Abstract
Pax3, a key myogenic regulator, is transiently expressed during activation of adult muscle stem cells, or satellite cells (SCs), and is also expressed in a subset of quiescent SCs (QSCs), but only in specific muscles. The mechanisms regulating these variations in expression are not well understood. Here we show that Pax3 levels are regulated by miR-206, a miRNA with a previously demonstrated role in myogenic differentiation. In most QSCs and activated SCs, miR-206 expression suppresses Pax3 expression. Paradoxically, QSCs that express high levels of Pax3 also express high levels of miR-206. In these QSCs, Pax3 transcripts are subject to alternative polyadenylation, resulting in transcripts with shorter 3′ untranslated regions (3′UTRs) that render them resistant to regulation by miR-206. Similar alternate polyadenylation of the Pax3 transcript also occurs in myogenic progenitors during development. Our findings may reflect a general role of alternative polyadenylation in circumventing miRNA-mediated regulation of stem cell function.
Keywords: Pax3, miRNA-206, 3′UTR, myogenesis, satellite cell
INTRODUCTION
Pax3 is a key regulator of myogenesis during development. In Splotch (Sp) mice, which carry a spontaneous mutation in the Pax3 locus, limb muscles are absent (Bober et al., 1994; Goulding et al., 1994). The formation of these muscles requires Pax3 for the induction of expression of c-Met, a tyrosine kinase receptor essential for the delamination and migration of muscle progenitor cells (Bladt et al., 1995; Epstein et al., 1995; Yang et al., 1996). Postnatally, Pax3 has been shown to be transiently expressed during SC activation to promote proliferation and inhibit differentiation (Boutet et al., 2007; Boutet et al., 2010) of a highly proliferative intermediate progenitor cell population (Conboy and Rando, 2002). During the subsequent myogenic differentiation, Pax3 is down-regulated both at the protein level by monoubiquitination and proteasomal degradation (Boutet et al., 2007; Boutet et al., 2010) and at the RNA level by at least two microRNAs (miRNA) (Crist et al., 2009; Hirai et al., 2010). However, postnatal regenerative myogenesis appears to be normal in mice in which Pax3 is conditionally deleted in SCs (Lepper et al., 2009). Therefore, the contexts in which Pax3 may regulate adult muscle stem and progenitor functions remain to be elucidated.
Interestingly, SCs exhibit heterogeneity in terms of Pax3 expression in quiescence (Montarras et al., 2005; Relaix et al., 2006), a difference that cannot be explained by differences in protein stability (Boutet et al., 2010). Whereas virtually all quiescent SCs in mosthind limb muscles do not express Pax3, those in the diaphragm, ventral trunk muscles and body wall muscle (e.g. serratus caudalis dorsalis), specific hindlimb muscles (e.g. gracilis), and about 50% of the forelimb muscles do express Pax3 (Montarras et al., 2005; Relaix et al., 2006). Remarkably, whenPax3+ve SCs are engrafted into the tibialis anterior (TA) muscle where the resident SCs do not express Pax3, they retain Pax3 expression, suggesting that Pax3 expression may be cell-autonomous (Montarras et al., 2005). Proliferating progeny of Pax3+ve and Pax3−ve SCs behave similarly in in vitro assays of differentiation (Montarras et al., 2005; Relaix et al., 2006). Overall, the mechanisms that differentially regulate Pax3 expression in the different SC populations remain to be determined as does the functional significance of Pax3 expression in the quiescent state. Given the fact that the Pax3 transcript is expressed in limb SCs that do not express the Pax3 protein, posttrancriptional regulation likely accounts for some of the spatial and temporal heterogeneity of Pax3 expression.
Several muscle-specific miRNAs have been shown to be important post-transcriptional regulators of different aspects of the myogenic program (Chen et al., 2006; Rao et al., 2006; Sweetman et al., 2008). Analysis of the 3′UTR of the Pax3 gene revealed target sites for miRNA-206 (miR-206)(Goljanek-Whysall et al., 2011; Hirai et al., 2010) and miRNA-27b (miR-27b) (Crist et al., 2009). miR-206 is a skeletal muscle-specific miRNA in mice (Kim et al., 2006), and its expression is regulated by the myogenic regulatory factors MyoD, Myf5 and Myogenin (MyoG) (Rao et al., 2006; Sweetman et al., 2008). miR-206 promotes terminal differentiation of myogenic progenitors by inhibiting the expression of the p180 subunit of DNA polymerase α as well as Id1-3, and MyoR (Kim et al., 2006). Pax3 and Pax7 are targeted by miR-206and miR-1 during terminal differentiation of SCs -derived myoblasts to enforce the differentiation program (Chen et al., 2010; Hirai et al., 2010). Likewise, miR-206 and miR-1 target Pax3 to promote differentiation of embryonic myogenic progenitors during development (Goljanek -Whysall et al., 2011). To date, no miRNAs have been shown to regulate the early stages of activation of quiescent SCs.
In this report, we show that miR-206 is unexpectedly highly expressed in quiescent SCs where it regulates the expression of Pax3. We further show that quiescent SCs in different muscles differentially process the Pax3 transcript through alternative polyadenylation to yield transcripts with different 3′UTR lengths, rendering them differentially susceptible to miR-206 regulation. This results in markedly different levels of Pax3 protein expression and a clear functional change in muscle progenitor behaviour. Our results suggest that alternative polyadenylation is an important process involved not only in the modulation of miRNA regulation but also in the control of stem cell function and in the determination of stem cell heterogeneity.
