Abstract
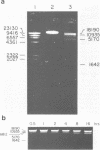
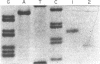
A Type II restriction endonuclease, designated FseI, has been partially purified from a Frankia species (NRRL 18528). This enzyme cleaves Adenovirus 2 DNA at three sites, but does not cleave the DNAs from bacteriophages lambda, T7, and phi X174, the animal virus SV40, pUC18 and pBR322. FseI recognizes the octanucleotide sequence 5' GGCCGG decreases CC 3' and cleaves as indicated by the arrow. The frequency of occurrence of FseI sites within sequenced regions of the human genome is similar to that for NotI sites.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brown N. L., Hutchison C. A., 3rd, Smith M. The specific non-symmetrical sequence recognized by restriction endonuclease MboII. J Mol Biol. 1980 Jun 15;140(1):143–148. doi: 10.1016/0022-2836(80)90360-5. [DOI] [PubMed] [Google Scholar]
- Gingeras T. R., Sciaky D., Gelinas R. E., Bing-Dong J., Yen C. E., Kelly M. M., Bullock P. A., Parsons B. L., O'Neill K. E., Roberts R. J. Nucleotide sequences from the adenovirus-2 genome. J Biol Chem. 1982 Nov 25;257(22):13475–13491. [PubMed] [Google Scholar]
- Mann M. B., Smith H. O. Specificity of Hpa II and Hae III DNA methylases. Nucleic Acids Res. 1977 Dec;4(12):4211–4221. doi: 10.1093/nar/4.12.4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland M., Hanish J., Nelson M., Patel Y. KGB: a single buffer for all restriction endonucleases. Nucleic Acids Res. 1988 Jan 11;16(1):364–364. doi: 10.1093/nar/16.1.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland M. Recognition sequences of type II restriction systems are constrained by the G + C content of host genomes. Nucleic Acids Res. 1988 Mar 25;16(5):2283–2294. doi: 10.1093/nar/16.5.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersson U., Sambrook J. Amount of viral DNA in the genome of cells transformed by adenovirus type 2. J Mol Biol. 1973 Jan;73(1):125–130. doi: 10.1016/0022-2836(73)90164-2. [DOI] [PubMed] [Google Scholar]
- Qiang B. Q., Schildkraut I. A type II restriction endonuclease with an eight nucleotide specificity from Streptomyces fimbriatus. Nucleic Acids Res. 1984 Jun 11;12(11):4507–4516. doi: 10.1093/nar/12.11.4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiang B. Q., Schildkraut I. NotI and SfiI: restriction endonucleases with octanucleotide recognition sequences. Methods Enzymol. 1987;155:15–21. doi: 10.1016/0076-6879(87)55005-4. [DOI] [PubMed] [Google Scholar]
- Roberts R. J. Restriction enzymes and their isoschizomers. Nucleic Acids Res. 1989;17 (Suppl):r347–r387. doi: 10.1093/nar/17.suppl.r347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz D. C., Cantor C. R. Separation of yeast chromosome-sized DNAs by pulsed field gradient gel electrophoresis. Cell. 1984 May;37(1):67–75. doi: 10.1016/0092-8674(84)90301-5. [DOI] [PubMed] [Google Scholar]
- Walder R. Y., Langtimm C. J., Chatterjee R., Walder J. A. Cloning of the MspI modification enzyme. The site of modification and its effects on cleavage by MspI and HpaII. J Biol Chem. 1983 Jan 25;258(2):1235–1241. [PubMed] [Google Scholar]