Abstract
Recent evidence suggests that survival of arctic-alpine organisms in peripheral or interior glacial refugia are not mutually exclusive and may both be involved in shaping an organism’s Pleistocene history, yet potentially at different time levels. Here, we test this hypothesis in a high-mountain plant (diploid lineage of Senecio carniolicus, Asteraceae) from the Eastern European Alps, in which patterns of morphological variation and current habitat requirements suggest survival in both types of refugia. To this end, we used AFLPs, nuclear and plastid DNA sequences and analysed them, among others, within a graph theoretic framework and using novel Bayesian methods of phylogeographic inference. On the basis of patterns of genetic diversity, occurrence of rare markers, distribution of distinct genetic lineages and patterns of range connectivity both interior refugia in the formerly strongly glaciated central Alps and peripheral refugia along the southern margin of the Alps were identified. The presence of refugia congruently inferred by markers resolving at different time levels suggests that these refugia acted as such throughout several glacial cycles. The high degree of range persistence together with gradual range expansion, which contrasts with the extent of range shifts implied for other Alpine species, is likely responsible for incipient lineage differentiation evident from the genetic data. Replacing a simplistic peripheral vs. interior refugia dualism by more complex models involving both types of refugia and considering different time levels will help identifying common phylogeographic patterns with respect to, for instance, location of refugia and colonization routes and elucidating their underlying genetic and/or ecological causes.
Keywords: AFLPs, Alps, geographic diffusion models, phylogeography, plastid sequences, Pleistocene refugia, range dynamics, Senecio carniolicus
Introduction
A central question of arctic-alpine phylogeography is concerned with Pleistocene range dynamics and the location of refugial areas (peripheral vs. interior refugia, so-called nunataks; reviewed in Stehlik 2000). Potentially governed by the inherent difficulty of providing convincing evidence for nunatak survival because of, for instance, extirpation of the likely small nunatak populations or genetic swamping by geographically close peripheral populations (Stehlik et al. 2001; Hewitt 2004), results of numerous phylogeographic studies within the last decades have diminished the importance of nunatak survival and identified peripheral refugia as the most relevant ones (Gabrielsen et al. 1997; Schönswetter et al. 2005). Thus, the discussion on peripheral vs. nunatak survival appears to have been settled in favour of the former (but see Holderegger et al. 2011). On the occasion of recent evidence for nunatak survival in arctic-alpine plants and animals (Lohse et al. 2011; Westergaard et al. 2011), it has been (re-)emphasized that nunatak and peripheral survival are not mutually exclusive and actually may both be involved in an organism’s Pleistocene history, yet potentially at different time levels (Schneeweiss & Schönswetter 2011).
The European Alps provide a suitable geographic model system for testing hypotheses on glacial survival of mountain species during the last glacial. On the basis of distributional (Merxmüller 1952, 1953, 1954; Pawłowski 1970) and molecular data (Paun et al. 2008; Burnier et al. 2009; Schneeweiss & Schönswetter 2010), peripheral refugia at the edges of the Alpine arc have been identified. In contrast, convincing evidence for nunatak survival in central parts of the Alps is much scarcer (Stehlik et al. 2002; Bettin et al. 2007; Parisod & Besnard 2007). Both results may, however, be misleading. Wide application (Schönswetter et al. 2002, 2003, 2004; Tribsch et al. 2002) of well-resolving, but rapidly homogenizing AFLP markers may cause underestimation of nunatak survival, especially in species with high colonizing capabilities that are particularly prone to genetic swamping (Gabrielsen et al. 1997; Schneeweiss & Schönswetter 2011). (In the context of AFLPs, homogenization refers to the rapid decay of signals of immigrant genotypes as a result of their repeated backcrossing with resident genotypes: Zhou et al. 2005). Geographically or phylogenetically restricted sampling (Stehlik et al. 2001; Bettin et al. 2007; Parisod & Besnard 2007) may result in overestimation of nunatak survival in case colonization from unstudied regions or interspecific gene flow from not sampled congeners remain unrecognized. Therefore, additional studies accounting for those potential pitfalls and using molecular markers resolving at different time levels are necessary for better assessment of modes of refugial survival in the Alps.
Identification of spatial genetic patterns, such as traces left by glacial refugia, remains an important objective of phylogeography, but yields only a snapshot of an organism’s history. Additional aspects of interest include, for instance, connectivity among different refugial areas or modes and directions of range expansions. Although such aspects may be tested using statistical phylogeographic approaches (Knowles & Maddison 2002; Knowles 2009), the sheer amount of plausible hypotheses that often need to be considered renders their application unfeasible except for simple settings (e.g. Spellman & Klicka 2006; Carstens & Richards 2007). Statistically rigorous methods for inferring phylogeographic history in a continuous landscape have been implemented in a maximum-likelihood framework (Lemmon & Lemmon 2008). More recently, this has been achieved in a Bayesian framework (Lemey et al. 2009, 2010) within the software beast (Drummond & Rambaut 2007), thus accommodating uncertainties in inferences of genealogy and of geographic diffusion. These methods allow establishing phylogeographic hypotheses with respect to range connectivity or ancestral locations in the absence of a priori knowledge, but to our knowledge, none of the Bayesian methods has been applied to plants yet.
A good model system for addressing the complexity of Pleistocene range dynamics in alpine to subnival species is the diploid lineage of Senecio carniolicus (Asteraceae). Although commonly growing together, diploid and polyploid (tetraploids and hexaploids: Suda et al. 2007) cytotypes of S. carniolicus are reproductively isolated by different habitat requirements (Schönswetter et al. 2007; Hülber et al. 2009; Sonnleitner et al. 2010) as well as strong crossing barriers (Sonnleitner et al. 2010; M. Sonnleitner et al., unpublished) and apparently constitute distinct and nonintermixing lineages. Therefore, it is justified to restrict ourselves here to the diploid cytotype. Several lines of evidence suggest that both peripheral and internal refugia may have played a role in its Pleistocene history. Diploid S. carniolicus occurs in essentially all peripheral refugia identified for silicicolous species in the Eastern Alps (Schönswetter et al. 2005; Thiel-Egenter et al. 2009; the locations of these refugia are shown in Fig. 1a). Furthermore, taxonomically acknowledged (as var. or subsp. insubricus) morphological differentiation between populations from the southwestern Eastern Alps and those elsewhere suggests (under a neutral model) sufficiently long and/or repeated isolation within the prominent and geographically stable refugium in the Southern Alps. On the other hand, diploid S. carniolicus grows in exposed, rocky habitats up to more than 3100 m a.s.l. (Hülber et al. 2009; Sonnleitner et al. 2010), rendering occurrence on steep south-exposed slopes, which likely served as microclimatically favourable habitats on nunataks (Parisod & Besnard 2007), a valid hypothesis. Additionally, nunatak survival has already been suggested for S. halleri (Bettin et al. 2007), a closely related endemic of the Western Alps with similar habitat requirements.
Fig. 1.
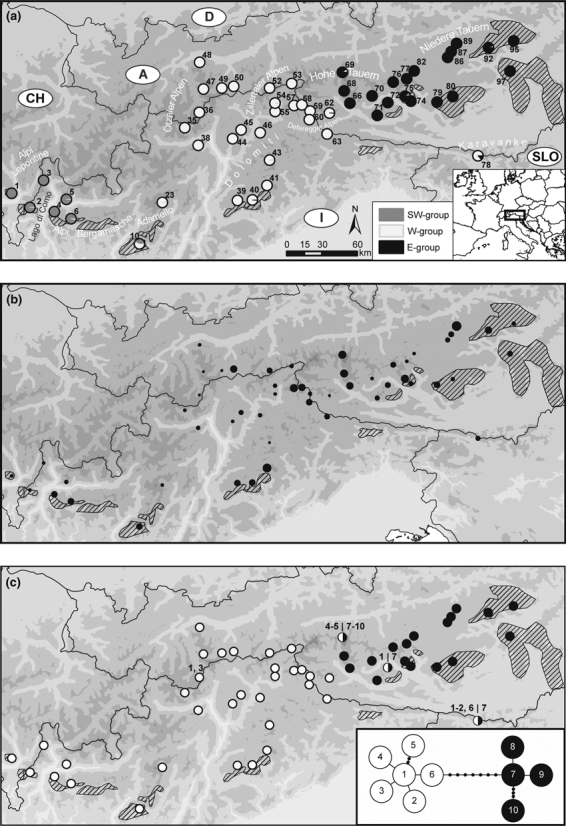
Physical map of the distribution of the analysed populations (a) and patterns of AFLP and ITS sequence variation in diploid Senecio carniolicus. (a) Phylogeographic grouping of populations according to Bayesian clustering analysis of AFLP phenotypes conducted with Structure. (b) Within-population rarity of AFLP markers (frequency-down-weighted marker values), its magnitude being proportional to dot size. (c) Distribution of ITS ribotypes, their relationships (inferred using statistical parsimony) shown in the insert; unless otherwise indicated, white and black dots represent ribotypes 1 and 7, respectively. In (a), populations are numbered as in Sonnleitner et al. (2010) and toponyms mentioned in the text are indicated. Hatched areas indicate potential glacial refugia on siliceous bedrock (modified from Schönswetter et al. 2005).