RESULTS
miRNA-206 is highly expressed in adult muscle stem cells
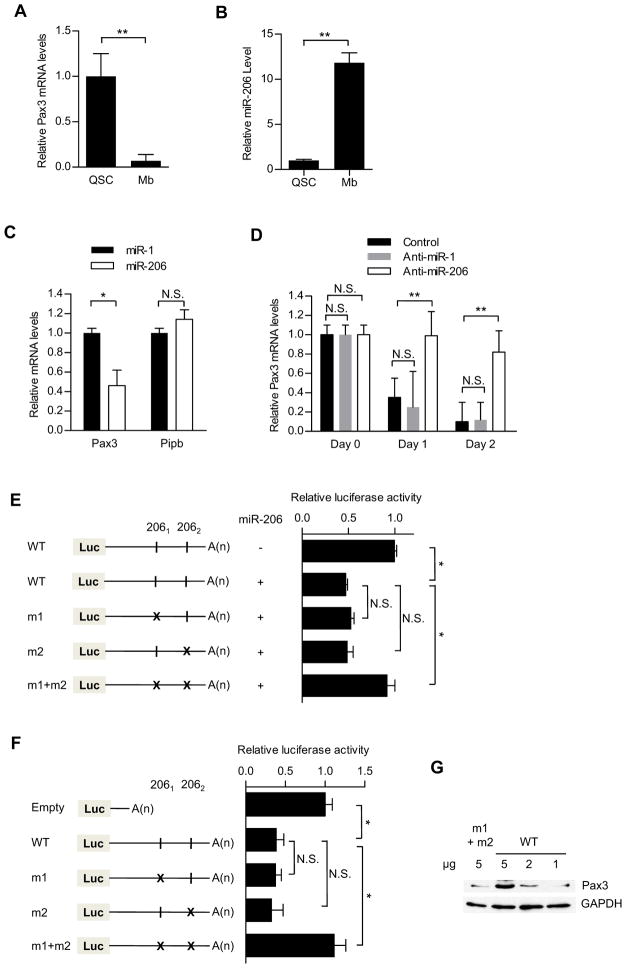
Pax3 transcript is present in QSCs from limb muscles, despite the absence of detectable protein (Figure 1A). Although there is a transient increase in Pax3 protein during SC activation (Conboy and Rando, 2002), both transcript and protein levels decline during the progression to fusion-competent myoblasts ((Boutet et al., 2007) and Figure 1A). Because of the discordance between Pax3 transcript and protein levels in limb QSCs, we hypothesized that Pax3 might be regulated post-transcriptionally by miRNAs in QSCs (Figure S1A). Indeed, we found that miR-206 is expressed at high levels in QSCs and increases even further during SC activation (Figure S1B, 1B). By contrast, miR-1 (which has the same seed sequence as miR206) and miR-27b are expressed at a much lower level in QSCs (Figure S1B). Thus, we explored the role of miR-206 in the regulation of Pax3 expression in QSCs and progenitor cells postnatally.
Figure 1. miR-206 is highly expressed in QSCs and regulates Pax3 transcript in SCs.
Quantitative RT-PCR analysis of Pax3 mRNA (A) or miR-206 (B) levels in QSCs sorted from uninjured muscle and from myoblasts (Mb) sorted from injured muscle 3.5 days after BaCl2 injection. (C) Quantitative analysis of mRNA levels of Pax3 and Cyclophilin B (Pipb) in primary myoblast cultures treated with miR -1 (black) or miR-206 (white) in growth medium. (D) Quantitative analysis of Pax3 mRNA in primary myoblast cultures treated with anti-miR-1 (black) or anti-miR-206 (white), then cultured in differentiation medium for 1 or 2 days.(E) Luciferase reporter assays showing the long form of Pax3 3′UTR repression by miR-206 in 293 cells. Luciferase constructs and miR-206-expressing plasmid were co-transfected in 293 cells, and luciferase activity was measured 48 hours post-transfection. Mutation of both target sites is necessary to abolish the repression of luciferase activity by miR-206 (m1+m2). (F) Luciferase reporter assays showing the long form of Pax3 3′UTR repression by miR-206 in C2C12 cells after differentiation. After transfection with luciferase constructs, C2C12 cells were cultured in differentiation medium for 48 hours to allow endogenous miR-206 upregulation. Mutation of both target sites is necessary to abolish the repression of luciferase activity by miR-206 (m1+m2). Pax3 murine long 3′UTR was appended to the luciferase ORF (Luc). The different luciferase constructs are indicated on the left of the graphs (WT, m1, m2, m1+m2). miR-206 complementary sites (2061and 2062) (vertical line) and mutated sites (cross) are indicated.(G) Competitive inhibition of miR-206 using the Pax3 3′UTR construct. Immunoblot analysis of Pax3 protein level in satellite cell-derived myoblasts, 48 hours after transfection with luciferase constructs containing either wild-type or mutated miR-206 target sites, then cultured in differentiation medium. Repression of Pax3 transcript by miR-206 was rescued by overexpressing wild-type Pax3 3′UTR construct, which acts as a competitive inhibitor (* p<0.05; ** p<0.001; N.S. - not significant; n=3). See also figure S1.
miRNA-206 regulates Pax3 in vitro
To test whether Pax3 is posttranscriptionally regulated by miR-206, we performed quantitative RT-PCR analysis in primary myoblasts transfected with miR-206. As a control we used miR-1, another miRNA expressed in differentiating myoblasts (Kim et al., 2006; Rao et al., 2006) that contains the same seed sequence as miR-206 (Figure S1C). Pax3 mRNA levels were reduced in cells treated with miR-206 but not in cells treated with miR-1 (Figure 1C). To test whether endogenous miR-206 controls Pax3 transcript levels, we assessed Pax3 levels in cells treated with anti-miR-206 during myogenic differentiation, when the levels of Pax3 transcript decline precipitously (Boutet et al., 2007). After 24 and 48 hours in differentiation medium, Pax3 mRNA levels were maintained at high levels in cells treated with anti-miR-206 but not with anti-miR-1 (Figures 1D, S1D). These results suggest that miR-206 is an endogenous regulator of Pax3 transcript levels. Consistent with previous studies in proliferative myoblasts (Chen et al., 2010), we found that Pax7 transcript levels, which are high in QSCs, were also reduced by miR-1 and miR-206 but, unlike Pax3, Pax7 was more susceptible to miR-1 than to miR-206 treatment (Figures S1E, S1F).
To analyze the functional roles of the two putative miR-206 target sites, we introduced the Pax3 3′UTR downstream of a luciferase reporter gene. We then generated Pax3 3′UTR constructs containing a mutation in either or both miR-206 target sites. To abolish miR-206 pairing, point mutations were introduced in the putative target sites at the bases corresponding to positions 2 and 4 of the miR-206 seed sequence (Figure S1C). In 293 cells, which do not express Pax3, miR-1, or miR-206, we co-transfected either miR-1 or miR-206 with each of the reporter constructs. miR-206, but not miR-1, reduced luciferase expression from the wild-type reporter and from reporter constructs bearing single miR-206 target site mutations with equal efficacy (Figures 1E, S1G). However, when miR-206 was co-transfected with the reporter bearing mutations in both target sites, the level of luciferase expression was identical to that in cells transfected with no miRNA. Using these constructs, we observed the same results in C2C12 cells undergoing differentiation (Figure 1F), when endogenous miR-206 is highly induced (Figure S1H). Taken together, these results suggest that Pax3 mRNA is targeted by miR-206 and that either of the two 3′UTR sites is sufficient to mediate the downregulation of Pax3 transcript.
