxin-3 genes by targeting expanded CAG repeats in mRNAs. Nat Biotechnol. 2009;27:478–484. doi: 10.1038/nbt.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huen NY, Chan HY. Dynamic regulation of molecular chaperone gene expression in polyglutamine disease. Biochem Biophys Res Commun. 2005;334:1074–1084. doi: 10.1016/j.bbrc.2005.07.008. [DOI] [PubMed] [Google Scholar]
- Ichikawa Y, Goto J, Hattori M, Toyoda A, Ishii K, Jeong SY, Hashida H, Masuda N, Ogata K, Kasai F, Hirai M, Maciel P, Rouleau GA, Sakaki Y, Kanazawa I. The genomic structure and expression of MJD, the Machado-Joseph disease gene. J Hum Genet. 2001;46:413–422. doi: 10.1007/s100380170060. [DOI] [PubMed] [Google Scholar]
- Igarashi S, Takiyama Y, Cancel G, Rogaeva EA, Sasaki H, Wakisaka A, Zhou YX, Takano H, Endo K, Sanpei K, Oyake M, Tanaka H, Stevanin G, Abbas N, Durr A, Rogaev EI, Sherrington R, Tsuda T, Ikeda M, Cassa E, Nishizawa M, Benomar A, Julien J, Weissenbach J, Wang GX, Agid Y, St George-Hyslop PH, Brice A, Tsuji S. Intergenerational instability of the CAG repeat of the gene for Machado-Joseph disease (MJD1) is affected by the genotype of the normal chromosome: implications for the molecular mechanisms of the instability of the CAG repeat. Hum Mol Genet. 1996;5:923–932. doi: 10.1093/hmg/5.7.923. [DOI] [PubMed] [Google Scholar]
- Ikeda H, Yamaguchi M, Sugai S, Aze Y, Narumiya S, Kakizuka A. Expanded polyglutamine in the Machado-Joseph disease protein induces cell death in vitro and in vivo. Nature Genetics. 1996;13:196–202. doi: 10.1038/ng0696-196. [DOI] [PubMed] [Google Scholar]
- Ikeda H, Yamaguchi M, Sugai S, Aze Y, Narumiya S, Kakizuka A. Expanded polyglutamine in the Machado-Joseph disease protein induces cell death in vitro and in vivo. Nat Genet. 1996;13:196–202. doi: 10.1038/ng0696-196. [DOI] [PubMed] [Google Scholar]
- Iwabuchi K, Tsuchiya K, Uchihara T, Yagishita S. Autosomal dominant spinocerebellar degenerations. Clinical, pathological, and genetic correlations. Rev Neurol (Paris) 1999;155:255–270. [PubMed] [Google Scholar]
- Jana NR, Dikshit P, Goswami A, Kotliarova S, Murata S, Tanaka K, Nukina N. Co-chaperone CHIP associates with expanded polyglutamine protein and promotes their degradation by proteasomes. J Biol Chem. 2005;280:11635–11640. doi: 10.1074/jbc.M412042200. [DOI] [PubMed] [Google Scholar]
- Jardim LB, Pereira ML, Silveira I, Ferro A, Sequeiros J, Giugliani R. Neurologic findings in Machado-Joseph disease: relation with disease duration, subtypes, and (CAG)n. Arch Neurol. 2001;58:899–904. doi: 10.1001/archneur.58.6.899. [DOI] [PubMed] [Google Scholar]
- Jeub M, Herbst M, Spauschus A, Fleischer H, Klockgether T, Wuellner U, Evert BO. Potassium channel dysfunction and depolarized resting membrane potential in a cell model of SCA3. Exp Neurol. 2006;201:182–192. doi: 10.1016/j.expneurol.2006.03.029. [DOI] [PubMed] [Google Scholar]
- Jung J, Xu K, Lessing D, Bonini NM. Preventing Ataxin-3 protein cleavage mitigates degeneration in a Drosophila model of SCA3. Hum Mol Genet. 2009;18:4843–4852. doi: 10.1093/hmg/ddp456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi Y, Okamoto T, Taniwaki M, Aizawa M, Inoue M, Katayama S, Kawakami H, Nakamura S, Nishimura M, Akiguchi I, et al. CAG expansions in a novel gene for Machado-Joseph disease at chromosome 14q32.1. Nat Genet. 1994;8:221–228. doi: 10.1038/ng1194-221. [DOI] [PubMed] [Google Scholar]
- Kawai Y, Takeda A, Abe Y, Washimi Y, Tanaka F, Sobue G. Cognitive impairments in Machado-Joseph disease. Arch Neurol. 2004;61:1757–1760. doi: 10.1001/archneur.61.11.1757. [DOI] [PubMed] [Google Scholar]
- Khan LA, Bauer PO, Miyazaki H, Lindenberg KS, Landwehrmeyer BG, Nukina N. Expanded polyglutamines impair synaptic transmission and ubiquitin-proteasome system in Caenorhabditis elegans. J Neurochem. 2006;98:576–587. doi: 10.1111/j.1471-4159.2006.03895.x. [DOI] [PubMed] [Google Scholar]
- Klockgether T, Skalej M, Wedekind D, Luft AR, Welte D, Schulz JB, Abele M, Burk K, Laccone F, Brice A, Dichgans J. Autosomal dominant cerebellar ataxia type I. MRI-based volumetry of posterior fossa structures and basal ganglia in spinocerebellar ataxia types 1, 2 and 3. Brain. 1998;121(Pt 9):1687–1693. doi: 10.1093/brain/121.9.1687. [DOI] [PubMed] [Google Scholar]
- Kuhlbrodt K, Janiesch PC, Kevei E, Segref A, Barikbin R, Hoppe T. The Machado-Joseph disease deubiquitylase ATX-3 couples longevity and proteostasis. Nat Cell Biol. 2011;13:273–281. doi: 10.1038/ncb2200. [DOI] [PubMed] [Google Scholar]
- La Spada AR, Wilson EM, Lubahn DB, Harding AE, Fischbeck KH. Androgen receptor gene mutations in X-linked spinal and bulbar muscular atrophy. Nature. 1991;352:77–79. doi: 10.1038/352077a0. [DOI] [PubMed] [Google Scholar]
- LaLonde DP, Bretscher A. The UBX protein SAKS1 negatively regulates endoplasmic reticulum-associated degradation and p97-dependent degradation. J Biol Chem. 2011;286:4892–4901. doi: 10.1074/jbc.M110.158030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landau WM, Schmidt RE, McGlennen RC, Reich SG. Hereditary spastic paraplegia and hereditary ataxia, Part 2: A family demonstrating various phenotypic manifestations with the SCA3 genotype. Arch Neurol. 2000;57:733–739. doi: 10.1001/archneur.57.5.733. [DOI] [PubMed] [Google Scholar]