Here, we test the hypothesis that Pleistocene range dynamics of diploid S. carniolicus were influenced by survival in both peripheral and interior refugia, even if these potentially acted at different time levels. To this end, based on comprehensive geographic sampling and taking closely related congeners into account, we employ several different molecular markers: rapidly homogenizing, biparentally inherited AFLPs, which often are used to resolve intraspecific relationships because of their ability to provide phylogenetic signal in young or rapidly evolving study systems (Després et al. 2003; McKinnon et al. 2008); biparentally inherited nuclear ITS sequences, which often provide enough information for addressing evolutionary questions at the species level, including hybridization (Nieto Feliner & Rosselló 2007); and maternally inherited (for Asteraceae: Corriveau & Coleman 1988) plastid DNA sequences, which are less likely to be genetically swamped owing to their higher propensity of introgressing into immigrants (Currat et al. 2008) and whose slower mutation rates (Petit et al. 2005; Shaw et al. 2005; Borsch & Quandt 2009) allow tracing of patterns pre-dating the last glacial maximum. We analysed the molecular data, among others, within a graph theoretic framework and using novel Bayesian methods of phylogeographic inference. Our first aim is to identify putative refugia based on patterns of genetic diversity, occurrence of rare markers, and distribution of distinct genetic lineages (Taberlet 1998; Hewitt 1999, 2000; Paun et al. 2008; Provan & Bennett 2008), including testing hypotheses of previously defined peripheral refugia for silicicolous species (Schönswetter et al. 2005) via their correspondence with genetic groups; in case both peripheral and interior refugia are found, we additionally assess whether these acted as refugia at the same time levels or not. Our second aim is to infer relationships among different refugia as well as the (re)colonization directionality, specifically, whether there is a general eastward migration trend as expected from the distribution of the closest relatives of S. carniolicus (S. halleri and S. incanus) in the Western Alps.
Materials and methods
Study species
Senecio carniolicus (syn. Jacobaea carniolica; Pelser et al. 2006) is a common and abundant acidophilic species of the Eastern Alps and the Carpathians, where it grows in alpine grasslands, moraines and stable screes up to 3300 m a.s.l. (Reisigl & Pitschmann 1958). Within the Eastern Alps, S. carniolicus comprises three main cytotypes (diploids with 2n = 2x = 40, tetraploids with 2n = 4x = 80, hexaploids with 2n = 6x = 120; Suda et al. 2007; Sonnleitner et al. 2010). Diploids are reproductively isolated from polyploids by different habitat requirements (Sonnleitner et al. 2010) as well as strong crossing barriers (crossings of diploids with polyploids produce nearly no seeds, and these show reduced viability; M. Sonnleitner et al., unpublished), and we only consider the diploid lineage here. Populations of S. carniolicus from the southwestern Eastern Alps have more deeply lobed and more densely hairy leaves, causing morphological resemblance to S. incanus, a closely related, but morphologically and genetically clearly distinct species (Hess et al. 1980; Pelser et al. 2003) from the Western Alps and the northern Apennines. These forms, initially described as S. carniolicus var. insubricus (Chenevard 1906), are no hybridogenic intermediates, but phylogenetically clearly belong to S. carniolicus (Pelser et al. 2003; Fig. S1).
Plant material
From those populations analysed by Sonnleitner et al. (2010), where diploids were found, two to eight diploid individuals per population were included. No plant material was available from their populations 24 and 96 (depleted after flow cytometry), resulting in 52 studied populations (Table S1), which cover the entire distribution range of diploid S. carniolicus. This sampling also included several populations of S. carniolicus var. insubricus (pops. 1–6). As outgroups for some analyses, we included samples of all members of the Incani clade (Pelser et al. 2003, 2004), that is, S. abrotanifolius, S. adonidifolius, S. boissieri, S. halleri, S. incanus, S. leucophyllus and S. personii (Appendix S1, Table S2). Leaf material was collected and immediately stored in silica gel. Voucher specimens are deposited at the Institute of Botany, University of Vienna, Austria (WU, voucher numbers and collecting details are given in Sonnleitner et al. 2010).
Molecular methods
Total genomic DNA was extracted from similar amounts of dried tissue (c. 10 mg) with the DNeasy 96 plant mini kit (Qiagen, Hilden, Germany) following the manufacturer’s protocol.
The AFLP procedure followed Vos et al. (1995) with the modifications described in Schönswetter et al. (2009). Initially, selective primers were screened using 16 primer combinations. The three final primer combinations for the selective PCR (fluorescent dye in brackets) were EcoRI (6-Fam)-ACA/MseI-CAT, EcoRI (VIC)-AGG/MseI-CTC, and EcoRI (NED)-ACA/MseI-CAC. Purification and visualization of PCR products as well as scoring were carried out as described in Rebernig et al. (2010b). Six plants were extracted twice to test the reproducibility of AFLP fingerprinting (Bonin et al. 2004). Seven samples were used as replicates between PCR plates and were replicated more than twice, resulting in a total of 75 replicates. The error rate (Bonin et al. 2004) was calculated as the ratio of mismatches (scoring of 0 vs. 1) over phenotypic comparisons in AFLP profiles of replicated individuals. Nonreproducible fragments were excluded from the analyses.
PCR products of the nrDNA ITS region and the plastid spacers trnL-rpl32, rps16-trnK, psbD-trnT and petL-psbE were obtained in reaction volumes of 13 μL including 4.5 μL REDTaq ReadyMix PCR mix (Sigma-Aldrich, Steinheim, Germany), 0.5 μL of 1 mg/mL bovine serum albumin (BSA; Promega, Madison, WI, USA), 0.5 μL 10 μm each of forward and reverse primers and about 25 ng of DNA suspended in 1 μL 1× TAE buffer. For ITS, primers and PCR conditions were as in Blöch et al. (2009). Plastid spacers were amplified using the primers of Shaw et al. (2007). Some samples from the outgroup taxa repeatedly failed to amplify and were amplified using newly designed internal primers listed in Table S3. PCR conditions were as follows: 95 °C for 1 min followed by 10 cycles each consisting of 30 s at 95 °C, 30 s at 47 °C, 1.5 min at 65 °C; 20 cycles each of 30 s at 95 °C, 30 s at 49 °C, 1.5 min at 65 °C; and a final elongation step of 8 min at 65 °C. PCR programs were run on GeneAmp PCR System 9700 thermocyclers (PE Applied Biosystems, Foster City, CA, USA). ITS PCR products from individuals in which length polymorphism made direct sequencing impossible were cloned using the pGEM-T-easy vector system and JM109 competent cells (Promega) following the manufacturer’s instructions. Inserts of 8–15 clones per individual were amplified using universal primers M13F(–47) and M13R(–48) and 1 μL of colony resuspended in 70 μL 1× TBE. PCR conditions were the same as described above except for annealing and elongation temperatures of 62 and 72 °C, respectively. PCR products were purified with E. coli Exonuclease I and FastAP Thermosensitive Alkaline Phosphatase (Fermentas, St. Leon-Rot, Germany) following the manufacturer’s instructions. Cycle sequencing reactions were performed using 5 μL of purified template and 1 μL BigDye Terminator (PE Applied Biosystems), then cleaned with Sephadex G-50 Fine (GE Healthcare Bio-Sciences, Uppsala, Sweden) and sequenced on an ABI 3770 DNA Analyzer (PE Applied Biosystems).
Data analysis
AFLP data
All monomorphic fragments and the ones present in all but one individual of the diploid data set were removed from the data set to avoid biased parameter estimates (Bonin et al. 2004). Fragments present in only one diploid individual were only removed if they were singular with respect to a data set including all cytotypes of S. carniolicus.
Population structure was inferred employing a Bayesian clustering approach developed for dominant markers (Structure 2.2; Pritchard et al. 2000; Falush et al. 2007) with an admixture model with uncorrelated allele frequencies and recessive alleles. Ten replicate runs for each K (number of groups) ranging from 1 to 10 were carried out at the Bioportal of the University of Oslo (http://www.bioportal.uio.no/), using a burn-in of 105 iterations followed by 106 additional MCMC iterations. Similarity among results of different runs for the same K was calculated according to Nordborg et al. (2005) using Structure-sum-2009 (Ehrich 2006). We identified the optimal number of groups as the value of K where the likelihood reached a plateau; the results of replicate runs were identical, and no empty groups were encountered. Replicate runs of the best K were merged with Clumpp 1.1.1 (Jakobsson & Rosenberg 2007) using the ‘Full Search’-algorithm. The relative ‘cluster membership coefficients’ of all individuals were then averaged for each population. A principal coordinate analysis (PCoA) based on a matrix of Nei72 distances (Nei 1972) among individuals was calculated using the modules ‘Simgend’, ‘Dcenter’ and ‘Eigen’ from NTSYS-pc 2.2 (Rohlf 1997).
The frequency of rare markers as frequency-down-weighted marker values was calculated according to Schönswetter & Tribsch (2005) using AFLPdat (available from http://www2.uit.no/ikbViewer/page/ansatte/organisasjon/ansatte/person?p_document_id=41186&p_dimension_id=88165). These individual values were averaged to obtain values for the within-population rarity of markers (in the following termed ‘DW’).
The genetic covariance structure among sampled populations was modelled within a graph theoretic framework (Population Graphs: Dyer & Nason 2004) using PopGraphs (available from http://dyerlab.bio.vcu.edu/software/). A population network is constructed, where populations, which constitute the nodes, are connected by edges only if there is significant genetic covariance between the populations after removing the covariation each population has with the remaining populations in the data set. The distribution of shortest paths through the Population Graph creates an expectation of proportional distances for populations. If genetic covariance is spatially structured, the physical distances should be proportional to the genetic distances. If not, then the populations are either closer (compressed edges) or further apart (extended edges) than expected given the genetic distances (χ2 tests at α = 0.05 significance level). Extended edges may indicate long-distance dispersal, whereas compressed edges point to the presence of topological, historical and/or ecological sources of vicariance (Garrick et al. 2009).