To test further whether endogenous miR-206 regulates endogenous Pax3 protein levels, we reduced miR-206 in SC-derived myoblasts by transfecting constructs expressing the Pax3 3′UTR to sequester endogenous miR-206 (Ebert et al., 2007). Indeed, in the presence of this miR-206 “sponge”, Pax3 protein levels increased in a dose-dependent fashion (Figure 1G). By contrast, Pax3 protein levels did not change in cells transfected with a construct expressing the Pax3 3′UTR in which both miR-206 target sites had been mutated. These results demonstrate that Pax3 protein level is regulated posttranscriptionally by miR-206 in a dose-dependent manner.
miRNA-206 regulates Pax3-mediated functions in adult muscle stem cells ex vivo
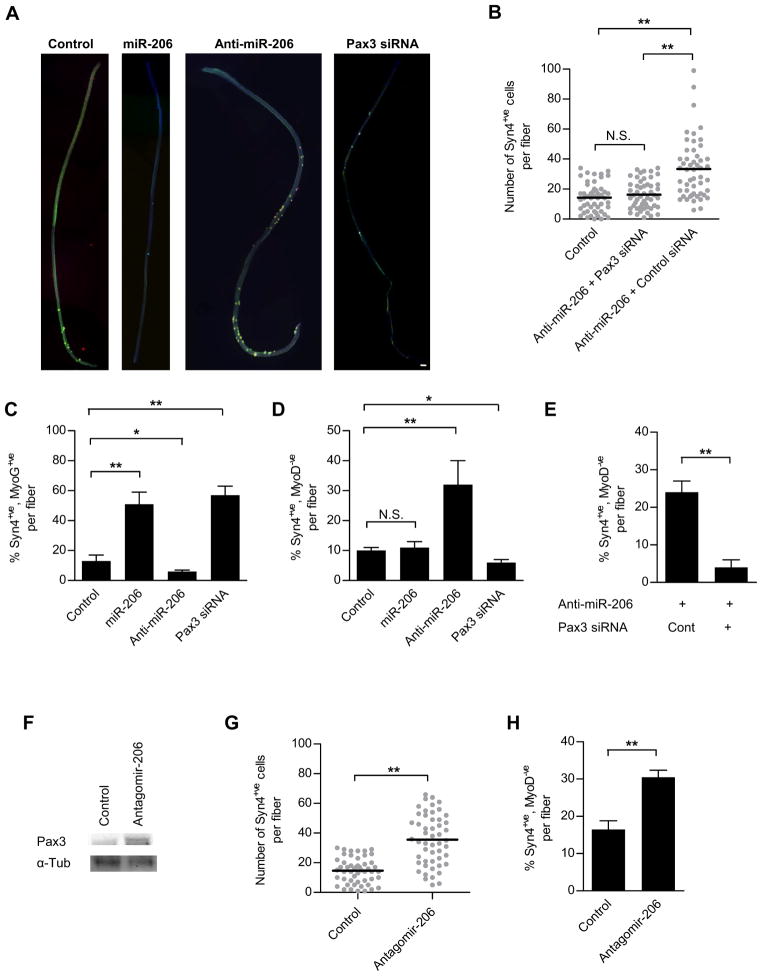
As previously reported by our group and others, Pax3 enhances proliferation and prevents differentiation of SCs (Boutet et al., 2007; Boutet et al., 2010; Crist et al., 2009). In order to assess the functional relevance of miR-206 regulation of Pax3, we modulated the level of miR-206 in SCs ex vivo. We transfected SCs associated with single fibers with miR-206, anti-miR-206, Pax3 siRNA or control miRNA (Figure 2A). To detect SCs on single fibers, we analyzed the expression of Syndecan4 which has been shown to be specific for QSCs in single fiber explants (Cornelison et al., 2001) and for activated SC progeny in regenerating muscle (LeGrand et al., 2009; Tanaka et al., 2009). SCs were transfected very efficiently as shown with Cy3-labeled oligonucleotides (Figure S2A). Compared to control, treatment of limb SCs with miR-206 resulted in a significantly reduced number of activated cells (Figure 2A; and see Figure 3D (left panel)). Conversely, transfection of SCs with anti-miR-206 resulted in an increase in the proliferative expansion of the population (Figure 2A, 3D (left panel)). The increased proliferation observed in fiber cultures treated with anti-miR-206 was abrogated by co-transfection with Pax3 siRNA (Figure 2B), suggesting that the regulation of cell proliferation by miR-206 occurs through the inhibition of Pax3. Although the proliferative expansion of SCs was inhibited by treatment with miR-206, it did not prevent SCs from breaking quiescence and entering the cell cycle, as evidenced by the incorporation of EdU, a process that was only enhanced by treatment with anti-miR-206 (Figure S2B). Given the fact that Pax7 is targeted by miR-206 to regulate myoblast differentiation (Chen et al., 2010), we tested whether Pax7 was contributing to the regulation of SC activation and proliferative expansion. As such, we transfected SCs associated with single fibers with Pax7 siRNA. Compared to control, treatment of limb SCs with Pax7 siRNA did not result in any significant change in SC activation and proliferation (Figure S2C, S2D). These results suggest that miR-206 controls proliferation during SC activation by regulating Pax3 levels and that Pax7, although a target of miR-206, does not play a major role in these processes.
Figure 2. miR-206 regulates Pax3-mediated proliferation and myogenic lineage progression during adult myogenesis ex vivo and in vivo.
(A) Low magnification (10x) images of single fibers treated with control miRNA, anti-miR-206, miR-206 or Pax3 siRNA, then stained for Syn4 (green) and DAPI (blue) 3 days after isolation (Bar: 40 μm). (B) Number ofSyn4+ve cells counted on single fiber explants cultured for 3 days. Fibers were treated with control, anti-miR-206 with control siRNA, or anti-miR-206 with Pax3 siRNA. Quantitative analysis of MyoG (C) or MyoD (D) expression in Syn4+ve SCs per fiber in single fiber explants treated with control miRNA, miR-206, anti-miR-206, or Pax3 siRNA and cultured for 3 days. (E) Quantitative analysis of MyoD expression inSyn4+ve SCs per fiber in single fiber explants treated with anti -miR-206 either with Control or Pax3 siRNA. (F) Western blot analysis of QSCs from limb muscle (except extensor digitorum longus (EDL)) in mice injected with control antagomirs (Control) or a nti-miR-206 antagomirs (Antagomir-206). (G) Number of Syn4+ve cells counted on EDL single fiber explants from mice injected with control antagomir (Control) or anti-miR-206 antagomir (Antagomir -206) and cultured for 3 days. (H) Quantitative analysis of MyoD expression in Syn4+ve SCs per fiber on single fiber explants (EDL) from mice injected with control antagomirs (Control) or anti-miR-206 antagomirs (Antagomir -206) and cultured for 3 days (Line indicates mean; * p<0.01; ** p<0.0001;N.S. -not significant; n=51). See also Figure S2.