- Lang AE, Rogaeva EA, Tsuda T, Hutterer J, St George-Hyslop P. Homozygous inheritance of the Machado-Joseph disease gene. Ann Neurol. 1994;36:443–447. doi: 10.1002/ana.410360318. [DOI] [PubMed] [Google Scholar]
- Lerer I, Merims D, Abeliovich D, Zlotogora J, Gadoth N. Machado-Joseph disease: correlation between the clinical features, the CAG repeat length and homozygosity for the mutation. Eur J Hum Genet. 1996;4:3–7. doi: 10.1159/000472162. [DOI] [PubMed] [Google Scholar]
- Li F, Macfarlan T, Pittman RN, Chakravarti D. Ataxin-3 is a histone-binding protein with two independent transcriptional corepressor activities. J Biol Chem. 2002;277:45004–45012. doi: 10.1074/jbc.M205259200. [DOI] [PubMed] [Google Scholar]
- Li LB, Yu Z, Teng X, Bonini NM. RNA toxicity is a component of ataxin-3 degeneration in Drosophila. Nature. 2008;453:1107–1111. doi: 10.1038/nature06909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J, Crespo-Barreto J, Jafar-Nejad P, Bowman AB, Richman R, Hill DE, Orr HT, Zoghbi HY. Opposing effects of polyglutamine expansion on native protein complexes contribute to SCA1. Nature. 2008 doi: 10.1038/nature06731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima L, Coutinho P. Clinical criteria for diagnosis of Machado-Joseph disease: report of a non-Azorena Portuguese family. Neurology. 1980;30:319–322. doi: 10.1212/wnl.30.3.319. [DOI] [PubMed] [Google Scholar]
- Lima M, Costa MC, Montiel R, Ferro A, Santos C, Silva C, Bettencourt C, Sousa A, Sequeiros J, Coutinho P, Maciel P. Population genetics of wild-type CAG repeats in the Machado-Joseph disease gene in Portugal. Hum Hered. 2005;60:156–163. doi: 10.1159/000090035. [DOI] [PubMed] [Google Scholar]
- Lima M, Kay T, Vasconcelos J, Mota-Vieira L, Gonzalez C, Peixoto A, Abade A, MacLeod P, Graca R, Santos J. Disease knowledge and attitudes toward predictive testing and prenatal diagnosis in families with Machado-Joseph disease from the Azores Islands (Portugal) Community Genet. 2001;4:36–42. doi: 10.1159/000051154. [DOI] [PubMed] [Google Scholar]
- Lindblad K, Lunkes A, Maciel P, Stevanin G, Zander C, Klockgether T, Ratzlaff T, Brice A, Rouleau GA, Hudson T, Auburger G, Schalling M. Mutation detection in Machado-Joseph disease using repeat expansion detection. Mol Med. 1996;2:77–85. [PMC free article] [PubMed] [Google Scholar]
- Linhartova I, Repitz M, Draber P, Nemec M, Wiche G, Propst F. Conserved domains and lack of evidence for polyglutamine length polymorphism in the chicken homolog of the Machado-Joseph disease gene product ataxin-3. Biochim Biophys Acta. 1999;1444:299–305. doi: 10.1016/s0167-4781(99)00004-4. [DOI] [PubMed] [Google Scholar]
- Livingstone IR, Sequeiros J. Machado-Joseph disease in an American-Italian family. J Neurogenet. 1984;1:185–188. doi: 10.3109/01677068409107084. [DOI] [PubMed] [Google Scholar]
- Lopes-Cendes I, Maciel P, Kish S, Gaspar C, Robitaille Y, Clark HB, Koeppen AH, Nance M, Schut L, Silveira I, Coutinho P, Sequeiros J, Rouleau GA. Somatic mosaicism in the central nervous system in spinocerebellar ataxia type 1 and Machado-Joseph disease. Ann Neurol. 1996;40:199–206. doi: 10.1002/ana.410400211. [DOI] [PubMed] [Google Scholar]
- Macedo-Ribeiro S, Cortes L, Maciel P, Carvalho AL. Nucleocytoplasmic shuttling activity of ataxin-3. PLoS One. 2009;4:e5834. doi: 10.1371/journal.pone.0005834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciel P, Costa MC, Ferro A, Rousseau M, Santos CS, Gaspar C, Barros J, Rouleau GA, Coutinho P, Sequeiros J. Improvement in the molecular diagnosis of Machado-Joseph disease. Arch Neurol. 2001;58:1821–1827. doi: 10.1001/archneur.58.11.1821. [DOI] [PubMed] [Google Scholar]
- Maciel P, Gaspar C, DeStefano AL, Silveira I, Coutinho P, Radvany J, Dawson DM, Sudarsky L, Guimaraes J, Loureiro JE, et al. Correlation between CAG repeat length and clinical features in Machado-Joseph disease. Am J Hum Genet. 1995;57:54–61. [PMC free article] [PubMed] [Google Scholar]
- Maciel P, Gaspar C, Guimaraes L, Goto J, Lopes-Cendes I, Hayes S, Arvidsson K, Dias A, Sequeiros J, Sousa A, Rouleau GA. Study of three intragenic polymorphisms in the Machado-Joseph disease gene (MJD1) in relation to genetic instability of the (CAG)n tract. Eur J Hum Genet. 1999;7:147–156. doi: 10.1038/sj.ejhg.5200264. [DOI] [PubMed] [Google Scholar]
- Maciel P, Lopes-Cendes I, Kish S, Sequeiros J, Rouleau GA. Mosaicism of the CAG repeat in CNS tissue in relation to age at death in spinocerebellar ataxia type 1 and Machado-Joseph disease patients. Am J Hum Genet. 1997;60:993–996. [PMC free article] [PubMed] [Google Scholar]