Genetic covariance among geographic groups as defined for the phylogeographic analyses of the plastid sequence data (see next section) was also analysed using Population Graphs networks as just described. Stability of edges among geographic groups was assessed using a bootstrap approach with 200 bootstrap replicates. Pseudoreplicate data sets were generated using seqboot from the Phylip package (Felsenstein 1989) and analysed like the original data set. The proportion of replicates where a certain edge is found constitutes its bootstrap support.
Sequence data
Sequences were assembled using SeqMan II (DNAStar, Madison, WI, USA), manually edited and aligned with BioEdit 7.0.5.2 (Hall 1999). Within-population haplotype diversity was estimated using π, the mean number of pairwise differences (Tajima 1983), calculated with Arlequin 3.11 (Excoffier et al. 2005).
Nuclear ITS and plastid sequence data (the four regions concatenated under the assumption that the plastid genome is a single linkage group) were analysed using statistical parsimony as implemented in TCS (Clement et al. 2000) with the connection limit set to 95%. As gaps were treated as fifth character state, indels longer than 1 bp were recoded as single characters by reducing them to single base pair columns. Nested indels were coded as missing data. Mononucleotide repeats were removed owing to their high degree of homoplasy over larger geographic scales (Ingvarsson et al. 2003; Vachon & Freeland 2011). Prior to the TCS analysis, monophyly of the S. carniolicus ribotypes with respect to those of other species of the Incani clade was assessed via phylogenetic analyses using maximum parsimony in paup* 4.0b10 (Swofford 2003) and Bayesian inference in MrBayes 3.1.2 (Ronquist & Huelsenbeck 2003; see Appendix S1 for details).
Phylogeographic analyses of the plastid data set (comprising the four concatenated regions) were conducted in beast 1.6 (Drummond & Rambaut 2007). Model-fit of nucleotide substitution models was assessed via the Akaike Information Criterion (AIC) and the Bayesian Information Criterion (BIC) as implemented in jModelTest 0.1.1 (Posada 2008). Model uncertainty was considerable and the sets of models with cumulative AIC and BIC weights of at least 0.95 contained models differing in the number of substitution rates (3–6 vs. 1–3). Therefore, we finally used a HKY model with rate heterogeneity modelled by a gamma distribution (with six rate categories) to avoid over-parameterization. As prior for the transition-transversion ratio κ, we used a normal distribution with mean 0.41 (derived from the model-averaged value for this parameter determined via AIC and BIC) and a deliberately wide standard deviation of 0.4. Rate evolution was modelled in a strict clock framework, because a relaxed clock model had an only slightly better marginal log-likelihood and the coefficient of rate variation had its highest posterior density around zero (data not shown). Because of the lack of external calibrations, we used a strong prior on the substitution rate modelled with a lognormal distribution with a mean of 7.5 × 10−3 substitutions per site per million years, a standard deviation of 0.6 and an offset at 0.0, thus ensuring a modal value of the distribution around 4 × 10−3 substitutions per site per million years in line with previously suggested values (Yamane et al. 2003; Smith et al. 2008). As these values were obtained for groups phylogenetically very distant from Asteraceae, their applicability to S. carniolicus remains uncertain, and, consequently, the obtained age estimates should be interpreted with appropriate caution. As population model, we used the Bayesian skyline plot (Drummond et al. 2005) with a group interval m = 3. Stationarity of the Markov chain was determined using Tracer 1.4 (available from http://tree.bio.ed.ac.uk/software/tracer/).
Spatial distribution through time was inferred employing a discrete model of geographic diffusion. Under this model, rates of diffusion between a priori defined discrete locations are estimated using a continuous-time Markov chain model (Lemey et al. 2009). Starting from the unobserved location at the root of the tree derived from a uniform distribution over all sampled locations, dispersal proceeds along each branch according to this model and gives rise to the observed locations at the tips. The geospatial model may be reversible, that is, the diffusion rate between regions is identical in both directions, or nonreversible, that is, the diffusion rate in one direction can differ from that in the reverse direction. Application of a continuous model of geographic diffusion (Lemey et al. 2010), apparently appropriate for the largely continuous range of S. carniolicus (Fig. 1a), was not possible because of insufficient signal in the data evident from simple reconstruction of the priors during analyses (data not shown).
Although geographically distinct sampling localities constitute intuitive discrete geographic units, the enormous number of possible rate parameters renders their use in S. carniolicus impossible. To be able to delimit a manageable number of discrete geographic units in a largely continuous distribution range in a repeatable manner, we applied agglomerative hierarchical clustering to populations using the function agnes with default options (Euclidean distances and upgma clustering) in the package ‘cluster’ (Maechler et al. 2005) in R 2.8.0 (R Development Core Team 2008). Groups were delineated applying a certain geographic distance cut-off. As we wanted to address questions concerning peripheral vs. interior refugia, the value of this cut-off was chosen to result in sufficient geographic resolution (i.e. peripheral and central populations should be in different geographic units) with as few groups as possible. Eventually, using a geographic distance cut-off of about 40 km, 13 geographic groups were distinguished (Fig. S2).
The full diffusion matrix of the simpler reversible model for 13 groups contains 78 rate parameters and therefore is still grossly over-parameterized. Hence, we used Bayesian stochastic variable selection to reduce the number of nonzero rates. Following Lemey et al. (2009), we used a truncated Poisson prior with a mean of 0.693 (i.e. ln2) and an offset corresponding to the number of rates necessary to minimally connect all regions (i.e. number of regions minus 1), which puts 50% prior probability on the minimal rate configuration. Sensitivity to the choice of prior was assessed via rerunning the analysis with different prior means (0.1, 1, 2, 5, 10). For the nonreversible model (comprising 156 rate parameters), the Poisson prior was parameterized with a mean of 12 (i.e. number of regions minus 1) and an offset of 0 (P. Lemey, pers. comm.). Sensitivity was assessed via rerunning the analysis with different prior means as above. In all cases, we used equal expectations for all rates, that is, the prior on the diffusion rates is not informed by the geographic distances among geographic units. Two runs per parameterization, each for 2 × 108 generations with sampling every 5000th generation, were conducted. As both runs converged on the same stationary distribution and effective sample size values safely exceeded 200, they were combined after removal of the first 10% of sampled generations as burn-in. All parameter estimates were based on these two runs combined (72 000 sampling points). To assess the robustness of the results with respect to different delimitation of discrete geographic units, a second analysis using eight regions (corresponding to a geographic distance of about 70 km; Fig. S2) was conducted, using the same prior parameterizations as above. Identification of well-supported rates was performed using the beast module RateIndicatorBF.
Results
AFLPs
The three AFLP primer combinations yielded 264 unambiguous polymorphic fragments after the removal of 16 nonreproducible and 31 invariable markers in 227 individuals. Six singular markers were retained because they occurred also in individuals of the data set comprising polyploid cytotypes, rendering it unlikely that these markers merely were artefacts. In the AFLP profiles from replicated samples, 294 differences were observed out of 23 925 phenotypic comparisons, resulting in an error rate of 1.23%.
The optimal number of groups in the Structure analysis was K = 3, comprising a southwestern, a western and an eastern group (Fig. 1a). The disjunct population 78 from the southeastern margin of the distribution area belonged to the western group. Only populations 1–6, 10, 75 and 78 showed admixture among the groups (using an arbitrary cut-off of 10% or more for the proportion of the minor group). The two-dimensional PCoA corroborated the pattern shown in the structure analysis (Fig. S3). The division between the eastern group and the other two groups was much more distinct with 50.4% of the variation explained, than the division between the western and the southwestern groups (7.4% of the variation explained).
DW values ranged from 0.91 in population 49 to 1.57 in population 89 (Table S1). DW values were highest in the western Niedere Tauern (population 89), in the southern Dolomiti (population 41), and in the central Alps (populations 57, 66, 68, 69, and 75; Fig. 1b).
Roughly one-fourth of the sampled populations remained unconnected in the Population Graphs network (Fig. S4). The covariance structure among populations of the southwestern group (populations 1–6) was characterized by compressed edges, indicating that population pairs were geographically closer to each other than expected given their genetic distances. The same was true for populations from the central Alps (populations 35, 47–49; 53 and 57; 69 and 71) and the southern Dolomiti (populations 40 and 43). Extended edges were predominantly oriented in west–east direction. Two edges connected the western and eastern group: a normal edge joined population 78 in the Karavanke and population 97 from the easternmost Central Alps, and the southern Dolomiti were connected with the Niedere Tauern by an extended edge (populations 40 and 87).
In the Population Graphs network of geographic regions, stable edges with a bootstrap support >50% occurred mainly among neighbouring regions (Fig. 3a), with the exception of edges connecting region R5 with R13 and R8 with R11. Two of the 13 regions (R1, R6) had no stable edges.
Fig. 3.