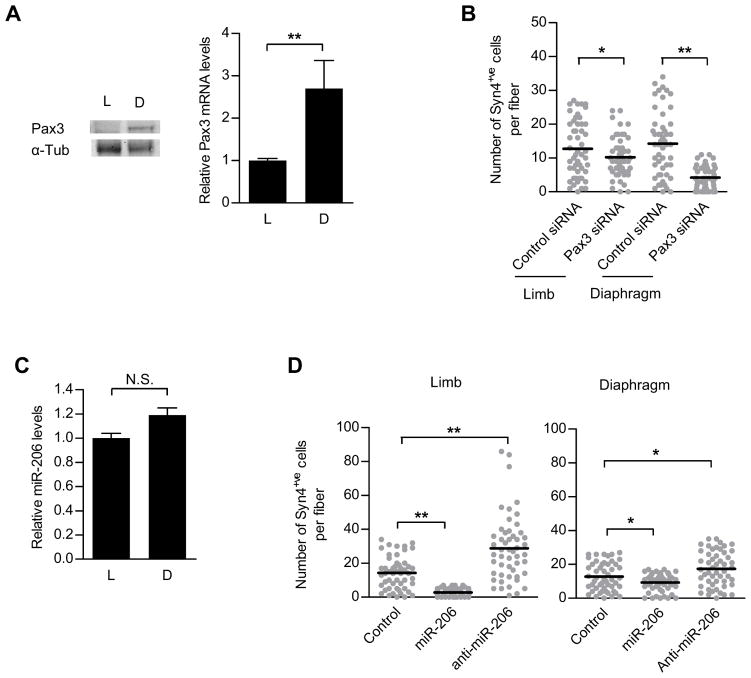
Figure 3. Pax3 mRNA is not susceptible to miR-206 regulation in diaphragm SCs.
(A) Western blot analysis of Pax3 protein level (left) and quantitative RT-PCR analysis of Pax3 mRNA level (right) in QSCs from limb (L) and diaphragm (D) muscles (n=3). (B) Number of Syn4+ve cells counted on single fiber explants from limb and from diaphragm cultured for 3 days. Fibers were treated with control or Pax3 siRNA (Line indicates mean, n=51). (C) Quantitative RT-PCR analysis of miR-206 levelin QSCs from limb (L) and diaphragm (D) muscles (n=3). (D) Number of Syn4+ve cells counted on single fiber explants from limb (EDL) and from diaphragm cultured for 3 days. Fibers were treated with control, miR-206 or anti-miR-206 (Line indicates mean;*p<0.05, ** p<0.0001; n=51). See also Figure S3.
Overexpression of miR-206 also significantly increased the proportion of cells undergoing myogenic lineage progression and differentiation as determined by high levels of MyoD or Myogenin (MyoG) expression, respectively, whereas treatment with anti-miR-206 had the opposite effects (Figures 2C, 2D, S2E, S2F). Maintenance of cells in an undifferentiated state (MyoD−ve/MyoG−ve) by anti-miR-206 was mediated by sustained Pax3 level since the effect could be abolished by co-transfection of Pax3 siRNA with anti-miR-206 (Figure 2E). Together, these results suggest that miR-206 strongly represses Pax3 in limb SCs, limits its ability to enhance proliferation, and delays myogenic differentiation in adult muscle stem cells.
Downregulation of miR-206 increases Pax3 protein and Pax3-mediated functions in limb SCs in vivo
To test for the regulation of Pax3 by miR-206 in limb SCs in vivo, we administered miR-206 antagomirs to 8-week-old mice by tail vein injection. We then assessed Pax3 protein expression in purified limb SCs and the proliferative expansion comparing SCs from EDL muscles of antagomir-treated and control mice. After a single miR-206 antagomir injection, SCs were purified by FACS and subject to western blot analysis. Higher levels of Pax3 protein were detected in SCs from antagomir-treated mice, suggesting miR-206 suppresses endogenous Pax3 protein level in QSCs (Figure 2F). When SC proliferation from antagomir-treated mice was analyzed ex vivo, more SCs were found on single fibers 3 days after isolation (Figure 2G), although no significant difference in SC was observed in freshly isolated fibers at day 0. This was also associated with an inhibition of myogenic commitment and differentiation in the population (Figure 2H). Therefore, down-regulation of miR-206 by anti-miR-206 treatment in vivo leads to increased Pax3 protein in QSCs and subsequent increased Pax3-mediated progenitor proliferation.
Pax3 transcripts are not susceptible to miRNA-206 in diaphragm SCs
These data indicate that miR-206 is a physiological regulator of Pax3 expression. As such, we hypothesized that different expression patterns of miR-206 in other myogenic stem cells or progenitors would account for patterns of Pax3 expression that differ from those in limb muscle SCs. We therefore examined the relationship between Pax3 and miR-206 expression in two distinct populations of adult SCs – those that express Pax3 protein in the quiescent state, such as SCs in the diaphragm muscle, and those that do not, such as SCs in nearly all limb muscles (Montarras et al., 2005; Relaix et al., 2006). Indeed, we found that Pax3 transcript and protein levels were much greater in SCs from diaphragm compared to limb SCs (Figure 3A). Consistent with this observation, knockdown of Pax3 in diaphragm SCs inhibited proliferative expansion much more than in limb SCs (Figure 3B). However, although we predicted that the higher level of Pax3 transcript would be associated with a lower level of miR-206 expression in SCs of the diaphragm compared to the limb, we were surprised to discover that miR-206 was expressed in similar levels in SCs from both muscles (Figure 3C). To test whether miR-206 regulates Pax3 levels in diaphragm SCs as it does in limb SCs, we compared proliferative expansion of limb and diaphragm SCs treated with miR-206 and anti-miR-206. Surprisingly, unlike in limb SCs, increasing or decreasing miR-206 levels had a negligible effect on Pax3-mediated proliferation of SCs from the diaphragm (Figure 3D). Likewise, miR-206 had less effect on SCs from the diaphragm than those from limb muscles in terms of inhibiting differentiation (Figures S3A–S3D; compare to Figures 2C, 2D). Thus, Pax3 transcript in diaphragm SCs appeared not to be subject to regulation by miR-206.