- Mao Y, Senic-Matuglia F, Di Fiore PP, Polo S, Hodsdon ME, De Camilli P. Deubiquitinating function of ataxin-3: insights from the solution structure of the Josephin domain. Proc Natl Acad Sci U S A. 2005;102:12700–12705. doi: 10.1073/pnas.0506344102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis RL. The spinocerebellar ataxias: order emerges from chaos. Curr Neurol Neurosci Rep. 2002;2:447–456. doi: 10.1007/s11910-002-0072-8. [DOI] [PubMed] [Google Scholar]
- Martins S, Calafell F, Gaspar C, Wong VC, Silveira I, Nicholson GA, Brunt ER, Tranebjaerg L, Stevanin G, Hsieh M, Soong BW, Loureiro L, Durr A, Tsuji S, Watanabe M, Jardim LB, Giunti P, Riess O, Ranum LP, Brice A, Rouleau GA, Coutinho P, Amorim A, Sequeiros J. Asian origin for the worldwide-spread mutational event in Machado-Joseph disease. Arch Neurol. 2007;64:1502–1508. doi: 10.1001/archneur.64.10.1502. [DOI] [PubMed] [Google Scholar]
- Martins S, Calafell F, Wong VC, Sequeiros J, Amorim A. A multistep mutation mechanism drives the evolution of the CAG repeat at MJD/SCA3 locus. Eur J Hum Genet. 2006;14:932–940. doi: 10.1038/sj.ejhg.5201643. [DOI] [PubMed] [Google Scholar]
- Martins S, Coutinho P, Silveira I, Giunti P, Jardim LB, Calafell F, Sequeiros J, Amorim A. Cis-acting factors promoting the CAG intergenerational instability in Machado-Joseph disease. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:439–446. doi: 10.1002/ajmg.b.30624. [DOI] [PubMed] [Google Scholar]
- Maruyama H, Nakamura S, Matsuyama Z, Sakai T, Doyu M, Sobue G, Seto M, Tsujihata M, Oh-i T, Nishio T, et al. Molecular features of the CAG repeats and clinical manifestation of Machado-Joseph disease. Hum Mol Genet. 1995;4:807–812. doi: 10.1093/hmg/4.5.807. [DOI] [PubMed] [Google Scholar]
- Masino L, Musi V, Menon RP, Fusi P, Kelly G, Frenkiel TA, Trottier Y, Pastore A. Domain architecture of the polyglutamine protein ataxin-3: a globular domain followed by a flexible tail. FEBS Lett. 2003;549:21–25. doi: 10.1016/s0014-5793(03)00748-8. [DOI] [PubMed] [Google Scholar]
- Masino L, Nicastro G, Calder L, Vendruscolo M, Pastore A. Functional interactions as a survival strategy against abnormal aggregation. FASEB J. 2011a;25:45–54. doi: 10.1096/fj.10-161208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masino L, Nicastro G, De Simone A, Calder L, Molloy J, Pastore A. The Josephin domain determines the morphological and mechanical properties of ataxin-3 fibrils. Biophys J. 2011b;100:2033–2042. doi: 10.1016/j.bpj.2011.02.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matilla T, McCall A, Subramony SH, Zoghbi HY. Molecular and clinical correlations in spinocerebellar ataxia type 3 and Machado-Joseph disease. Ann Neurol. 1995;38:68–72. doi: 10.1002/ana.410380113. [DOI] [PubMed] [Google Scholar]
- Matos CA, de Macedo-Ribeiro S, Carvalho AL. Polyglutamine diseases: The special case of ataxin-3 and Machado-Joseph disease. Prog Neurobiol. 2011;95:26–48. doi: 10.1016/j.pneurobio.2011.06.007. [DOI] [PubMed] [Google Scholar]
- Matsumoto M, Yada M, Hatakeyama S, Ishimoto H, Tanimura T, Tsuji S, Kakizuka A, Kitagawa M, Nakayama KI. Molecular clearance of ataxin-3 is regulated by a mammalian E4. EMBO J. 2004;23:659–669. doi: 10.1038/sj.emboj.7600081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauri PL, Riva M, Ambu D, De Palma A, Secundo F, Benazzi L, Valtorta M, Tortora P, Fusi P. Ataxin-3 is subject to autolytic cleavage. Febs J. 2006;273:4277–4286. doi: 10.1111/j.1742-4658.2006.05419.x. [DOI] [PubMed] [Google Scholar]
- Mazzucchelli S, De Palma A, Riva M, D'Urzo A, Pozzi C, Pastori V, Comelli F, Fusi P, Vanoni M, Tortora P, Mauri P, Regonesi ME. Proteomic and biochemical analyses unveil tight interaction of ataxin-3 with tubulin. Int J Biochem Cell Biol. 2009;41:2485–2492. doi: 10.1016/j.biocel.2009.08.003. [DOI] [PubMed] [Google Scholar]
- Menzies FM, Huebener J, Renna M, Bonin M, Riess O, Rubinsztein DC. Autophagy induction reduces mutant ataxin-3 levels and toxicity in a mouse model of spinocerebellar ataxia type 3. Brain. 2010;133:93–104. doi: 10.1093/brain/awp292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra A, Dikshit P, Purkayastha S, Sharma J, Nukina N, Jana NR. E6-AP promotes misfolded polyglutamine proteins for proteasomal degradation and suppresses polyglutamine protein aggregation and toxicity. J Biol Chem. 2008;283:7648–7656. doi: 10.1074/jbc.M706620200. [DOI] [PubMed] [Google Scholar]
- Mori F, Nishie M, Piao YS, Kito K, Kamitani T, Takahashi H, Wakabayashi K. Accumulation of NEDD8 in neuronal and glial inclusions of neurodegenerative disorders. Neuropathol Appl Neurobiol. 2005;31:53–61. doi: 10.1111/j.1365-2990.2004.00603.x. [DOI] [PubMed] [Google Scholar]
- Mueller T, Breuer P, Schmitt I, Walter J, Evert BO, Wullner U. CK2-dependent phosphorylation determines cellular localization and stability of ataxin-3. Hum Mol Genet. 2009;18:3334–3343. doi: 10.1093/hmg/ddp274. [DOI] [PubMed] [Google Scholar]
- Murata Y, Yamaguchi S, Kawakami H, Imon Y, Maruyama H, Sakai T, Kazuta T, Ohtake T, Nishimura M, Saida T, Chiba S, Oh-i T, Nakamura S. Characteristic magnetic resonance imaging findings in Machado-Joseph disease. Arch Neurol. 1998;55:33–37. doi: 10.1001/archneur.55.1.33. [DOI] [PubMed] [Google Scholar]