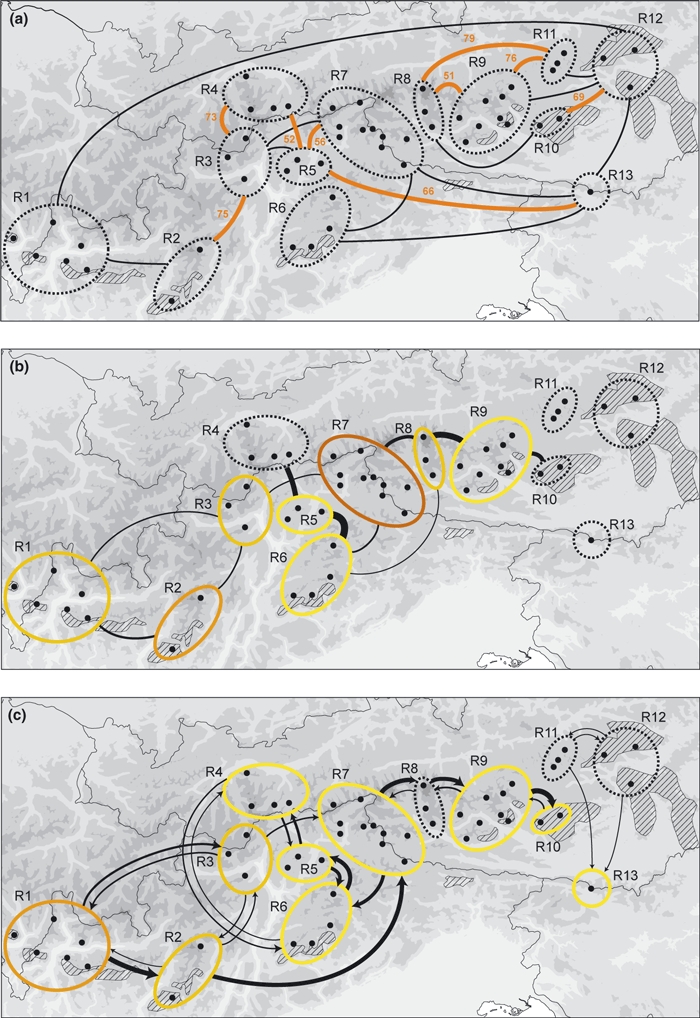
Range connectivity among discrete geographic regions (their designations R1–R13 are indicated) in diploid Senecio carniolicus. (a) Population Graphs network illustrating the genetic covariance structure among regions based on AFLP fingerprints; connections between regions indicated in orange are stable edges (with a bootstrap support >50% given at the edges), other edges are drawn in black. (b–c) Range connectivity inferred from plastid sequence data using (b) reversible and (c) nonreversible models of geographic diffusion between discrete geographic regions, the thickness of the connections being proportional to their support by Bayes Factors (see text for details); decreasing posterior probability (shown in four classes from 0.05 to 0.19) of a region to be the ancestral area is indicated by increasingly lighter colours. Hatched areas in (a) to (c) as in Fig. 1.
ITS
GenBank accession numbers and ITS sequence statistics are provided in Tables S1 and S4, respectively. Ribotypes of S. carniolicus constituted a monophyletic clade (60% bootstrap support; 0.97 posterior probability; Fig. S1). Direct sequencing yielded unambiguous sequences in all cases except for populations 69, 72 and 78, which displayed within-individual ITS sequence length variation and required cloning. Chimeric ITS clones may be the result of PCR recombination and were removed. The statistical parsimony network of ITS ribotypes (Fig. 1c) recovered two groups separated by eight mutational steps. The circumscription of these groups, which were also recovered in the larger data set (Fig. S1), was congruent with the southwestern plus the western AFLP groups on the one hand and the eastern AFLP group on the other (Fig. 1a,c).
Plastid DNA sequences
GenBank accession numbers and sequence statistics are given in Tables S1, S2 and S4, respectively. No inversions were detected. Phylogenetic analysis using beast revealed three clades: haplotype group 1 (posterior probability 0.96) comprising H1–H10, haplotype group 2 (posterior probability 1.00) consisting of H11 and haplotype group 3 (posterior probability 0.99) with H12–H35. Within haplotype group 3, a moderately supported (posterior probability 0.96) subgroup constituted by H20–H35 could be distinguished. The earliest differentiation among these three lineages (age given as median and its 95% highest posterior density interval) is estimated to have occurred 1.15 (0.28–2.34) mya, subsequent diversifications falling safely within the Pleistocene (data not shown).
A statistical parsimony network constituted of 35 plastid DNA haplotypes, and their geographic distribution are presented in Fig. 2a,b. The parsimony network of plastid DNA haplotypes revealed strong reciprocal differentiation among haplotypes pertaining to haplotype groups 1 and 3. The mean number of pairwise differences (π) calculated for sets of haplotypes of haplotype groups 1 and 3 amounted to 5.99 and 1.23, respectively (haplotype group 2 contains a single haplotype). The most frequent haplotype was H35 found in 53% of the investigated individuals throughout the distribution range except for its western third (Fig. 2b). Twelve further haplotypes (H17, H20, H21, H23, H25, H26, H28–H33) differed only in a single mutational step from H35 (together with H35 encompassing 71% of the investigated individuals). Most divergent were H1–H11, which were separated by at least 10 mutational steps from the other haplotypes. These haplotypes were present in 11% of the sampled individuals (Fig. 2b). Most outgroup haplotypes were similar to haplotypes H1–H11, but three haplotypes present in S. incanus and S. halleri were close to haplotypes H14 and H17 of haplotype group 3. No outgroup haplotypes were shared with the ingroup (Fig. 2a).
Fig. 2.
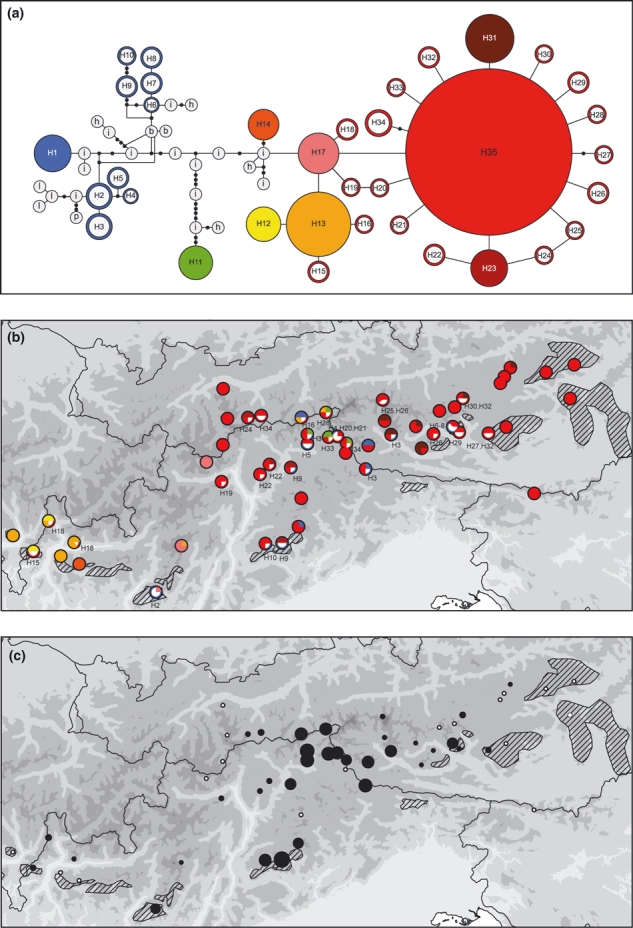
Patterns of plastid DNA variation in diploid Senecio carniolicus. (a) Statistical parsimony network of plastid haplotypes (blue, green and yellow to red colours correspond to haplotype groups 1, 2 and 3, respectively, identified by beast), a circle’s size being proportional to the square-root transformed frequency of the respective haplotype; haplotypes found in other species of the Incani clade are indicated by grey circles (b, S. boissieri; h, S. halleri; i, S. incanus; l, S. leucophyllus; p, S. persoonii) without indication of these haplotypes’ frequencies; unsampled haplotypes are represented by black dots. (b) Geographic distribution of plastid haplotypes; colours as in (a), haplotypes occurring in fewer than four individuals are marked with their number. (c) Within-population plastid haplotype diversity calculated as the mean number of pairwise differences (π), its magnitude being proportional to dot size (invariable populations indicated as white dots). Hatched areas in (b) and (c) as in Fig. 1.
The mean number of pairwise differences within populations ranged from 0.0 (in 19 populations throughout the range) to 10.0 (population 40). The most diverse regions were the southern Dolomiti (populations 39–41) and the central Alps (populations 52–55, 57, 58, and 63; Fig. 2c, Table S1).
Qualitative inferences of geographic diffusion using a reversible discrete model were robust against prior parameterizations. Differences were of quantitative nature, as Bayes Factors (BF) usually increased (up to fourfold) with increasing prior means (Table S5a), which might cause BFs for some rates to eventually pass the applied threshold of 3 (the number of supported rates increased from 11 to 16 with prior means of 0.1 and 10, respectively: Table S5a). The composition of the set of rates congruently supported under all prior parameterizations essentially did not change and in two of the three cases, where rates showed decreasing BFs with increasing prior means, BFs dropped down to 2.7 (Table S5a). Qualitatively similar results were obtained with eight regions, even if trends in changes of BFs with changes of prior means were less clear (Table S5b).