Alternative polyadenylation allows Pax3 transcripts to escape miR-206 targeting in diaphragm muscle stem cells
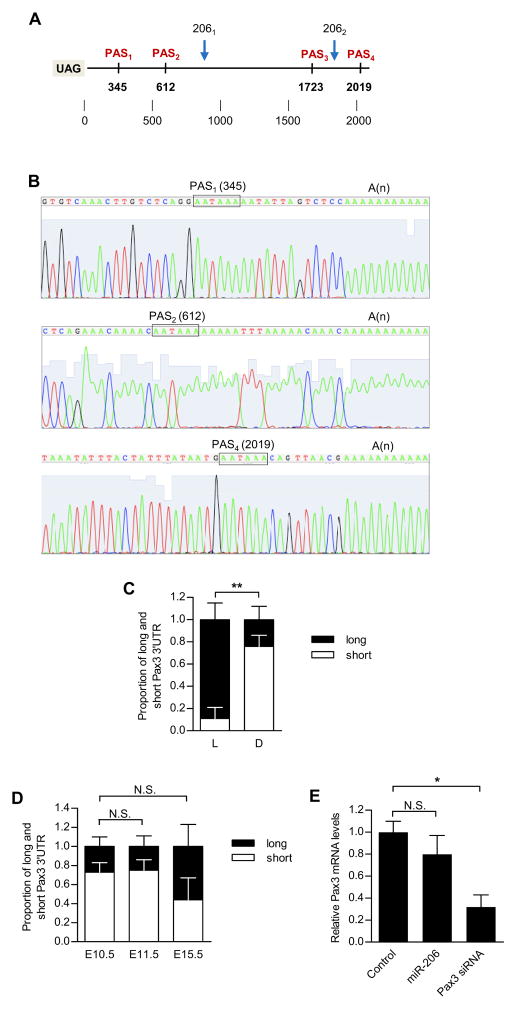
To address this conundrum of the apparent lack of regulation of Pax3 by miR-206 in diaphragm SCs, we examined the Pax3 3′UTR in more detail. Interestingly, four putative polyadenylation signals (PAS) were identified at positions 345, 612, 1723 and 2019 in the 3′UTR downstream from the stop codon. Importantly, the Pax3 transcript could contain both or neither of the miR-206 target sites depending on the selection of PAS (Figure 4A). We performed 3′ rapid amplification of cDNA ends (RACE) and confirmed that three PAS were functional (Figure 4B). The longest form (PAS at position 2019) contained both miR-206 target sites whereas the shorter forms (PAS at position 345 or 612) contained neither (Figure 4A). As such, the long form would be susceptible to miR-206 regulation whereas the short forms would be resistant. We performed quantitative RT-PCR using Taqman probes specific to the longest and shortest forms of the 3′UTR to determine their relative abundance (Figures S4A, S4B). In limb SCs, the long form of the Pax3 transcript, susceptible to miR-206 regulation, was the predominant form expressed (Figure 4C). By contrast, the short form that lacks the miRNA-206 sites was abundantly expressed in diaphragm SCs (Figure 4C). Therefore, these data suggested that diaphragm SCs are able to express Pax3 at high levels even in the presence of high levels of miR-206 because the form of the Pax3 transcript that is expressed is not susceptible to regulation by miR-206 by virtue of choice of PAS, resulting in 3′UTR without any known miRNA target sites.
Figure 4. Differential polyadenylation of Pax3 mRNA 3′UTR in limb and diaphragm QSCs and in embryonic limb progenitors.
(A) Graphical representation indicating the positions of the putative alternative polyadenylation sites (PAS1, PAS2, PAS3, PAS4) and putative miR-206 targeted sites (2061, 2062) in the Pax3 3′UTR.(B) DNA sequencing of 3′UTRs of three different isoforms of Pax3 detected by 3′RACE. PAS consensus sequence (PAS) and polyadenylation tail region (A(n)) are indicated. (C) Quantitative RT-PCR analysis of Pax3 transcripts bearing short and long 3′UTRs in limb (L) and in diaphragm (D) QSCs. (D) Ratios of long and short Pax3 mRNA isoforms in limb buds at E10.5 and E11.5. In (C) and (D), the proportions of the short form in L and D were compared. (E) Quantitative RT-PCR analysis of Pax3 mRNA levels in E10.5 myogenic progenitors treated with control or miR-206. Pax3 siRNA was used as a positive control (**p<0.001; N.S. - not significant; n=3). See also Figure S4.
Alternative polyadenylation allows Pax3 transcripts to escape miR-206 targeting in embryonic progenitors
We hypothesized that early embryogenesis might represent another context in which the regulation of Pax3 by miR-206 in myogenic progenitors is determined by 3′UTR length. Between E10.5 and E15.5, there is only a gradual decline in Pax3 expression in myogenic progenitors, particularly with high sustained levels through E10.5 and E11.5 (Figure S4C), despite an exponential increase in the expression of miR-206 (Figure S4D). As such, we analyzed the relative abundance of short and long forms of Pax3 3′UTR at these different stages. Indeed, the short form was the predominant form expressed at E10.5 and E11.5, representing ~80% of the total at both time points and still representing ~50% of the total at E15.5 (Figure 4D). Thus, in embryonic progenitors as in diaphragm SCs, the expression of Pax3 with a short 3′UTR allowed for persistently high levels of Pax3 protein expression despite exponential increases in miR-206 expression (Figure 3A). To test this directly, we transfected miR-206 into E10.5 progenitors and analyzed the levels of Pax3 expression. As with SCs from diaphragm, the expression of Pax3 in embryonic progenitors was negligibly affected by high levels of miR-206, whereas Pax3 siRNA knocked down Pax3 transcript to 30% of control levels (Figure 4E). Taken together, our data indicate that miR-206 targets and down-regulates the Pax3 transcript in adult SCs from limb muscles, but has negligible effects on embryonic muscle progenitors as well as SCs from the diaphragm.
DISCUSSION
Our results provide a molecular mechanism underlying the heterogeneity of satellite cells with regard to Pax3 expression in the quiescent state. SC heterogeneity has been described based on functional characterizations including developmental origin, functional characteristics, and properties of associated myofibers (Biressi and Rando, 2010). Our studies shed light on the peculiar heterogeneity of SCs in terms of Pax3 expression, which does not follow any known physiological, structural or developmental pattern. Pax3+ve SCs are observed in hindlimb gracilis muscle, in about 50% forelimb muscles, in subsets of the trunk muscles, and in the diaphragm (Relaix et al., 2006). Interestingly, Pax3 expression is preserved when Pax3+ve SCs are transplanted into Pax3−ve limb muscle, suggesting that Pax3 expression is cell-autonomous (Montarras et al., 2005). Our data present the first evidence of a molecular mechanism, based on the propensity of certain SCs to produce different Pax3 transcripts through alternative polyadenylation (APA), underlying any functional heterogeneity. One interesting aspect of our data is the finding that different ratios of Pax3 transcript variants with regard to 3′UTR length are found in different muscle precursor populations. Clarifying whether the observation of different ratios of Pax3 isoforms is due to molecular heterogeneity in individual cells or cellular heterogeneity within the population (with molecular homogeneity within individual cells) will necessarily await analysis of Pax3 isoforms on a single cell level.
Almost all eukaryotic mRNAs are polyadenylated through a process that involves cleavage at a specific site followed by the synthesis of a polyA tail (Lutz, 2008; Maniatis and Reed, 2002), and more than half of the mammalian genes generate transcripts that are subjected to APA (Tian et al., 2005). While several studies have examined polyadenylation signals and patterns, the molecular mechanisms involved in the choice of polyadenylation sites are poorly understood (Licatalosi and Darnell, 2010; Lutz, 2008; Millevoi and Vagner, 2010; Zhao et al., 1999). Three types of APA (I, II, and III) have been defined with regard to the location of the different PAS and whether the APA is coupled with an alternative splicing (Lutz, 2008; Millevoi and Vagner, 2010; Zhao et al., 1999; Edwalds-Gilbert et al., 1997). In human and mouse cells, Pax3 transcripts display a complex pattern of alternative splicing with at least eight different transcript variants reported (Barber et al., 1999; Pritchard et al., 2003; Wang et al., 2006; Parker et al., 2004). Considering the complex alternative splicing of Pax3 transcripts, it is likely that Pax3 APA falls into the type III category in which splicing events affect the 3′ end processing of Pax3 mRNA.