- Nagai Y, Inui T, Popiel HA, Fujikake N, Hasegawa K, Urade Y, Goto Y, Naiki H, Toda T. A toxic monomeric conformer of the polyglutamine protein. Nat Struct Mol Biol. 2007;14:332–340. doi: 10.1038/nsmb1215. [DOI] [PubMed] [Google Scholar]
- Nakano KK, Dawson DM, Spence A. Machado disease. A hereditary ataxia in Portuguese emigrants to Massachusetts. Neurology. 1972;22:49–55. doi: 10.1212/wnl.22.1.49. [DOI] [PubMed] [Google Scholar]
- Nascimento-Ferreira I, Santos-Ferreira T, Sousa-Ferreira L, Auregan G, Onofre I, Alves S, Dufour N, Colomer Gould VF, Koeppen A, Deglon N, Pereira de Almeida L. Overexpression of the autophagic beclin-1 protein clears mutant ataxin-3 and alleviates Machado-Joseph disease. Brain. 2011;134:1400–1415. doi: 10.1093/brain/awr047. [DOI] [PubMed] [Google Scholar]
- Natalello A, Frana AM, Relini A, Apicella A, Invernizzi G, Casari C, Gliozzi A, Doglia SM, Tortora P, Regonesi ME. A major role for side-chain polyglutamine hydrogen bonding in irreversible ataxin-3 aggregation. PLoS One. 2011;6:e18789. doi: 10.1371/journal.pone.0018789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicastro G, Habeck M, Masino L, Svergun DI, Pastore A. Structure validation of the Josephin domain of ataxin-3: conclusive evidence for an open conformation. J Biomol NMR. 2006;36:267–277. doi: 10.1007/s10858-006-9092-z. [DOI] [PubMed] [Google Scholar]
- Nicastro G, Masino L, Esposito V, Menon RP, De Simone A, Fraternali F, Pastore A. Josephin domain of ataxin-3 contains two distinct ubiquitin-binding sites. Biopolymers. 2009;91:1203–1214. doi: 10.1002/bip.21210. [DOI] [PubMed] [Google Scholar]
- Nicastro G, Menon RP, Masino L, Knowles PP, McDonald NQ, Pastore A. The solution structure of the Josephin domain of ataxin-3: structural determinants for molecular recognition. Proc Natl Acad Sci U S A. 2005;102:10493–10498. doi: 10.1073/pnas.0501732102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicastro G, Todi SV, Karaca E, Bonvin AM, Paulson HL, Pastore A. Understanding the role of the Josephin domain in the PolyUb binding and cleavage properties of ataxin-3. PLoS One. 2010;5:e12430. doi: 10.1371/journal.pone.0012430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onodera O, Idezuka J, Igarashi S, Takiyama Y, Endo K, Takano H, Oyake M, Tanaka H, Inuzuka T, Hayashi T, Yuasa T, Ito J, Miyatake T, Tsuji S. Progressive atrophy of cerebellum and brainstem as a function of age and the size of the expanded CAG repeats in the MJD1 gene in Machado-Joseph disease. Ann Neurol. 1998;43:288–296. doi: 10.1002/ana.410430305. [DOI] [PubMed] [Google Scholar]
- Padiath QS, Srivastava AK, Roy S, Jain S, Brahmachari SK. Identification of a novel 45 repeat unstable allele associated with a disease phenotype at the MJD1/SCA3 locus. Am J Med Genet B Neuropsychiatr Genet. 2005;133B:124–126. doi: 10.1002/ajmg.b.30088. [DOI] [PubMed] [Google Scholar]
- Paulson H. Spinocerebellar Ataxia Type 3. In: B T Pagon RA, Dolan CR, Stephens K, editors. GeneReviews [Internet] Seattle (WA): University of Washington, Seattle; 1998. Oct 10, p. 1993. [updated 2011 March 17] [PubMed] [Google Scholar]
- Paulson HL. Dominantly inherited ataxias: lessons learned from Machado-Joseph disease/spinocerebellar ataxia type 3. Semin Neurol. 2007;27:133–142. doi: 10.1055/s-2007-971172. [DOI] [PubMed] [Google Scholar]
- Paulson HL, Das SS, Crino PB, Perez MK, Patel SC, Gotsdiner D, Fischbeck KH, Pittman RN. Machado-Joseph disease gene product is a cytoplasmic protein widely expressed in brain. Ann Neurol. 1997a;41:453–462. doi: 10.1002/ana.410410408. [DOI] [PubMed] [Google Scholar]
- Paulson HL, Perez MK, Trottier Y, Trojanowski JQ, Subramony SH, Das SS, Vig P, Mandel JL, Fischbeck KH, Pittman RN. Intranuclear inclusions of expanded polyglutamine protein in spinocerebellar ataxia type 3. Neuron. 1997b;19:333–344. doi: 10.1016/s0896-6273(00)80943-5. [DOI] [PubMed] [Google Scholar]
- Perez MK, Paulson HL, Pendse SJ, Saionz SJ, Bonini NM, Pittman RN. Recruitment and the role of nuclear localization in polyglutamine-mediated aggregation. J Cell Biol. 1998;143:1457–1470. doi: 10.1083/jcb.143.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perutz MF, Finch JT, Berriman J, Lesk A. Amyloid fibers are water-filled nanotubes. Proc Natl Acad Sci U S A. 2002;99:5591–5595. doi: 10.1073/pnas.042681399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozzi C, Valtorta M, Tedeschi G, Galbusera E, Pastori V, Bigi A, Nonnis S, Grassi E, Fusi P. Study of subcellular localization and proteolysis of ataxin-3. Neurobiol Dis. 2008;30:190–200. doi: 10.1016/j.nbd.2008.01.011. [DOI] [PubMed] [Google Scholar]
- Reina CP, Zhong X, Pittman RN. Proteotoxic stress increases nuclear localization of ataxin-3. Hum Mol Genet. 2010;19:235–249. doi: 10.1093/hmg/ddp482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riess O, Rub U, Pastore A, Bauer P, Schols L. SCA3: Neurological features, pathogenesis and animal models. The Cerebellum. 2008:125–137. doi: 10.1007/s12311-008-0013-4. [DOI] [PubMed] [Google Scholar]