In contrast, inferences using a nonreversible discrete model were highly sensitive to prior parameterizations. Specifically, the number of rates supported by BF ≥3 increased with decreasing prior mean from 25 to 133 rates (i.e. from 16% to 85% of possible rates) with prior means of 12 and 0.1, respectively (Fig. S5a). In the case of eight delimited regions, a similar increase was found from 13 to 48 rates (i.e. from 23% to 86% of possible rates) with prior means of 10 and 0.1, respectively (Fig. S5b). Furthermore, BFs of some rates increased enormously with decreasing means (Table S6); for instance that for R1→R2 increased more than 90-fold (Fig. S5a) compared to more than 400-fold in case of eight delimited regions (Fig. S5b). Finally, with decreasing prior means some results concerning rate support became dubious. Most conspicuously, rates involving the region hosting the single population in the southeastern Alps (pop. 78: R13) became frequently supported with decreasing prior means (2 vs. 23 of 24 possible rates with prior means of 12 vs. 0.1, respectively: Fig. S5a), although it harboured only a single widespread haplotype. Similar patterns were observed in case of eight delimited regions (1 vs. 11 of 14 possible rates with prior means of 10 vs. 0.1, respectively: Fig. S5b). Reasons for this odd behaviour may include convergence issues when using lower Poisson prior means (P. Lemey, pers. comm.), but a proper assessment would require extensive simulation studies going beyond the scope of this study. Consequently, results from the nonreversible model will be interpreted with caution taking into account the results from the more robust reversible model.
The reversible model identified 11 significant connections mostly between adjacent regions (Fig. 3b). The three easternmost regions remained unconnected (i.e. received BF support <3). The number of significant connections inferred from the nonreversible model was higher (five unidirectional and ten bidirectional ones: Fig. 3c), but this set included all rates identified with the reversible model except one (southern Dolomiti to central Hohe Tauern: R6→R8). The three easternmost regions were connected among each other, albeit only weakly supported, but remained unconnected from the remaining regions. In case of asymmetrical rates, eastward rates were better supported than westward ones. This is also the case for the connection from the southern Adamello region to the central Alps (R2→R7), whose support under the reversible model (2.99) is slightly below the applied threshold. A well-supported westward direction was inferred from the western Hohe Tauern and Zillertaler Alpen to the southern Dolomiti (R7→R6). Whereas the overall eastward directionality was insensitive to the number of delimited regions, the northward directionality weakly suggested for the central part (Fig. 3c) disappeared and was actually reversed with fewer regions [i.e. R(3–5)→R6 obtained higher support than the reverse rate: Fig. S5b].
Whereas under the reversible model the ancestral location was inferred to be in the central Alps (western Hohe Tauern and Zillertaler Alpen), under the nonreversible model it was situated in the southwestern region (Alpi Lepontine and Alpi Bergamasche). However, in both cases support was low (posterior probabilities <0.2, BF for single regions 0.35–2.8). The sets of regions with cumulative posterior probabilities of at least 0.8 included 8–10 regions (reversible and nonreversible model, respectively), only the two northeastern regions being congruently excluded.
Discussion
Against previous, often confrontational assertions, glacial survival in peripheral refugia (massifs de refuge) does not preclude survival in interior refugia (nunataks), and both may contribute to shaping Pleistocene range dynamics of alpine biota (Stehlik et al. 2002; Lohse et al. 2011). This is also the case for diploid Senecio carniolicus, where several peripheral and interior refugia have been identified based on increased frequency of rare AFLP markers (Fig. 1b), elevated haplotype diversity and the distribution patterns of the oldest and hence most divergent haplotype lineages (Fig. 2). Suggested refugial areas are both along the formerly not or only weakly glaciated southern margin of the Alps (Alpi Bergamasche, southern Adamello and southern Dolomiti), where major refugia for silicicolous plants have been proposed before (Schönswetter et al. 2005), and within formerly strongly glaciated central parts of the cytotype’s range (Deferegger and Zillertaler Alpen). Remarkably, there is no evidence for survival of S. carniolicus in the easternmost Alps, which acted as a refugium for most silicicolous species investigated so far (e.g. Saponaria pumila and Androsace wulfeniana: Tribsch et al. 2002; Schönswetter et al. 2003). Instead, low frequency of rare AFLP markers (Fig. 1b), the near exclusive presence of the most widespread haplotype H35 (Fig. 2b) and inferred range expansions (Fig. 3) suggest recent, possibly postglacial, eastward range expansion into this area. The presence of refugia congruently inferred by markers resolving at different time levels suggests that these refugia acted as such throughout several glacial cycles and that diploid S. carniolicus responded to the Quaternary climatic oscillations in a cyclic manner (Rebernig et al. 2010a; Stewart et al. 2010).
An unexpectedly high degree of range persistence is evident not only from the presence of several refugia and their stability over time, but is also well reflected in patterns of range connectivity (Fig. 3) as usually only immediately adjacent areas are significantly connected. (It is worth emphasizing that neither PopGraphs nor beast, as used here, take distances between the geographic units into account; consequently any connections are deduced solely from the genetic data.) This pattern suggests that any range expansion occurred mostly gradually to nearby ranges; this also involves adjacent refugia (Fig. 3c). If sufficiently slow, this mode of (re-)colonization may be responsible for the preservation of the signal for interior refugia in the AFLP data, because populations in these refugia are particularly prone to genetic swamping because of their likely small population sizes (Stehlik 2000). Likewise, prolonged isolation in refugia and nonsaltatory range shifts are conducive for lineage differentiation, as is evident from the presence of a western and an eastern genetic group (AFLP and nuclear ITS data: Fig. 1a,c and Figs S1 and S3). The distribution areas of these groups nearly abut in the Hohe Tauern region, which roughly coincides with genetic discontinuities in other alpine taxa (Schönswetter et al. 2002, 2004, 2005; Thiel-Egenter et al. 2009; Fig. 1a). Only a few western populations of the eastern group (pops. 69, 72) show signs of admixture (Fig. 1c, Table S1) in line with low levels of gene flow across this contact zone. This main genetic split into a western and an eastern group does not coincide with the morphology-based taxonomic differentiation of S. carniolicus var. insubricus (i.e. the southwestern populations within the western group, which are also, but less strongly genetically differentiated: Fig. 1a and Fig. S3) from the nominal variety. However, a morphological re-evaluation of diploid S. carniolicus indicates that var. insubricus is morphologically similar to the remaining populations of the western lineage, but clearly divergent from the eastern group (Flatscher 2010). Finally, slow range shifts conferring potentially still incomplete range filling (Svenning et al. 2008) may also be responsible for the lack of diploid S. carniolicus in the ecologically suitable northwesternmost central Alps, although alternative hypotheses, such as niche pre-emption by the here abundant hexaploid cytotype, cannot be ruled out and require further studies.
Range persistence, however, does not imply range stasis, and both long-distance dispersal and range expansion contributed to the range dynamics of diploid S. carniolicus. This is in line with the presence of traits conducive to dispersal over longer distances exhibited by the diaspores, including low terminal velocities (determined on most likely hexaploid S. carniolicus: Tackenberg & Stöcklin 2008). Evidence for long-distance dispersal is most prominently found in the disjunct population 78 from the southeastern Alps (Fig. 1a). Although this population is spatially isolated, it displays unambiguous traces of admixture between the western and the eastern groups (Fig. 1a,c; Table S1; plastid data are not informative, because the population is fixed for the widespread haplotype H35: Fig. 2b). This is also reflected in the Population Graph analysis both at the level of populations (genetic covariance with pop. 57 from the western and pop. 97 from the eastern group: Fig. S4) and at the level of geographic regions (genetic covariance with the central and the easternmost Alps: Fig. 3a).
Unambiguous evidence for range expansion is found for the eastern half of the distribution area of diploid S. carniolicus, which likely has been (re-)colonized from the interior refugium around Deferegger and Zillertaler Alpen. This strictly eastward (re-)colonization is supported by the lack of putative refugial areas in the easternmost Alps (see above), the restriction of the most ancestral plastid DNA haplotypes (i.e. haplotype groups 1 and 2 that intermix with haplotypes found in the outgroup species) to the central parts of the entire distribution range (around western Deferegger and Zillertaler Alpen: Fig. 2), and the asymmetry in favour of eastward diffusion rates (Fig. 3c). Range expansions into northwestern regions, such as the Ötztaler Alpen, probably involved source areas corresponding to interior as well as peripheral refugia at the southern and southwestern part of the distribution range (Fig. 3c). Although (re-)colonization within the Alps from different refugia has been identified before (e.g. Schönswetter et al. 2005; Parisod & Besnard 2007), only the application of spatially explicit methods, as used here, allows a more detailed characterization of the (re-)colonization routes.
A general eastward directionality is expected from the distribution of S. carniolicus’ closest relatives from the Incani clade (Pelser et al. 2003). These are distributed throughout the western half of the southern European mountain systems from the Sierra Nevada and the Cordillera Cantabrica (S. boissieri) over the Pyrenees and the Massif Central (S. leucophyllus) to the Western Alps (S. halleri, S. incanus, S. persoonii), the Eastern Alpine and Carpathian S. carniolicus being the easternmost representative of the group. This hypothesis finds support in the nonreversible discrete diffusion model, where the most likely ancestral location is inferred in the southwesternmost portion of the distribution area (Alpi Bergamasche and Alpi Lepontine) and eastward diffusion rates generally predominate (Fig. 3c). This contrasts with westward directionality out from an ancestral location in the central part of the distribution range indirectly suggested by the location of the most likely ancestral area inferred with the reversible discrete diffusion model (Fig. 3b). The reasons for this contradiction are unclear, but the fact that the most likely ancestral area (Fig. 3b) harbours many of the ancestral haplotypes (Fig. 2) suggests an influence of the distribution of character states (i.e. geographic regions) on a genealogy on ancestral area reconstruction under the reversible model. The current restriction of ancestral haplotypes to central parts of the distribution area (Fig. 2) might be due to long-lasting isolation of populations within interior refugia. Such isolation may delay the possibilities of genetic replacement by immigrants from the periphery (similar to island populations: Hargreaves et al. 2010), thus assisting retention of ancestral haplotypes beyond the high propensity of organellar genomes to introgress into invading genomes (Currat et al. 2008). An alternative explanation for the presence of haplotype groups 1 and 2 in diploid S. carniolicus might be hybridization with closely related congeners. This is, however, unlikely based on geographic considerations (populations of S. carniolicus harbouring haplotype groups 1 and 2 are not geographically close to the potential source species S. incanus or S. halleri), rendering incomplete lineage sorting, possibly dating back to the late Pliocene, the more plausible hypothesis.