The importance of APA for regulation of protein expression, and the fact that APA may also be coordinated with alternative promoter choice (Costessi et al., 2006; Winter et al., 2007), raises fundamental questions about the extrapolation of endogenous protein expression from transgenic reporter mice in which endogenous PAS selection, splicing, and promoter choice may be altered at the genomic level. For Pax3 reporter mice, the temporal and spatial expression of the reporter exhibits both concordant and discordant patterns compared with those reported for the endogenous Pax3 protein (Boutet et al., 2007; Boutet et al., 2010; Montarras et al., 2005; Relaix et al., 2006) including the results reported here. Therefore, extrapolating the endogenous gene expression levels, especially when multiple isoforms exist with regard to different promoters and different 3′ UTRs, from studies of knock-in mice is risky because of the possibility of disruption of regulatory mechanisms associated with the genetic recombination associated with the knock-in.
Our studies demonstrate that APA can lead to transcripts with different 3′UTR lengths even within a specific population of quiescent stem cells. APA leading to longer or shorter 3′UTR lengths has recently been described as a more global phenomenon both in terms of transcripts affected (i.e. at the transcriptome in general, not specific transcripts) and in terms of biological context (e.g. during development, in cancer, and in response to induction of proliferation) (Ji et al., 2009; Mayr and Bartel, 2009; Sandberg et al., 2008). In these cases, specific transcripts were not analyzed to assess the functional relevance of global shortening or lengthening of 3′UTRs, but it was postulated that such changes could affect multiple aspects of transcript function including localization, stability, and translation, some of which could be due to changes in miRNA targeting as we have shown specifically here for Pax3 transcripts in quiescent SCs. Specific examples of APA leading to transcripts of specific genes with different 3′UTR lengths, rendering them more or less susceptible to regulation by miRNAs, are just beginning to be identified. In studies of the regulation of the Hsp70.3 gene in the setting of cardiac ischemia, it was found that ischemic preconditioning leads to APA of Hsp70.3 transcript resulting in a transcript with a shorter 3′UTR and lacking a miR-378 target site (albeit while still retaining a miR-711 target site) (Tranter et al., 2011). The resulting transcript may allow for higher levels of Hsp70.3 protein expression to promote protection from ischemic damage. In the current study, we present evidence of APA of a specific gene leading to shortened transcripts lacking any known miRNA binding site and leading to functional changes in stem cells that are determined specifically by PAS choice.
Studies of the regulation of Pax3 by miRNAs have yielded divergent conclusions. Hirai et al. showed that miR-206, a direct target of MyoD, controls the level of Pax3 expression to promote myoblast survival (Hirai et al., 2010). Our study shows that while miR-206 is significantly upregulated during myogenic differentiation, it is also expressed at high levels in specific populations of QSCs as well as in SC progeny and therefore plays a significant role in SC activation prior to the onset of differentiation. miR-1 has been shown to downregulate Pax3 expression in myoblasts and glioma cells (Goljanek-Whysall et al., 2011; Hirai et al., 2010). However, neither we nor Crist et al. (Crist et al., 2009) detected a downregulation of Pax3 mRNA when miR-1 was overexpressed in SCs, suggesting that the regulation of Pax3 expression by miR-1 may be limited to specific cell types and stages of the myogenic program. Clearly, the quiescent state represents a unique cell stage in which regulation by miRNAs may be distinct from the regulation as a cell is undergoing terminal differentiation. In that context, we have recently identified a miRNA, miR-489, that is important for maintenance of SC quiescence (Cheung et al., 2011).
Regulation of Pax3 expression by miRNAs has also been studied in the context of development. Myogenic progenitors during embryonic development express high level of miR-27b that also targets Pax3 (Crist et al., 2009). Overexpression of miR-27b led to the down-regulation of Pax3 and suppression of Pax3-mediated migration and proliferation, resulting in premature differentiation. Similarly, miR-206 was shown to regulate Pax3 functions in the somite of the developing embryos and was proposed to be required to stabilize myoblast commitment and subsequent differentiation (Goljanek-Whysall et al., 2011). Interestingly, overexpression of miR-27b was able to reduce Pax3 expression by only about 50% (Crist et al., 2009), consistent with our data that the short Pax3 isoform lacking both miR-206 and miR-27b target sites represents 70% of the total Pax3 transcripts in myogenic progenitors during this stage of development. Our data highlights the importance of understanding the processing of transcript 3′UTRs to be able to understand both qualitatively and quantitatively the regulation of that transcript by miRNAs. Certainly the differences that underlie the variations in PAS choice and the resulting effect on miRNA-mediated regulation of expression of a key myogenic lineage gene suggests that the post-transcriptional regulation of Pax3, even in the quiescent state, is part of an important regulatory network.
EXPERIMENTAL PROCEDURES
Mouse lines
C57BL6 mice were maintained in accordance with an approved Institutional Animal Care and Use Committee protocol at the Veterinary Medical Unit guidelines at the VA Palo Alto Health Care Systems.
Satellite cell isolation
Hindlimb muscles or diaphragms were dissected, digested with collagenase and dispase and then triturated. Mononucleated cells were stained with anti-VCAM, anti-CD31, anti-CD45 and anti-Sca-1 antibodies (BD-Pharmingen) and sorted using a BD-FACS Aria II. To obtain activated satellite cells, tibialis anterior muscles were injured with BaCl2 before cell isolation.
Primary myoblast cultures and single fiber isolation
Primary myoblast cultures were isolated and maintained as previously described (Quach and Rando, 2006). Single myofibers from extensor digitorum longus muscles or diaphragms were prepared as previously described (Rosenblatt et al., 1995).
Quantitative RT-PCR and analysis of alternative polyadenylation sites
Total RNA from SCs and embryonic tissues was extracted using Trizol reagent (Invitrogen). For expression analysis, total RNA was reverse transcribed and qPCR was carried out on a LightCycler 480 system (Roche) using Taqman probes (Applied Biosystems). Relative quantification of gene expression were carried out using the comparative CT method (Pfaffl, 2001). For 3′UTR quantification, reverse transcription was carried out with specific oligos for short and long form UTRs. Absolute quantification of gene expression was performed using standard curves to establish the amounts of long transcript and total transcript. The amount of short transcript was obtained by substraction. For the identification of alternative polyadenylation sites, total RNA was analyzed with SMARTer RACE cDNA amplification kit (Clontech). Amplified fragments were subcloned into pGEM-T-Easy (Promega) and sequenced.
cDNA cloning and constructs
Total RNA was extracted from primary myoblasts using TRIzol (Invitrogen). Pax3 3′UTR cDNA was generated with Qiagen one-step RT-PCR Kit (Qiagen), subcloned into pGEM-T-Easy (Promega), sequenced and subcloned into pMIR-REPORT vector (Applied Biosystems). To disrupt miR-206 binding sites, mutations were generated using Quickchange PCR directed mutagenesis Kit (Stratagene).
miRNA and siRNA transfection of primary myoblasts and single fibers
Primary myoblasts or single fibers were transfected with 50 pmol of miRNA, siRNA or anti-miRNA. Primary cells were switched to differentiation medium and harvested after 0, 24 or 48 hours while fibers were cultured for 72 hours then fixed.