- Robertson AL, Headey SJ, Saunders HM, Ecroyd H, Scanlon MJ, Carver JA, Bottomley SP. Small heat-shock proteins interact with a flanking domain to suppress polyglutamine aggregation. Proc Natl Acad Sci U S A. 2010;107:10424–10429. doi: 10.1073/pnas.0914773107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues AJ, Coppola G, Santos C, Costa MC, Ailion M, Sequeiros J, Geschwind DH, Maciel P. Functional genomics and biochemical characterization of the C. elegans orthologue of the Machado-Joseph disease protein ataxin-3. Faseb Journal. 2007;21:1126–1136. doi: 10.1096/fj.06-7002com. [DOI] [PubMed] [Google Scholar]
- Rodrigues AJ, do Carmo Costa M, Silva TL, Ferreira D, Bajanca F, Logarinho E, Maciel P. Absence of ataxin-3 leads to cytoskeletal disorganization and increased cell death. Biochim Biophys Acta. 2010;1803:1154–1163. doi: 10.1016/j.bbamcr.2010.07.004. [DOI] [PubMed] [Google Scholar]
- Rodrigues AJ, Neves-Carvalho A, Teixeira-Castro A, Rokka A, Corthals G, Logarinho E, Maciel P. Absence of ataxin-3 leads to enhanced stress response in C. elegans. PLoS One. 2011;6:e18512. doi: 10.1371/journal.pone.0018512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolim L, Leite A, Ledo S, Paneque M, Sequeiros J, Fleming M. Psychological aspects of pre-symptomatic testing for Machado-Joseph disease and familial amyloid polyneuropathy type I. Clin Genet. 2006;69:297–305. doi: 10.1111/j.1399-0004.2006.00606.x. [DOI] [PubMed] [Google Scholar]
- Romanul FC, Fowler HL, Radvany J, Feldman RG, Feingold M. Azorean disease of the nervous system. N Engl J Med. 1977;296:1505–1508. doi: 10.1056/NEJM197706302962606. [DOI] [PubMed] [Google Scholar]
- Rosenberg RN. Machado-Joseph disease: an autosomal dominant motor system degeneration. Mov Disord. 1992;7:193–203. doi: 10.1002/mds.870070302. [DOI] [PubMed] [Google Scholar]
- Rosenberg RN, Nyhan WL, Bay C, Shore P. Autosomal dominant striatonigral degeneration. A clinical, pathologic, and biochemical study of a new genetic disorder. Neurology. 1976;26:703–714. doi: 10.1212/wnl.26.8.703. [DOI] [PubMed] [Google Scholar]
- Ross CA. When more is less: pathogenesis of glutamine repeat neurodegenerative diseases. Neuron. 1995;15:493–496. doi: 10.1016/0896-6273(95)90138-8. [DOI] [PubMed] [Google Scholar]
- Rub U, Brunt ER, Deller T. New insights into the pathoanatomy of spinocerebellar ataxia type 3 (Machado-Joseph disease) Curr Opin Neurol. 2008;21:111–116. doi: 10.1097/WCO.0b013e3282f7673d. [DOI] [PubMed] [Google Scholar]
- Rüb U, de Vos RA, Brunt ER, Sebesteny T, Schols L, Auburger G, Bohl J, Ghebremedhin E, Gierga K, Seidel K, den Dunnen W, Heinsen H, Paulson H, Deller T. Spinocerebellar Ataxia Type 3 (SCA3): Thalamic Neurodegeneration Occurs Independently from Thalamic Ataxin-3 Immunopositive Neuronal Intranuclear Inclusions. Brain Pathol. 2006;16:218–227. doi: 10.1111/j.1750-3639.2006.00022.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rub U, Seidel K, Ozerden I, Gierga K, Brunt ER, Schols L, de Vos RA, den Dunnen W, Schultz C, Auburger G, Deller T. Consistent affection of the central somatosensory system in spinocerebellar ataxia type 2 and type 3 and its significance for clinical symptoms and rehabilitative therapy. Brain Res Brain Res Rev. 2007;53:235–249. doi: 10.1016/j.brainresrev.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Sakai T, Kawakami H. Machado-Joseph disease: A proposal of spastic paraplegic subtype. Neurology. 1996;46:846–847. [PubMed] [Google Scholar]
- Sakai T, Ohta M, Ishino H. Joseph disease in a non-Portuguese family. Neurology. 1983;33:74–80. doi: 10.1212/wnl.33.1.74. [DOI] [PubMed] [Google Scholar]
- Saunders HM, Gilis D, Rooman M, Dehouck Y, Robertson AL, Bottomley SP. Flanking domain stability modulates the aggregation kinetics of a polyglutamine disease protein. Protein Sci. 2011 doi: 10.1002/pro.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaglione KM, Zavodszky E, Todi SV, Patury S, Xu P, Rodriguez-Lebron E, Fischer S, Konen J, Djarmati A, Peng J, Gestwicki JE, Paulson HL. Ube2w and Ataxin-3 Coordinately Regulate the Ubiquitin Ligase CHIP. Mol Cell. 2011;43:599–612. doi: 10.1016/j.molcel.2011.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheel H, Tomiuk S, Hofmann K. Elucidation of ataxin-3 and ataxin-7 function by integrative bioinformatics. Hum Mol Genet. 2003;12:2845–2852. doi: 10.1093/hmg/ddg297. [DOI] [PubMed] [Google Scholar]
- Schmidt T, Landwehrmeyer GB, Schmitt I, Trottier Y, Auburger G, Laccone F, Klockgether T, Volpel M, Epplen JT, Schols L, Riess O. An isoform of ataxin-3 accumulates in the nucleus of neuronal cells in affected brain regions of SCA3 patients. Brain Pathol. 1998;8:669–679. doi: 10.1111/j.1750-3639.1998.tb00193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt I, Brattig T, Gossen M, Riess O. Characterization of the rat spinocerebellar ataxia type 3 gene. Neurogenetics. 1997;1:103–112. doi: 10.1007/s100480050015. [DOI] [PubMed] [Google Scholar]