Using spatially explicit phylogeographic methods including novel Bayesian approaches has greatly advanced our understanding of Pleistocene range dynamics in a widespread Alpine plant species. Specifically, range dynamics of diploid S. carniolicus are characterized by remarkable range persistence in both peripheral (massifs de refuge) and interior refugia (nunataks), which likely acted as such recurrently through several glaciations cycles, and by independent range expansions out from these refugia in a manner determined mainly by geographic adjacency. This study emphasizes that our understanding of Pleistocene range dynamics even in a comparatively well-studied region like the Alps is still limited, negatively affecting identification of the most relevant patterns and processes. Future comparative studies will be necessary not only to identify common phylogeographic patterns with respect to, for instance, location of refugia and colonization routes (Carstens et al. 2004; Victoriano et al. 2008), but also to elucidate their underlying genetic and/or ecological causes (Álvarez et al. 2009).
Acknowledgments
Financial support by the Austrian Science Fund (FWF, project P20736-B16 to PS), the Commission for Interdisciplinary Ecological Studies (KIÖS; grant P2008-01 to GMS) and the Association for the Advancement of Plant Sciences (supporting RF) is gratefully acknowledged. We thank Marco Caccianiga, Božo Frajman, Christian Gilli, Manfred Schmucker and Daniela Stawik for help with collecting plant material, Philippe Lemey for help with the phylogeographic models in beast and three anonymous reviewers for their helpful comments.
Footnotes
P.E.G. is mainly interested in plant biodiversity and evolution in the Mediterranean Region, with focus on phylogeography. M.W. is interested in plant population dynamics, including population genetics and phylogeography. R.F. is interested in plant evolutionary biology, speciation and adaptation mechanisms and the related morphological divergence. M.S. is interested in ecological aspects of polyploid evolution, with a special focus on habitat segregation and reproductive isolation mechanisms. J.K. is interested in polyploid evolution and in application of flow cytometry in plant population studies. J.S. is interested in polyploid evolution, including patterns and processes in mixed-ploidy populations, and in multivariate morphometrics. K.H. is interested in metapopulation dynamics in alpine biomes. G.M.S. is interested in different aspects of plant evolution, including genome evolution of parasitic plants, phylogeography, polyploid evolution and speciation. P.S. is interested in polyploid evolution and in the spatio-temporal diversification of European alpine plants.
Data accessibility
DNA sequences have been deposited in GenBank under accession nos FR796701–FR797793 and nos HE614296–HE614583. Details regarding individual samples are available in Tables S1 and S2. Phylogenetic data (data matrices of ITS sequences and of coded plastid sequences, phylogenetic tree based on ITS sequences) are available at TreeBase (http://purl.org/phylo/treebase/phylows/study/TB2:S12125). AFLP data matrix, alignment of uncoded plastid sequences and exemplary beast input files are available at Dryad: doi: 10.5061/dryad.6v89fj76.
Supporting information
Additional supporting information may be found in the online version of this article.
Appendix S1 Details of the phylogeneticanalyses of the Incani clade.
Fig. S1 Phylogenetic relationships of Senecio carniolicus and related species (i.e., the Incani clade).
Fig. S2 Population dendrogram based on agglomerative hierarchical clustering using geographic distances.
Fig. S3 Principal Coordinate Analysis of AFLP data.
Fig. S4 Population graphs network of AFLP data at the population level.
Fig. S5 Bayes Factor support for diffusion rates in the discrete non-reversible geographic diffusion model under different parameterizations.
Table S1 Population numbers, sampling locations and their coordinates, number of analysed individuals, population diversity measures, ITS ribotypes, plastid haplotypes, and GenBank accession numbers
Table S2 Sampling localities and their coordinates as well as GenBank accession numbers of four plastid regions for outgroup species of the Incani clade
Table S3 Newly designed internal primers for three plastid spacer regions
Table S4 Sequence statistics
Table S5 Bayes Factor support for geographic diffusion rates under the reversible discrete model
Table S6 Bayes Factor support for geographic diffusion rates under the non-reversible discrete model
References
- Álvarez N, Thiel-Egenter C, Tribsch A, et al. History or ecology? Substrate type as a major driver of spatial genetic structure in Alpine plants. Ecology Letters. 2009;12:632–640. doi: 10.1111/j.1461-0248.2009.01312.x. [DOI] [PubMed] [Google Scholar]
- Bettin O, Cornejo C, Edwards PJ, Holderegger R. Phylogeography of the high alpine plant Senecio halleri (Asteraceae) in the European Alps: in situ glacial survival with postglacial stepwise dispersal into peripheral areas. Molecular Ecology. 2007;16:2517–2524. doi: 10.1111/j.1365-294X.2007.03273.x. [DOI] [PubMed] [Google Scholar]
- Blöch C, Weiss-Schneeweiss H, Schneeweiss GM, et al. Molecular phylogenetic analyses of nuclear and plastid DNA sequences support dysploid and polyploid chromosome number changes and reticulate evolution in the diversification of Melampodium (Millerieae, Asteraceae) Molecular Phylogenetics and Evolution. 2009;53:220–233. doi: 10.1016/j.ympev.2009.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonin A, Bellemain E, Bronken Eidesen P, et al. How to track and assess genotyping errors in population genetics studies. Molecular Ecology. 2004;13:3261–3273. doi: 10.1111/j.1365-294X.2004.02346.x. [DOI] [PubMed] [Google Scholar]
- Borsch T, Quandt D. Mutational dynamics and phylogenetic utility of noncoding chloroplast DNA. Plant Systematics and Evolution. 2009;282:169–199. [Google Scholar]
- Burnier J, Buerki S, Arrigo N, Küpfer P, Álvarez N. Genetic structure and evolution of Alpine polyploid complexes: Ranunculus kuepferi (Ranunculaceae) as a case study. Molecular Ecology. 2009;18:3730–3744. doi: 10.1111/j.1365-294X.2009.04281.x. [DOI] [PubMed] [Google Scholar]
- Carstens BC, Richards CL. Integrating coalescent and ecological niche modeling in comparative phylogeography. Evolution. 2007;61:1439–1454. doi: 10.1111/j.1558-5646.2007.00117.x. [DOI] [PubMed] [Google Scholar]
- Carstens BC, Stevenson AL, Degenhardt JD, Sullivan J. Testing nested phylogenetic and phylogeographic hypotheses in the Plethodon vandykei species group. Systematic Biology. 2004;53:781–792. doi: 10.1080/10635150490522296. [DOI] [PubMed] [Google Scholar]
- Chenevard P. Notes floristiques alpines. Bulletin de l’Herbier Boissier 2me série. 1906;6:365–370. [Google Scholar]
- Clement M, Posada D, Crandall KA. TCS: a computer program to estimate gene genealogies. Molecular Ecology. 2000;9:1657–1659. doi: 10.1046/j.1365-294x.2000.01020.x. [DOI] [PubMed] [Google Scholar]
- Corriveau J, Coleman A. Rapid screening method to detect potential biparental inheritance of plastid DNA and results for over 200 angiosperm species. American Journal of Botany. 1988;75:1443–1458. [Google Scholar]
- Currat M, Ruedi M, Petit RJ, Excoffier L. The hidden side of invasions: massive introgression by local genes. Evolution. 2008;62:1908–1920. doi: 10.1111/j.1558-5646.2008.00413.x. [DOI] [PubMed] [Google Scholar]
- Després L, Gielly L, Redoutet B, Taberlet P. Using AFLP to resolve phylogenetic relationships in a morphologically diversified plant species complex when nuclear and chloroplast sequences fail to reveal variability. Molecular Phylogenetics and Evolution. 2003;27:185–196. doi: 10.1016/s1055-7903(02)00445-1. [DOI] [PubMed] [Google Scholar]
- Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evolutionary Biology. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond A, Rambaut A, Shapiro B, Pybus O. Bayesian coalescent inference of past population dynamics from molecular sequences. Molecular Biology and Evolution. 2005;22:1185–1192. doi: 10.1093/molbev/msi103. [DOI] [PubMed] [Google Scholar]
- Dyer RJ, Nason JD. Population graphs: the graph theoretic shape of genetic structure. Molecular Ecology. 2004;13:1713–1727. doi: 10.1111/j.1365-294X.2004.02177.x. [DOI] [PubMed] [Google Scholar]
- Ehrich D. AFLPdat: a collection of R functions for convenient handling of AFLP data. Molecular Ecology Notes. 2006;6:603–604. [Google Scholar]