Immunofluorescence and quantitative microscopy
Single fibers were fixed, permeabilized, blocked and then stained with anti -MyoD antibody (Dako) or anti-MyoG antibody (BD-Pharmingen), and anti-Syndecan4 antibody and 4′-6-diamidino-2-phenylindole (DAPI). Imaging was performed with a Zeiss Observer Z1 fluorescent microscope (Carl Zeiss). Maximum pixel intensity in each cell was subtracted with the pixel intensity on the supporting fiber. Threshold for low and high intensity was defined for each cell.
Luciferase assays
HEK293 cells were transfected with 3′UTR luciferase, control Renilla and miRNA expression vectors and cultured for 24 hours. To study the effects of endogenous miRNA on Pax3 3′UTR, C2C12 cells were transfected with 3′UTR luciferase and control Renilla vectors using Lipofectamine 2000(Invitrogen), and then cultured in differentiation medium. Cells were lysed and luciferase activity was quantified with Dual Luciferase Assay System (Promega).
Limb explants culture and transfection
Limb buds from E10.5 and E11.5 mouse embryos were placed in culture medium, transfected with miR-206, Pax3 or control siRNA, then lysed in TRIzol (Invitrogen).
In vivo antagomir treatment
Synthetic anti-miR-206 and mutant anti-miR-206 antagomirs (ThermoFisher Scientific) were administered by tail vein injection in CD1 mice. Hindlimb muscles were harvested 4.5 days after injection for single fiber or satellite cell isolation.
Supplementary Material
Acknowledgments
We thank Dr. B. Olwin for generously providing the chicken anti-Syndecan4 antibody. This work was supported by a Muscular Dystrophy Association Development Grant to S.C.B., and by grants from the Glenn Foundation for Medical Research, the NIH (P01 AG036695, R01 AG023806 (R37MERIT Award), R01 AR056849, R01 AR062185, and DP1 OD000392 (an NIH Director’s Pioneer Award)), and the Department of Veterans Affairs (Merit Review) to T.A.R.
Footnotes
Supplemental information for this article includes Figures S1-S4, experimental procedures and supplemental references, and can be found with this article online at (weblink).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barber TD, Barber MC, Cloutier TE, Friedman TB. PAX3 gene structure, alternative splicing and evolution. Gene. 1999;237:311–319. doi: 10.1016/s0378-1119(99)00339-x. [DOI] [PubMed] [Google Scholar]
- Biressi S, Rando TA. Heterogeneity in the muscle satellite cell population. Semin Cell Dev Biol. 2010;21:845–854. doi: 10.1016/j.semcdb.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bladt F, Riethmacher D, Isenmann S, Aguzzi A, Birchmeier C. Essential role for the c-met receptor in the migration of myogenic precursor cells into the limb bud. Nature. 1995;376:768–771. doi: 10.1038/376768a0. [DOI] [PubMed] [Google Scholar]
- Bober E, Franz T, Arnold HH, Gruss P, Tremblay P. Pax-3 is required for the development of limb muscles: a possible role for the migration of dermomyotomal muscle progenitor cells. Development. 1994;120:603–612. doi: 10.1242/dev.120.3.603. [DOI] [PubMed] [Google Scholar]
- Boutet SC, Biressi S, Iori K, Natu V, Rando TA. Taf1 regulates Pax3 protein by monoubiquitination in skeletal muscle progenitors. Mol Cell. 2010;40:749–761. doi: 10.1016/j.molcel.2010.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutet SC, Disatnik MH, Chan LS, Iori K, Rando TA. Regulation of Pax3 by proteasomal degradation of monoubiquitinated protein in skeletal muscle progenitors. Cell. 2007;130:349–362. doi: 10.1016/j.cell.2007.05.044. [DOI] [PubMed] [Google Scholar]
- Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, Conlon FL, Wang DZ. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet. 2006;38:228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JF, Tao Y, Li J, Deng Z, Yan Z, Xiao X, Wang DZ. microRNA-1 and microRNA-206 regulate skeletal muscle satellite cell proliferation and differentiation by repressing Pax7. J Cell Biol. 2010;190:867–879. doi: 10.1083/jcb.200911036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung TH, Quach NL, Charville GW, Liu L, Park L, Edalati A, Yoo B, Hoang P, Rando TA. Maintenance of muscle stem cell quiescence by microRNA-489. 2011 doi: 10.1038/nature10834. in preparation. Ref Type: Generic. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conboy IM, Rando TA. The regulation of Notch signaling controls satellite cell activation and cell fate determination in postnatal myogenesis. Dev Cell. 2002;3:397–409. doi: 10.1016/s1534-5807(02)00254-x. [DOI] [PubMed] [Google Scholar]
- Cornelison DD, Filla MS, Stanley HM, Rapraeger AC, Olwin BB. Syndecan-3 and syndecan-4 specifically mark skeletal muscle satellite cells and are implicated in satellite cell maintenance and muscle regeneration. Dev Biol. 2001;239:79–94. doi: 10.1006/dbio.2001.0416. [DOI] [PubMed] [Google Scholar]
- Costessi L, Devescovi G, Baralle FE, Muro AF. Brain-specific promoter and polyadenylation sites of the beta-adducin pre-mRNA generate an unusually long 3′-UTR. Nucleic Acids Res. 2006;34:243–253. doi: 10.1093/nar/gkj425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crist CG, Montarras D, Pallafacchina G, Rocancourt D, Cumano A, Conway SJ, Buckingham M. Muscle stem cell behavior is modified by microRNA-27 regulation of Pax3 expression. Proc Natl Acad Sci U S A. 2009;106:13383–13387. doi: 10.1073/pnas.0900210106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods. 2007;4:721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwalds-Gilbert G, Veraldi KL, Milcarek C. Alternative poly (A) site selection in complex transcription units: means to an end? Nucleic Acids Res. 1997;25:2547–2561. doi: 10.1093/nar/25.13.2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein JA, Lam P, Jepeal L, Maas RL, Shapiro DN. Pax3 inhibits myogenic differentiation of cultured myoblast cells. J Biol Chem. 1995;270:11719–11722. doi: 10.1074/jbc.270.20.11719. [DOI] [PubMed] [Google Scholar]
- Goljanek-Whysall K, Sweetman D, bu-Elmagd M, Chapnik E, Dalmay T, Hornstein E, Munsterberg A. MicroRNA regulation of the paired-box transcription factor Pax3 confers robustness to developmental timing of myogenesis. Proc Natl Acad Sci U S A. 2011;108:11936–11941. doi: 10.1073/pnas.1105362108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulding M, Lumsden A, Paquette AJ. Regulation of Pax-3 expression in the dermomyotome and its role in muscle development. Development. 1994;120:957–971. doi: 10.1242/dev.120.4.957. [DOI] [PubMed] [Google Scholar]