- Schmitt I, Evert BO, Khazneh H, Klockgether T, Wuellner U. The human MJD gene: genomic structure and functional characterization of the promoter region. Gene. 2003;314:81–88. doi: 10.1016/s0378-1119(03)00706-6. [DOI] [PubMed] [Google Scholar]
- Schmitt I, Linden M, Khazneh H, Evert BO, Breuer P, Klockgether T, Wuellner U. Inactivation of the mouse Atxn3 (ataxin-3) gene increases protein ubiquitination. Biochem Biophys Res Commun. 2007;362:734–739. doi: 10.1016/j.bbrc.2007.08.062. [DOI] [PubMed] [Google Scholar]
- Schols L, Amoiridis G, Epplen JT, Langkafel M, Przuntek H, Riess O. Relations between genotype and phenotype in German patients with the Machado-Joseph disease mutation. J Neurol Neurosurg Psychiatry. 1996;61:466–470. doi: 10.1136/jnnp.61.5.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schols L, Bauer P, Schmidt T, Schulte T, Riess O. Autosomal dominant cerebellar ataxias: clinical features, genetics, and pathogenesis. Lancet Neurol. 2004;3:291–304. doi: 10.1016/S1474-4422(04)00737-9. [DOI] [PubMed] [Google Scholar]
- Seidel K, den Dunnen WF, Schultz C, Paulson H, Frank S, de Vos RA, Brunt ER, Deller T, Kampinga HH, Rub U. Axonal inclusions in spinocerebellar ataxia type 3. Acta Neuropathol. 2010;120:449–460. doi: 10.1007/s00401-010-0717-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sequeiros J, Coutinho P. Epidemiology and clinical aspects of Machado-Joseph disease. Adv Neurol. 1993;61:139–153. [PubMed] [Google Scholar]
- Sequeiros J, Maciel P, Taborda F, Ledo S, Rocha JC, Lopes A, Reto F, Fortuna AM, Rousseau M, Fleming M, Coutinho P, Rouleau GA, Jorge CS. Prenatal diagnosis of Machado-Joseph disease by direct mutation analysis. Prenat Diagn. 1998;18:611–617. [PubMed] [Google Scholar]
- Sequeiros J, Suite ND. Spinopontine atrophy disputed as a separate entity: the first description of Machado-Joseph disease. Neurology. 1986;36:1408. [PubMed] [Google Scholar]
- Shakkottai VG, do Carmo Costa M, Dell'orco JM, Sankaranarayanan A, Wulff H, Paulson HL. Early changes in cerebellar physiology accompany motor dysfunction in the polyglutamine disease spinocerebellar ataxia type 3. J Neurosci. 2011;31:13002–13014. doi: 10.1523/JNEUROSCI.2789-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski P, Atkins E. New model for crystalline polyglutamine assemblies and their connection with amyloid fibrils. Biomacromolecules. 2005;6:425–432. doi: 10.1021/bm0494388. [DOI] [PubMed] [Google Scholar]
- Silva-Fernandes A, Costa Mdo C, Duarte-Silva S, Oliveira P, Botelho CM, Martins L, Mariz JA, Ferreira T, Ribeiro F, Correia-Neves M, Costa C, Maciel P. Motor uncoordination and neuropathology in a transgenic mouse model of Machado-Joseph disease lacking intranuclear inclusions and ataxin-3 cleavage products. Neurobiol Dis. 2010;40:163–176. doi: 10.1016/j.nbd.2010.05.021. [DOI] [PubMed] [Google Scholar]
- Sobue G, Doyu M, Nakao N, Shimada N, Mitsuma T, Maruyama H, Kawakami S, Nakamura S. Homozygosity for Machado-Joseph disease gene enhances phenotypic severity. J Neurol Neurosurg Psychiatry. 1996;60:354–356. doi: 10.1136/jnnp.60.3.354-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soong BW, Liu RS. Positron emission tomography in asymptomatic gene carriers of Machado-Joseph disease. J Neurol Neurosurg Psychiatry. 1998;64:499–504. doi: 10.1136/jnnp.64.4.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowa ME, Bennett EJ, Gygi SP, Harper JW. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009;138:389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiura A, Yonashiro R, Fukuda T, Matsushita N, Nagashima S, Inatome R, Yanagi S. A mitochondrial ubiquitin ligase MITOL controls cell toxicity of polyglutamine-expanded protein. Mitochondrion. 2010;11:139–146. doi: 10.1016/j.mito.2010.09.001. [DOI] [PubMed] [Google Scholar]
- Tait D, Riccio M, Sittler A, Scherzinger E, Santi S, Ognibene A, Maraldi NM, Lehrach H, Wanker EE. Ataxin-3 is transported into the nucleus and associates with the nuclear matrix. Hum Mol Genet. 1998;7:991–997. doi: 10.1093/hmg/7.6.991. [DOI] [PubMed] [Google Scholar]
- Takahashi J, Tanaka J, Arai K, Funata N, Hattori T, Fukuda T, Fujigasaki H, Uchihara T. Recruitment of nonexpanded polyglutamine proteins to intranuclear aggregates in neuronal intranuclear hyaline inclusion disease. J Neuropathol Exp Neurol. 2001;60:369–376. doi: 10.1093/jnen/60.4.369. [DOI] [PubMed] [Google Scholar]
- Takiyama Y, Igarashi S, Rogaeva EA, Endo K, Rogaev EI, Tanaka H, Sherrington R, Sanpei K, Liang Y, Saito M, et al. Evidence for inter-generational instability in the CAG repeat in the MJD1 gene and for conserved haplotypes at flanking markers amongst Japanese and Caucasian subjects with Machado-Joseph disease. Hum Mol Genet. 1995;4:1137–1146. doi: 10.1093/hmg/4.7.1137. [DOI] [PubMed] [Google Scholar]