- Excoffier L, Laval G, Schneider S. Arlequin ver. 3.11: an integrated software package for population genetics data analysis. Evolutionary Bioinformatics Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: dominant markers and null alleles. Molecular Ecology Notes. 2007;7:574–578. doi: 10.1111/j.1471-8286.2007.01758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP-phylogeny inference package (version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- Flatscher R. Morphological Differentiation of the Sympatric Di-, Tetra- and Hexaploid Cytotypes of Senecio carniolicus s.l. (syn. Jacobaea carniolica, Asteraceae) Vienna: University of Vienna; 2010. Diploma Thesis. [Google Scholar]
- Gabrielsen TM, Bachmann K, Jakobsen KS, Brochmann C. Glacial survival does not matter: RAPD phylogeography of Nordic Saxifraga oppositifolia. Molecular Ecology. 1997;6:831–842. [Google Scholar]
- Garrick R, Nason J, Meadows C, Dyer R. Not just vicariance: phylogeography of a Sonoran Desert euphorb indicates a major role of range expansion along the Baja peninsula. Molecular Ecology. 2009;18:1916–1931. doi: 10.1111/j.1365-294X.2009.04148.x. [DOI] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series. 1999;41:95–98. [Google Scholar]
- Hargreaves S, Maxted N, Hirano R, et al. Islands as refugia of Trifolium repens genetic diversity. Conservation Genetics. 2010;11:1317–1326. [Google Scholar]
- Hess HE, Landolt E, Hirzel R. Flora der Schweiz und angrenzender Gebiete III. Basel: Birkhäuser; 1980. [Google Scholar]
- Hewitt GM. Post-glacial re-colonization of European biota. Biological Journal of the Linnean Society. 1999;68:87–112. [Google Scholar]
- Hewitt G. The genetic legacy of the Quaternary ice ages. Nature. 2000;405:907–913. doi: 10.1038/35016000. [DOI] [PubMed] [Google Scholar]
- Hewitt GM. Genetic consequences of climatic oscillations in the Quaternary. Philosophical Transactions of the Royal Society London B. 2004;359:183–195. doi: 10.1098/rstb.2003.1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holderegger R, Thiel-Egenter C, Parisod C. Marie Brockmann-Jerosch and her influence on Alpine phylogeography. Alpine Botany. 2011;121:5–10. [Google Scholar]
- Hülber K, Sonnleitner M, Flatscher R, et al. Ecological segregation drives fine-scale cytotype distribution of Senecio carniolicus in the Eastern Alps. Preslia. 2009;81:309–319. [PMC free article] [PubMed] [Google Scholar]
- Ingvarsson PK, Ribstein S, Taylor DR. Molecular evolution of insertions and deletion in the chloroplast genome of Silene. Molecular Biology and Evolution. 2003;20:1737–1740. doi: 10.1093/molbev/msg163. [DOI] [PubMed] [Google Scholar]
- Jakobsson M, Rosenberg NA. CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics. 2007;23:1801–1806. doi: 10.1093/bioinformatics/btm233. [DOI] [PubMed] [Google Scholar]
- Knowles LL. Statistical phylogeography. Annual Review of Ecology, Evolution, and Systematics. 2009;40:593–612. [Google Scholar]
- Knowles LL, Maddison WP. Statistical phylogeography. Molecular Ecology. 2002;11:2623–2635. doi: 10.1046/j.1365-294x.2002.01637.x. [DOI] [PubMed] [Google Scholar]
- Lemey P, Rambaut A, Drummond AJ, Suchard MA. Bayesian phylogeography finds its roots. PLoS Computational Biology. 2009;5:e1000520. doi: 10.1371/journal.pcbi.1000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemey P, Rambaut A, Welch JJ, Suchard MA. Phylogeography takes a relaxed random walk in continuous space and time. Molecular Biology and Evolution. 2010;27:1877–1885. doi: 10.1093/molbev/msq067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon AR, Lemmon EM. A likelihood framework for estimating phylogeographic history on a continuous landscape. Systematic Biology. 2008;57:544–561. doi: 10.1080/10635150802304761. [DOI] [PubMed] [Google Scholar]
- Lohse K, Nicholls JA, Stone GN. Inferring the colonization of a mountain range—refugia vs. nunatak survival in high alpine ground beetles. Molecular Ecology. 2011;20:394–408. doi: 10.1111/j.1365-294X.2010.04929.x. [DOI] [PubMed] [Google Scholar]
- Maechler M, Rousseeuw P, Struyf A, Hubert M. 2005. Cluster analysis basics and extensions. http://cran.r-project.org/web/packages/cluster/
- McKinnon GE, Vaillancourt RE, Steane DA, Potts BM. An AFLP marker approach to lower-level systematics in Eucalyptus (Myrtaceae) American Journal of Botany. 2008;95:368–380. doi: 10.3732/ajb.95.3.368. [DOI] [PubMed] [Google Scholar]
- Merxmüller H. Untersuchungen zur Sippengliederung und Arealbildung in den Alpen. I. Jahrbuch des Vereins zum Schutze der Alpenpflanzen und –tiere. 1952;17:96–133. [Google Scholar]
- Merxmüller H. Untersuchungen zur Sippengliederung und Arealbildung in den Alpen. II. Jahrbuch des Vereins zum Schutze der Alpenpflanzen und –tiere. 1953;18:138–158. [Google Scholar]
- Merxmüller H. Untersuchungen zur Sippengliederung und Arealbildung in den Alpen. III. Jahrbuch des Vereins zum Schutze der Alpenpflanzen und –tiere. 1954;19:97–139. [Google Scholar]
- Nei M. Genetic distance between populations. American Naturalist. 1972;106:283–292. [Google Scholar]
- Nieto Feliner G, Rosselló JA. Better the devil you know? Guidelines for insightful utilization of nrDNA ITS in species-level evolutionary studies in plants. Molecular Phylogenetics and Evolution. 2007;44:911–919. doi: 10.1016/j.ympev.2007.01.013. [DOI] [PubMed] [Google Scholar]
- Nordborg M, Hu TT, Ishino Y, et al. The pattern of polymorphism in Arabidopsis thaliana. PLoS Biology. 2005;3:e196. doi: 10.1371/journal.pbio.0030196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisod C, Besnard G. Glacial in situ survival in the Western Alps and polytopic autopolyploidy in Biscutella laevigata L. (Brassicaceae) Molecular Ecology. 2007;16:2755–2767. doi: 10.1111/j.1365-294X.2007.03315.x. [DOI] [PubMed] [Google Scholar]
- Paun O, Schönswetter P, Winkler M, Intrabiodiv Consortium. Tribsch A. Historical divergence vs. contemporary gene flow: evolutionary history of the calcicole Ranunculus alpestris group (Ranunculaceae) in the European Alps and the Carpathians. Molecular Ecology. 2008;17:4263–4275. doi: 10.1111/j.1365-294x.2008.03908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawłowski B. Remarques sur l’endémisme dans la flore des Alpes et des Carpates. Vegetatio. 1970;21:181–243. [Google Scholar]
- Pelser P, Gravendeel B, van der Meijden R. Phylogeny reconstruction in the gap between too little and too much divergence: the closest relatives of Senecio jacobaea (Asteraceae) according to DNA sequences and AFLPs. Molecular Phylogenetics and Evolution. 2003;29:613–628. doi: 10.1016/s1055-7903(03)00139-8. [DOI] [PubMed] [Google Scholar]
- Pelser PB, van den Hof K, Gravendeel B, van der Meijden R. The systematic value of morphological characters in Senecio sect. Jacobaea (Asteraceae) as compared to DNA sequences. Systematic Botany. 2004;29:790–805. [Google Scholar]
- Pelser PB, Veldkamp JF, van der Meijden R. New combinations in Jacobaea Mill. (Asteraceae–Senecioneae) Compositae Newsletter. 2006;44:1–11. [Google Scholar]
- Petit RJ, Duminil J, Fineschi S, et al. Comparative organization of chloroplast, mitochondrial and nuclear diversity in plant populations. Molecular Ecology. 2005;14:689–701. doi: 10.1111/j.1365-294X.2004.02410.x. [DOI] [PubMed] [Google Scholar]
- Posada D. jModelTest: phylogenetic model averaging. Molecular Biology and Evolution. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provan J, Bennett KD. Phylogeographic insights into cryptic glacial refugia. Trends in Ecology and Evolution. 2008;23:564–571. doi: 10.1016/j.tree.2008.06.010. [DOI] [PubMed] [Google Scholar]
- Rebernig CA, Schneeweiss GM, Bardy KE, et al. Multiple Pleistocene refugia and Holocene range expansion of an abundant southwestern American desert plant species (Melampodium leucanthum, Asteraceae) Molecular Ecology. 2010a;19:3421–3443. doi: 10.1111/j.1365-294X.2010.04754.x. [DOI] [PubMed] [Google Scholar]
- Rebernig CA, Weiss-Schneeweiss H, Schneeweiss GM, et al. Quaternary range dynamics and polyploid evolution in an arid brushland plant species (Melampodium cinereum, Asteraceae) Molecular Phylogenetics and Evolution. 2010b;54:594–606. doi: 10.1016/j.ympev.2009.10.010. [DOI] [PubMed] [Google Scholar]
- Reisigl H, Pitschmann H. Obere Grenzen von Flora und Vegetation in der Nivalstufe der zentralen Ötztaler Alpen (Tirol) Plant Ecology. 1958;8:93–129. [Google Scholar]