- Hirai H, Verma M, Watanabe S, Tastad C, Asakura Y, Asakura A. MyoD regulates apoptosis of myoblasts through microRNA-mediated down-regulation of Pax3. J Cell Biol. 2010;191:347–365. doi: 10.1083/jcb.201006025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Z, Lee JY, Pan, Jiang B, Tian B. Progressive lengthening of 3′ untranslated regions of mRNAs by alternative polyadenylation during mouse embryonic development. Proc Natl Acad Sci U S A. 2009;106:7028–7033. doi: 10.1073/pnas.0900028106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HK, Lee YS, Sivaprasad U, Malhotra A, Dutta A. Muscle-specific microRNA miR-206 promotes muscle differentiation. J Cell Biol. 2006;174:677–687. doi: 10.1083/jcb.200603008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- LeGrand F, Jones AE, Seale V, Scime A, Rudnicki MA. Wnt7a activates the planar cell polarity pathway to drive the symmetric expansion of satellite stem cells. Cell Stem Cell. 2009;4:535–547. doi: 10.1016/j.stem.2009.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepper C, Conway SJ, Fan CM. Adult satellite cells and embryonic muscle progenitors have distinct genetic requirements. Nature. 2009;460:627–631. doi: 10.1038/nature08209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi DD, Darnell RB. RNA processing and its regulation: global insights into biological networks. Nat Rev Genet. 2010;11:75–87. doi: 10.1038/nrg2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz CS. Alternative polyadenylation: a twist on mRNA 3′ end formation. ACS Chem Biol. 2008;3:609–617. doi: 10.1021/cb800138w. [DOI] [PubMed] [Google Scholar]
- Maniatis T, Reed R. An extensive network of coupling among gene expression machines. Nature. 2002;416:499–506. doi: 10.1038/416499a. [DOI] [PubMed] [Google Scholar]
- Mayr C, Bartel DP. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138:673–684. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millevoi S, Vagner S. Molecular mechanisms of eukaryotic pre-mRNA 3′ end processing regulation. Nucleic Acids Res. 2010;38:2757–2774. doi: 10.1093/nar/gkp1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montarras D, Morgan J, Collins C, Relaix F, Zaffran S, Cumano A, Partridge T, Buckingham M. Direct isolation of satellite cells for skeletal muscle regeneration. Science. 2005;309:2064–2067. doi: 10.1126/science.1114758. [DOI] [PubMed] [Google Scholar]
- Parker CJ, Shawcross SG, Li H, Wang QY, Herrington CS, Kumar S, MacKie RM, Prime W, Rennie IG, Sisley K, Kumar P. Expression of PAX 3 alternatively spliced transcripts and identification of two new isoforms in human tumors of neural crest origin. Int J Cancer. 2004;108:314–320. doi: 10.1002/ijc.11527. [DOI] [PubMed] [Google Scholar]
- Pritchard C, Grosveld G, Hollenbach AD. Alternative splicing of Pax3 produces a transcriptionally inactive protein. Gene. 2003;305:61–69. doi: 10.1016/s0378-1119(02)01186-1. [DOI] [PubMed] [Google Scholar]
- Quach NL, Rando TA. Focal adhesion kinase is essential for costamerogenesis in cultured skeletal muscle cells. Dev Biol. 2006;293:38–52. doi: 10.1016/j.ydbio.2005.12.040. [DOI] [PubMed] [Google Scholar]
- Rao PK, Kumar RM, Farkhondeh M, Baskerville S, Lodish HF. Myogenic factors that regulate expression of muscle-specific microRNAs. Proc Natl Acad Sci U S A. 2006;103:8721–8726. doi: 10.1073/pnas.0602831103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relaix F, Montarras D, Zaffran S, Gayraud-Morel B, Rocancourt D, Tajbakhsh S, Mansouri A, Cumano A, Buckingham M. Pax3 and Pax7 have distinct and overlapping functions in adult muscle progenitor cells. J Cell Biol. 2006;172:91–102. doi: 10.1083/jcb.200508044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenblatt JD, Lunt AI, Parry DJ, Partridge TA. Culturing satellite cells from living single muscle fiber explants. In Vitro Cell Dev Biol Anim. 1995;31:773–779. doi: 10.1007/BF02634119. [DOI] [PubMed] [Google Scholar]
- Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science. 2008;320:1643–1647. doi: 10.1126/science.1155390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweetman D, Goljanek K, Rathjen T, Oustanina S, Braun T, Dalmay T, Munsterberg A. Specific requirements of MRFs for the expression of muscle specific microRNAs, miR-1, miR-206 and miR-133. Dev Biol. 2008;321:491–499. doi: 10.1016/j.ydbio.2008.06.019. [DOI] [PubMed] [Google Scholar]
- Tanaka KK, Hall JK, Troy AA, Cornelison DD, Majka SM, Olwin BB. Syndecan-4-expressing muscle progenitor cells in the SP engraft as satellite cells during muscle regeneration. Cell Stem Cell. 2009;4:217–225. doi: 10.1016/j.stem.2009.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian H, Chen HJ, Cross TH, Edenberg HJ. Alternative splicing and promoter use in the human GABRA2 gene. Brain Res Mol Brain Res. 2005;137:174–183. doi: 10.1016/j.molbrainres.2005.03.001. [DOI] [PubMed] [Google Scholar]
- Tranter M, Helsley RN, Paulding WR, McGuinness M, Brokamp C, Haar L, Liu Y, Ren X, Jones WK. Coordinated post-transcriptional regulation of Hsp70.3 gene expression by microRNA and alternative polyadenylation. J Biol Chem. 2011;286:29828–29837. doi: 10.1074/jbc.M111.221796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Kumar S, Slevin M, Kumar P. Functional analysis of alternative isoforms of the transcription factor PAX3 in melanocytes in vitro. Cancer Res. 2006;66:8574–8580. doi: 10.1158/0008-5472.CAN-06-0947. [DOI] [PubMed] [Google Scholar]
- Winter J, Kunath M, Roepcke S, Krause S, Schneider R, Schweiger S. Alternative polyadenylation signals and promoters act in concert to control tissue-specific expression of the Opitz Syndrome gene MID1. BMC Mol Biol. 2007;8:105. doi: 10.1186/1471-2199-8-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XM, Vogan K, Gros P, Park M. Expression of the met receptor tyrosine kinase in muscle progenitor cells in somites and limbs is absent in Splotch mice. Development. 1996;122:2163–2171. doi: 10.1242/dev.122.7.2163. [DOI] [PubMed] [Google Scholar]
- Zhao J, Hyman L, Moore C. Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol Mol Biol Rev. 1999;63:405–445. doi: 10.1128/mmbr.63.2.405-445.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.