- Takiyama Y, Nishizawa M, Tanaka H, Kawashima S, Sakamoto H, Karube Y, Shimazaki H, Soutome M, Endo K, Ohta S, et al. The gene for Machado-Joseph disease maps to human chromosome 14q. Nat Genet. 1993;4:300–304. doi: 10.1038/ng0793-300. [DOI] [PubMed] [Google Scholar]
- Takiyama Y, Sakoe K, Nakano I, Nishizawa M. Machado-Joseph disease: cerebellar ataxia and autonomic dysfunction in a patient with the shortest known expanded allele (56 CAG repeat units) of the MJD1 ene. Neurology. 1997;49:604–606. doi: 10.1212/wnl.49.2.604. [DOI] [PubMed] [Google Scholar]
- Taniguchi R, Konigsmark BW. Dominant spino-pontine atrophy. Report of a family through three generations. Brain. 1971;94:349–358. doi: 10.1093/brain/94.2.349. [DOI] [PubMed] [Google Scholar]
- Taniwaki T, Sakai T, Kobayashi T, Kuwabara Y, Otsuka M, Ichiya Y, Masuda K, Goto I. Positron emission tomography (PET) in Machado-Joseph disease. J Neurol Sci. 1997;145:63–67. doi: 10.1016/s0022-510x(96)00242-0. [DOI] [PubMed] [Google Scholar]
- Teixeira-Castro A, Ailion M, Jalles A, Brignull HR, Vilaca JL, Dias N, Rodrigues P, Oliveira JF, Neves-Carvalho A, Morimoto RI, Maciel P. Neuron-specific proteotoxicity of mutant ataxin-3 in C. elegans: rescue by the DAF-16 and HSF-1 pathways. Hum Mol Genet. 2011;20:2996–3009. doi: 10.1093/hmg/ddr203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todi SV, Laco MN, Winborn BJ, Travis SM, Wen HM, Paulson HL. Cellular turnover of the polyglutamine disease protein ataxin-3 is regulated by its catalytic activity. J Biol Chem. 2007;282:29348–29358. doi: 10.1074/jbc.M704126200. [DOI] [PubMed] [Google Scholar]
- Todi SV, Paulson HL. Balancing act: deubiquitinating enzymes in the nervous system. Trends Neurosci. 2011 doi: 10.1016/j.tins.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todi SV, Scaglione KM, Blount JR, Basrur V, Conlon KP, Pastore A, Elenitoba-Johnson K, Paulson HL. Activity and cellular functions of the deubiquitinating enzyme and polyglutamine disease protein ataxin-3 are regulated by ubiquitination at lysine 117. J Biol Chem. 2010;285:39303–39313. doi: 10.1074/jbc.M110.181610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todi SV, Winborn BJ, Scaglione KM, Blount JR, Travis SM, Paulson HL. Ubiquitination directly enhances activity of the deubiquitinating enzyme ataxin-3. Embo J. 2009;28:372–382. doi: 10.1038/emboj.2008.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torashima T, Koyama C, Iizuka A, Mitsumura K, Takayama K, Yanagi S, Oue M, Yamaguchi H, Hirai H. Lentivector-mediated rescue from cerebellar ataxia in a mouse model of spinocerebellar ataxia. EMBO Rep. 2008;9:393–399. doi: 10.1038/embor.2008.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trottier Y, Cancel G, An-Gourfinkel I, Lutz Y, Weber C, Brice A, Hirsch E, Mandel JL. Heterogeneous intracellular localization and expression of ataxin-3. Neurobiol Dis. 1998;5:335–347. doi: 10.1006/nbdi.1998.0208. [DOI] [PubMed] [Google Scholar]
- Tsai HF, Tsai HJ, Hsieh M. Full-length expanded ataxin-3 enhances mitochondrial-mediated cell death and decreases Bcl-2 expression in human neuroblastoma cells. Biochem Biophys Res Commun. 2004;324:1274–1282. doi: 10.1016/j.bbrc.2004.09.192. [DOI] [PubMed] [Google Scholar]
- Tsai YC, Fishman PS, Thakor NV, Oyler GA. Parkin facilitates the elimination of expanded polyglutamine proteins and leads to preservation of proteasome function. J Biol Chem. 2003;278:22044–22055. doi: 10.1074/jbc.M212235200. [DOI] [PubMed] [Google Scholar]
- van Alfen N, Sinke RJ, Zwarts MJ, Gabreels-Festen A, Praamstra P, Kremer BP, Horstink MW. Intermediate CAG repeat lengths (53,54) for MJD/SCA3 are associated with an abnormal phenotype. Ann Neurol. 2001;49:805–807. doi: 10.1002/ana.1089. [DOI] [PubMed] [Google Scholar]
- van Schaik IN, Jobsis GJ, Vermeulen M, Keizers H, Bolhuis PA, de Visser M. Machado-Joseph disease presenting as severe asymmetric proximal neuropathy. J Neurol Neurosurg Psychiatry. 1997;63:534–536. doi: 10.1136/jnnp.63.4.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Ide K, Nukina N, Goto J, Ichikawa Y, Uchida K, Sakamoto T, Kanazawa I. Machado-Joseph disease gene product identified in lymphocytes and brain. Biochem Biophys Res Commun. 1997;233:476–479. doi: 10.1006/bbrc.1997.6484. [DOI] [PubMed] [Google Scholar]
- Wang G, Sawai N, Kotliarova S, Kanazawa I, Nukina N. Ataxin-3, the MJD1 gene product, interacts with the two human homologs of yeast DNA repair protein RAD23, HHR23A and HHR23B. Hum Mol Genet. 2000;9:1795–1803. doi: 10.1093/hmg/9.12.1795. [DOI] [PubMed] [Google Scholar]
- Wang H, Jia N, Fei E, Wang Z, Liu C, Zhang T, Fan J, Wu M, Chen L, Nukina N, Zhou J, Wang G. p45, an ATPase subunit of the 19S proteasome, targets the polyglutamine disease protein ataxin-3 to the proteasome. J Neurochem. 2007 doi: 10.1111/j.1471-4159.2007.04460.x. [DOI] [PubMed] [Google Scholar]