- Rohlf FJ. NTSYS-pc: Numerical Taxonomy and Multivariate Analysis System, Version 2.2. New York: Applied Biostatistics; 1997. [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Schneeweiss GM, Schönswetter P. The wide but disjunct range of the European mountain plant Androsace lactea L. (Primulaceae) reflects Late Pleistocene range fragmentation and post-glacial distributional stasis. Journal of Biogeography. 2010;37:2016–2025. [Google Scholar]
- Schneeweiss GM, Schönswetter P. A re-appraisal of nunatak survival in arctic-alpine phylogeography. Molecular Ecology. 2011;20:190–192. doi: 10.1111/j.1365-294x.2010.04927.x. [DOI] [PubMed] [Google Scholar]
- Schönswetter P, Tribsch A. Vicariance and dispersal in the alpine perennial Bupleurum stellatum L. (Apiaceae) Taxon. 2005;54:725–732. [Google Scholar]
- Schönswetter P, Tribsch A, Barfuss M, Niklfeld H. Several Pleistocene refugia detected in the high alpine plant Phyteuma globulariifolium Sternb. & Hoppe (Campanulaceae) in the European Alps. Molecular Ecology. 2002;11:2637–2647. doi: 10.1046/j.1365-294x.2002.01651.x. [DOI] [PubMed] [Google Scholar]
- Schönswetter P, Tribsch A, Schneeweiss GM, Niklfeld H. Disjunctions in relict alpine plants: phylogeography of Androsace brevis and A. wulfeniana (Primulaceae) Botanical Journal of the Linnean Society. 2003;141:437–446. [Google Scholar]
- Schönswetter P, Tribsch A, Stehlik I, Niklfeld H. Glacial history of high alpine Ranunculus glacialis (Ranunculaceae) in the European Alps in a comparative phylogeographical context. Biological Journal of the Linnean Society. 2004;81:183–195. [Google Scholar]
- Schönswetter P, Stehlik I, Holderegger R, Tribsch A. Molecular evidence for glacial refugia of mountain plants in the European Alps. Molecular Ecology. 2005;14:3547–3555. doi: 10.1111/j.1365-294X.2005.02683.x. [DOI] [PubMed] [Google Scholar]
- Schönswetter P, Lachmayer M, Lettner C, et al. Sympatric diploid and hexaploid cytotypes of Senecio carniolicus (Asteraceae) in the Eastern Alps are separated along an altitudinal gradient. Journal of Plant Research. 2007;120:721–725. doi: 10.1007/s10265-007-0108-x. [DOI] [PubMed] [Google Scholar]
- Schönswetter P, Solstad H, Escobar García P, Elven R. A combined molecular and morphological approach to the taxonomically intricate European mountain plant Papaver alpinum s.l. (Papaveraceae). Taxa or informal phylogeographical groups? Taxon. 2009;17:1326–1343. [Google Scholar]
- Shaw J, Lickey EB, Beck JT, et al. The tortoise and the hare II: relative utility of 21 noncoding chloroplast DNA sequences for phylogenetic analysis. American Journal of Botany. 2005;92:142–166. doi: 10.3732/ajb.92.1.142. [DOI] [PubMed] [Google Scholar]
- Shaw J, Lickey EB, Schilling EE, Small RL. Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in angiosperms: the tortoise and the hare III. American Journal of Botany. 2007;94:275–288. doi: 10.3732/ajb.94.3.275. [DOI] [PubMed] [Google Scholar]
- Smith CI, Pellmyr O, Althoff DM, et al. Pattern and timing of diversification in Yucca (Agavaceae): specialized pollination does not escalate rates of diversification. Proceedings of the Royal Society B: Biological Sciences. 2008;275:249–258. doi: 10.1098/rspb.2007.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnleitner M, Flatscher R, Escobar García P, et al. Distribution and habitat segregation on different spatial scales among diploid, tetraploid and hexaploid cytotypes of Senecio carniolicus (Asteraceae) in the Eastern Alps. Annals of Botany. 2010;106:967–978. doi: 10.1093/aob/mcq192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spellman GM, Klicka J. Testing hypotheses of Pleistocene population history using coalescent simulations: phylogeography of the pygmy nuthatch (Sitta pygmaea. Proceedings of the Royal Society B: Biological Sciences. 2006;273:3057–3063. doi: 10.1098/rspb.2006.3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehlik I. Nunataks and peripheral refugia for alpine plants during Quaternary glaciation in the middle part of the Alps. Botanica Helvetica. 2000;110:25–30. [Google Scholar]
- Stehlik I, Schneller JJ, Bachmann K. Resistance or emigration: response of the high-alpine plant Eritrichium nanum (L.) Gaudin to the ice age within the Central Alps. Molecular Ecology. 2001;10:357–370. doi: 10.1046/j.1365-294x.2001.01179.x. [DOI] [PubMed] [Google Scholar]
- Stehlik I, Blattner F, Holderegger R, Bachmann K. Nunatak survival of the high Alpine plant Eritrichium nanum (L.) Gaudin in the central Alps during the ice ages. Molecular Ecology. 2002;11:2027–2036. doi: 10.1046/j.1365-294x.2002.01595.x. [DOI] [PubMed] [Google Scholar]
- Stewart JR, Lister AM, Barnes I, Dalén L. Refugia revisited: individualistic responses of species in space and time. Proceedings of the Royal Society B: Biological Sciences. 2010;277:661–671. doi: 10.1098/rspb.2009.1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suda J, Weiss-Schneeweiss H, Tribsch A, et al. Complex distribution patterns of di-, tetra-, and hexaploid cytotypes in the European high mountain plant Senecio carniolicus (Asteraceae) American Journal of Botany. 2007;94:1391–1401. doi: 10.3732/ajb.94.8.1391. [DOI] [PubMed] [Google Scholar]
- Svenning JC, Normand S, Skov F. Postglacial dispersal limitation of widespread forest plant species in nemoral Europe. Ecography. 2008;31:316–326. [Google Scholar]
- Swofford DL. PAUP*: Phylogenetic Analysis Using Parsimony, Version 4.0 b10. Sunderland, Massachusetts: Sinauer; 2003. [Google Scholar]
- Taberlet P. Biodiversity at the intraspecific level: the comparative phylogeographic approach. Journal of Biotechnology. 1998;64:91–100. [Google Scholar]
- Tackenberg O, Stöcklin J. Wind dispersal of alpine plant species: a comparison with lowland species. Journal of Vegetation Science. 2008;19:109–118. [Google Scholar]
- Tajima F. Evolutionary relationship of DNA sequences in finite populations. Genetics. 1983;105:437–460. doi: 10.1093/genetics/105.2.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel-Egenter C, Holderegger R, Brodbeck S, Gugerli F. Concordant genetic breaks, identified by combining clustering and tessellation methods, in two co-distributed alpine plant species. Molecular Ecology. 2009;18:4495–4507. doi: 10.1111/j.1365-294X.2009.04360.x. [DOI] [PubMed] [Google Scholar]
- Tribsch A, Schönswetter P, Stuessy TF. Saponaria pumila (Caryophyllaceae) and the ice age in the European Alps. American Journal of Botany. 2002;89:2024–2033. doi: 10.3732/ajb.89.12.2024. [DOI] [PubMed] [Google Scholar]
- Vachon N, Freeland JR. Phylogeographic inferences from chloroplast DNA: quantifying the effects of mutations in repetitive and non-repetitive sequences. Molecular Ecology Resources. 2011;11:279–285. doi: 10.1111/j.1755-0998.2010.02921.x. [DOI] [PubMed] [Google Scholar]
- Victoriano PF, Ortiz JC, Benavides E, Adams BJ, Sites JW., Jr Comparative phylogeography of codistributed species of Chilean Liolaemus (Squamata: Tropiduridae) from the central-southern Andean range. Molecular Ecology. 2008;17:2397–2416. doi: 10.1111/j.1365-294X.2008.03741.x. [DOI] [PubMed] [Google Scholar]
- Vos P, Hogers R, Bleeker M, et al. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Research. 1995;23:4407–4414. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westergaard KB, Alsos IG, Popp M, et al. Glacial survival may matter after all: nunatak signatures in the rare European populations of two west-arctic species. Molecular Ecology. 2011;20:376–393. doi: 10.1111/j.1365-294X.2010.04928.x. [DOI] [PubMed] [Google Scholar]
- Yamane K, Yasui Y, Ohnishi O. Intraspecific cpDNA variations of diploid and tetraploid perennial buckwheat, Fagopyrum cymosum (Polygonaceae) American Journal of Botany. 2003;90:339–346. doi: 10.3732/ajb.90.3.339. [DOI] [PubMed] [Google Scholar]
- Zhou R, Shi S, Wu CI. Molecular criteria for determining new hybrid species – an application to the Sonneratia hybrids. Molecular Phylogenetics and Evolution. 2005;35:595–601. doi: 10.1016/j.ympev.2005.03.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
DNA sequences have been deposited in GenBank under accession nos FR796701–FR797793 and nos HE614296–HE614583. Details regarding individual samples are available in Tables S1 and S2. Phylogenetic data (data matrices of ITS sequences and of coded plastid sequences, phylogenetic tree based on ITS sequences) are available at TreeBase (http://purl.org/phylo/treebase/phylows/study/TB2:S12125). AFLP data matrix, alignment of uncoded plastid sequences and exemplary beast input files are available at Dryad: doi: 10.5061/dryad.6v89fj76.


