Abstract
The wound healing assay is a commonly used technique to measure cell motility and migration. Traditional methods of performing the wound healing assay suffer from low throughput and a lack of quantitative data analysis. We have developed a new method to perform a high-throughput wound healing assay that produces quantitative data using the LEAP™ instrument. The LEAP™ instrument is used to create reproducible wounds in each well of a 96-well plate by laser ablation. The LEAP™ then records bright field images of each well at several time points. A custom texture segmentation algorithm is used to determine the wound area of each well at each time point. This texture segmentation analysis can provide faster and more accurate image analysis than traditional methods. Experimental results show that reproducible wounds are created by laser ablation with a wound area that varies by less than 10%. This method was tested by confirming that neuregulin-2β increases the rate of wound healing by MCF7 cells in a dose dependent manner. This automated wound healing assay has greatly improved the speed and accuracy, making it a suitable high-throughput method for drug screening.
Key terms: wound healing assay, quantitative image analysis, image cytometry, high-throughput screening, interactive imaging, LEAP
The wound healing or “scratch” assay is a traditional method that has been used for decades to study cell proliferation and migration (1–3). In a traditional wound healing assay, cells are seeded into a vessel (typically a small Petri dish or 6- to 24-well plates) and allowed to proliferate until they form a monolayer. A pipette tip is then used to scratch this monolayer to create a wound area that is free of cells. The cultures are then imaged over time using bright field or fluorescence microscopy to monitor the growth and migration of cells into the wound.
The most common way to measure wound healing is to manually measure the distance between edges of the wound and calculate the wound area (4,5). This method has many drawbacks. First, the method is manual and very tedious which limits the ability to perform the wound healing assay on many samples. The second drawback is that the manual selection of the edge of the wound is very subjective, and can vary depending on the person performing the measurement. A third problem is that the area calculation assumes that the wound has a rectangular shape with smooth edges, which is almost never the case. Because of these problems, in most cases, wound healing assays are low throughput and the data is subjective and provides mostly qualitative results.
There have been several attempts to fix these problems. An electrical wound healing assay has been developed that wounds a cell monolayer by lethal electroporation, and monitors the wound healing by measuring the surface resistance using microelectrodes (6). This technique is quantitative and highly reproducible, but the throughput is low and this assay requires specialized equipment that is expensive and not common in most laboratories. There are high-throughput methods that use fluorescence scanners to perform wound healing assays in 96- and 384-well plates. However, these assays require that the cells be labeled with a fluorescent probe (7). Ibidi produces a cell culture insert which allows cells to be grown in two cultures separated by a small gap. A very smooth and reproducible wound is created when the culture insert is removed (8,9). The downside of this kit is that the inserts are large and designed to be used in petri dishes, so they are not optimal for use in a high throughput assay. The software program TScratch uses an advanced edge detection method to perform automated image analysis to find the wound area in digitized photomicrographs (10,11). This program uses an algorithm based on curvelet transform to define the wound areas, and is able to reproducibly quantify the wound area. Even though this method is automated and increases throughput over conventional manual analysis, the detection can miss smaller features of the wound and is sensitive to the imaging conditions such as uneven illumination and slight changes in focus.
We have developed an automated high-throughput wound healing assay that uses the laser enabled analysis and processing (LEAP™, Cyntellect, San Diego, CA) instrument to create reproducible wounds and perform automated bright field imaging. The LEAP™ instrument is a high-throughput automated microscope that contains a pulsed laser that can be used to eliminate targeted cells by laser ablation (12,13). This capability is used to create reproducible wounds by laser ablation in each well of a multi-well plate. The LEAP™ can then be used to capture bright field images of each well for subsequent image analysis. We have developed an automated image analysis algorithm based on texture segmentation that is able to rapidly distinguish between areas of the image covered by cells, and the bare wound area (14). This algorithm can be performed using bright field images, so fluorescence staining is not required. Bright field microscopy permits the same wound to be monitored over many time points, with data normalized to the initial wound size. This data analysis method makes no assumptions about the size or morphology of the wound area, so the true wound area and any variety of initial wound shapes can be measured. The algorithm can process any wound healing image in any format and does not require that images be spatially registered. This permits wound tracking at different time points.
Materials and Methods
Cell Culture
The breast cancer cell line MCF7 (ATCC, Manassas, VA) was used for these proof of concept experiments. These cells were incubated at 37°C with 95% humidity and 5% CO2 in minimum essential medium (MEM, Hyclone) supplemented with 1 mM sodium pyruvate (MP Biomedicals), 10% fetal bovine serum (PAA Laboratories), and 10 µg/mL insulin (US Pharmacopeia). Cells were passaged at 90% confluence at a 1:4 dilution ratio.
Automated Wound Creation Using LEAP™
MCF7 cells were seeded into a plastic, flat-bottom ,96-well cell culture plate (C-Lect Stem Cell Manager, Cyntellect, San Diego, CA) and allowed to grow to confluence. Cells were starved in basal medium for 48 h, after which the plate was loaded into the LEAP™ instrument. The wound pattern was defined to be a circle with a diameter equal to 25% percent of the well diameter. This wound pattern was created in each well by laser pulses with an energy of 7 µJ/pulse and 25 µm grid spacing. The plate was then removed from the LEAP™ instrument, the wells were washed, and the medium was replaced with fresh basal medium. Immediately after the media was replaced, the whole plate was imaged in bright field at 3× using the LEAP™ instrument. The plate was then imaged in bright field at 3× magnification every 24 h for 96 h. The plate was incubated at 37°C with 95% humidity and 5% CO2 between time points. The wound creation method is demonstrated in Figure 1.