- Wang Q, Li L, Ye Y. Regulation of retrotranslocation by p97-associated deubiquitinating enzyme ataxin-3. J Cell Biol. 2006;174:963–971. doi: 10.1083/jcb.200605100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YG, Du J, Wang JL, Chen J, Chen C, Luo YY, Xiao ZQ, Jiang H, Yan XX, Xia K, Pan Q, Tang BS, Shen L. Six cases of SCA3/MJD patients that mimic hereditary spastic paraplegia in clinic. J Neurol Sci. 2009;285:121–124. doi: 10.1016/j.jns.2009.06.027. [DOI] [PubMed] [Google Scholar]
- Warrick JM, Morabito LM, Bilen J, Gordesky-Gold B, Faust LZ, Paulson HL, Bonini NM. Ataxin-3 suppresses polyglutamine neurodegeneration in Drosophila by a ubiquitin-associated mechanism. Molecular Cell. 2005;18:37–48. doi: 10.1016/j.molcel.2005.02.030. [DOI] [PubMed] [Google Scholar]
- Warrick JM, Chan HY, Gray-Board GL, Chai Y, Paulson HL, Bonini NM. Suppression of polyglutamine-mediated neurodegeneration in Drosophila by the molecular chaperone HSP70. Nat Genet. 1999;23:425–428. doi: 10.1038/70532. [DOI] [PubMed] [Google Scholar]
- Warrick JM, Morabito LM, Bilen J, Gordesky-Gold B, Faust LZ, Paulson HL, Bonini NM. Ataxin-3 suppresses polyglutamine neurodegeneration in Drosophila by a ubiquitin-associated mechanism. Mol Cell. 2005;18:37–48. doi: 10.1016/j.molcel.2005.02.030. [DOI] [PubMed] [Google Scholar]
- Warrick JM, Paulson HL, Gray-Board GL, Bui QT, Fischbeck KH, Pittman RN, Bonini NM. Expanded polyglutamine protein forms nuclear inclusions and causes neural degeneration in Drosophila. Cell. 1998;93:939–949. doi: 10.1016/s0092-8674(00)81200-3. [DOI] [PubMed] [Google Scholar]
- Wellington CL, Ellerby LM, Hackam AS, Margolis RL, Trifiro MA, Singaraja R, McCutcheon K, Salvesen GS, Propp SS, Bromm M, Rowland KJ, Zhang T, Rasper D, Roy S, Thornberry N, Pinsky L, Kakizuka A, Ross CA, Nicholson DW, Bredesen DE, Hayden MR. Caspase cleavage of gene products associated with triplet expansion disorders generates truncated fragments containing the polyglutamine tract. J Biol Chem. 1998;273:9158–9167. doi: 10.1074/jbc.273.15.9158. [DOI] [PubMed] [Google Scholar]
- Williams AJ, Paulson HL. Polyglutamine neurodegeneration: protein misfolding revisited. Trends Neurosci. 2008;31:521–528. doi: 10.1016/j.tins.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winborn BJ, Travis SM, Todi SV, Scaglione KM, Xu P, Williams AJ, Cohen RE, Peng J, Paulson HL. The deubiquitinating enzyme ataxin-3, a polyglutamine disease protein, edits Lys63 linkages in mixed linkage ubiquitin chains. J Biol Chem. 2008;283:26436–26443. doi: 10.1074/jbc.M803692200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woelk T, Sigismund S, Penengo L, Polo S. The ubiquitination code: a signalling problem. Cell Div. 2007;2:11. doi: 10.1186/1747-1028-2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods BT, Schaumburg HH. Nigro-spino-dentatal degeneration with nuclear ophthalmoplegia. A unique and partially treatable clinico-pathological entity. J Neurol Sci. 1972;17:149–166. doi: 10.1016/0022-510x(72)90137-2. [DOI] [PubMed] [Google Scholar]
- Wullner U, Reimold M, Abele M, Burk K, Minnerop M, Dohmen BM, Machulla HJ, Bares R, Klockgether T. Dopamine transporter positron emission tomography in spinocerebellar ataxias type 1, 2, 3, and 6. Arch Neurol. 2005;62:1280–1285. doi: 10.1001/archneur.62.8.1280. [DOI] [PubMed] [Google Scholar]
- Yamada M, Sato T, Tsuji S, Takahashi H. CAG repeat disorder models and human neuropathology: similarities and differences. Acta Neuropathol. 2008;115:71–86. doi: 10.1007/s00401-007-0287-5. [DOI] [PubMed] [Google Scholar]
- Yamada M, Tan CF, Inenaga C, Tsuji S, Takahashi H. Sharing of polyglutamine localization by the neuronal nucleus and cytoplasm in CAG-repeat diseases. Neuropathol Appl Neurobiol. 2004;30:665–675. doi: 10.1111/j.1365-2990.2004.00583.x. [DOI] [PubMed] [Google Scholar]
- Yamada M, Tsuji S, Takahashi H. Involvement of lysosomes in the pathogenesis of CAG repeat diseases. Ann Neurol. 2002;52:498–503. doi: 10.1002/ana.10328. [DOI] [PubMed] [Google Scholar]
- Ying Z, Wang H, Fan H, Zhu X, Zhou J, Fei E, Wang G. Gp78, an ER associated E3, promotes SOD1 and ataxin-3 degradation. Hum Mol Genet. 2009;18:4268–4281. doi: 10.1093/hmg/ddp380. [DOI] [PubMed] [Google Scholar]
- Yoshizawa T, Watanabe M, Frusho K, Shoji S. Magnetic resonance imaging demonstrates differential atrophy of pontine base and tegmentum in Machado-Joseph disease. J Neurol Sci. 2003;215:45–50. doi: 10.1016/s0022-510x(03)00185-0. [DOI] [PubMed] [Google Scholar]
- Yu YC, Kuo CL, Cheng WL, Liu CS, Hsieh M. Decreased antioxidant enzyme activity and increased mitochondrial DNA damage in cellular models of Machado-Joseph disease. J Neurosci Res. 2009;87:1884–1891. doi: 10.1002/jnr.22011. [DOI] [PubMed] [Google Scholar]
- Zhong X, Pittman RN. Ataxin-3 binds VCP/p97 and regulates retrotranslocation of ERAD substrates. Hum Mol Genet. 2006;15:2409–2420. doi: 10.1093/hmg/ddl164. [DOI] [PubMed] [Google Scholar]
- Zoghbi HY, Orr HT. Pathogenic mechanisms of a polyglutamine-mediated neurodegenerative disease, spinocerebellar ataxia type 1. J Biol Chem. 2009;284:7425–7429. doi: 10.1074/jbc.R800041200. [DOI] [PMC free article] [PubMed] [Google Scholar]