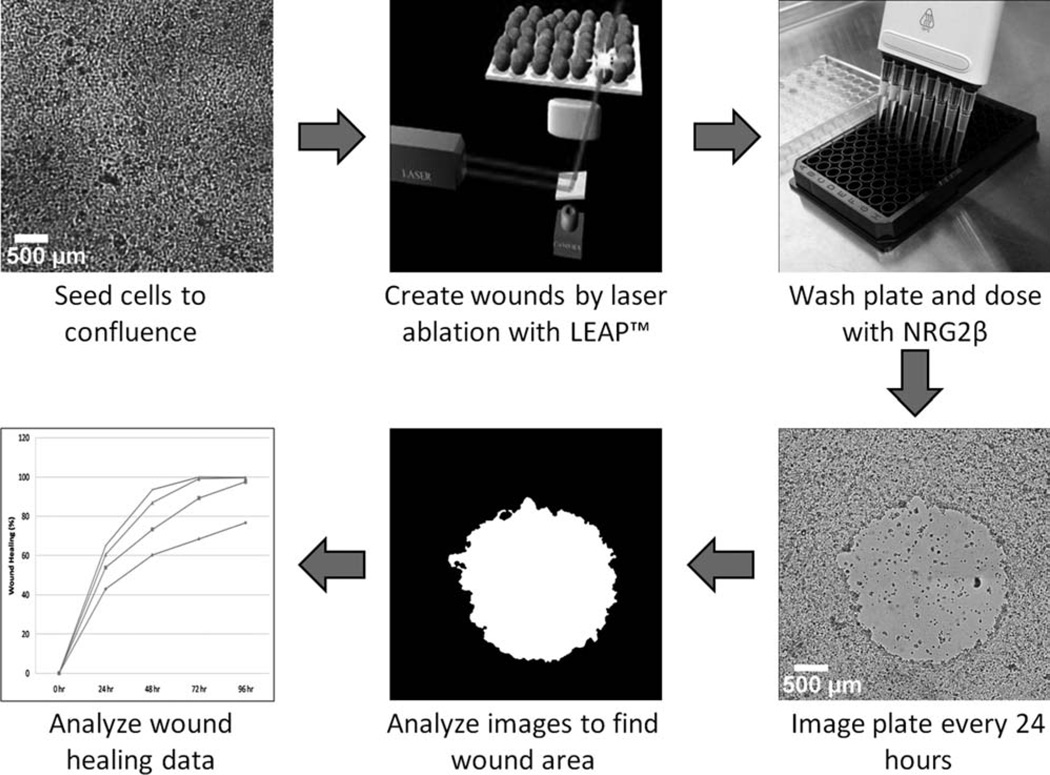
Figure 1.
Workflow of the automated wound healing analysis (clockwise from top left). First, cells are seeded and allowed to proliferate to confluence in a multi-well tissue culture plate. Then LEAP™ is used to create wounds in each well by laser ablation. The plate is then washed to remove cell debris, and if necessary the wells are dosed with a drug or compound. The LEAP™ is then used to image every well in bright field mode every 24 h. Image analysis is performed by our texture segmentation algorithm to determine the wound area for each well and time point. The wound healing is then calculated and analyzed to determine the effect of a given drug or compound.
Wound Area Determination Using Texture Segmentation Based Image Analysis
The LEAP instrument records 16 bright field images for each well in a 96-well plate. The whole well images were reconstructed using a custom MATLAB script. A different algorithm was written in MATLAB that performs texture segmentation to measure the wound area. Each image is first cropped to a user-defined size that only encompasses the wound immediately after wound creation. This step reduces the size of the images, thereby enabling the processing algorithm to run more quickly. A texture filter is then applied to the cropped image. In this case, the entropy filter measures the local disorder of an 11 × 11 field of pixels surrounding each pixel. Areas with large pixel intensity variation (cells) will appear bright, where smooth areas of the image (wound) will appear dark in the entropy image. The entropy image is then converted to a binary black and white image by applying a simple pixel threshold, typically between 0.5 and 0.85 (MATLAB grayscale images have an intensity range of 0–1). The thresholded binary image is then inverted so that the bare wound region is white and the cell monolayer region is black. The wound region is then morphologically opened by performing an erosion operation followed by a dilation operation. This removes small areas that are typically noise without affecting the larger wound region because the erosion and dilation operations have the same kernel size. The image is then dilated to smooth out the outer surface of the wound. A morphological close is then applied to produce a continuous wound area, this operation first dilates and then erodes the binary image by the same structural element (a 5-pixel disk), this operation functions to fill in the outer edges of the wound that were distorted during the previous morphological opening process. White regions that touch the border of the image are removed. To produce the final wound area, the binary image is morphologically filled to remove black spots in the wound area. The wound area is the white region that remains, and the wound area is determined by counting the number of white pixels in the final binary image. This process is demonstrated in Figure 2.
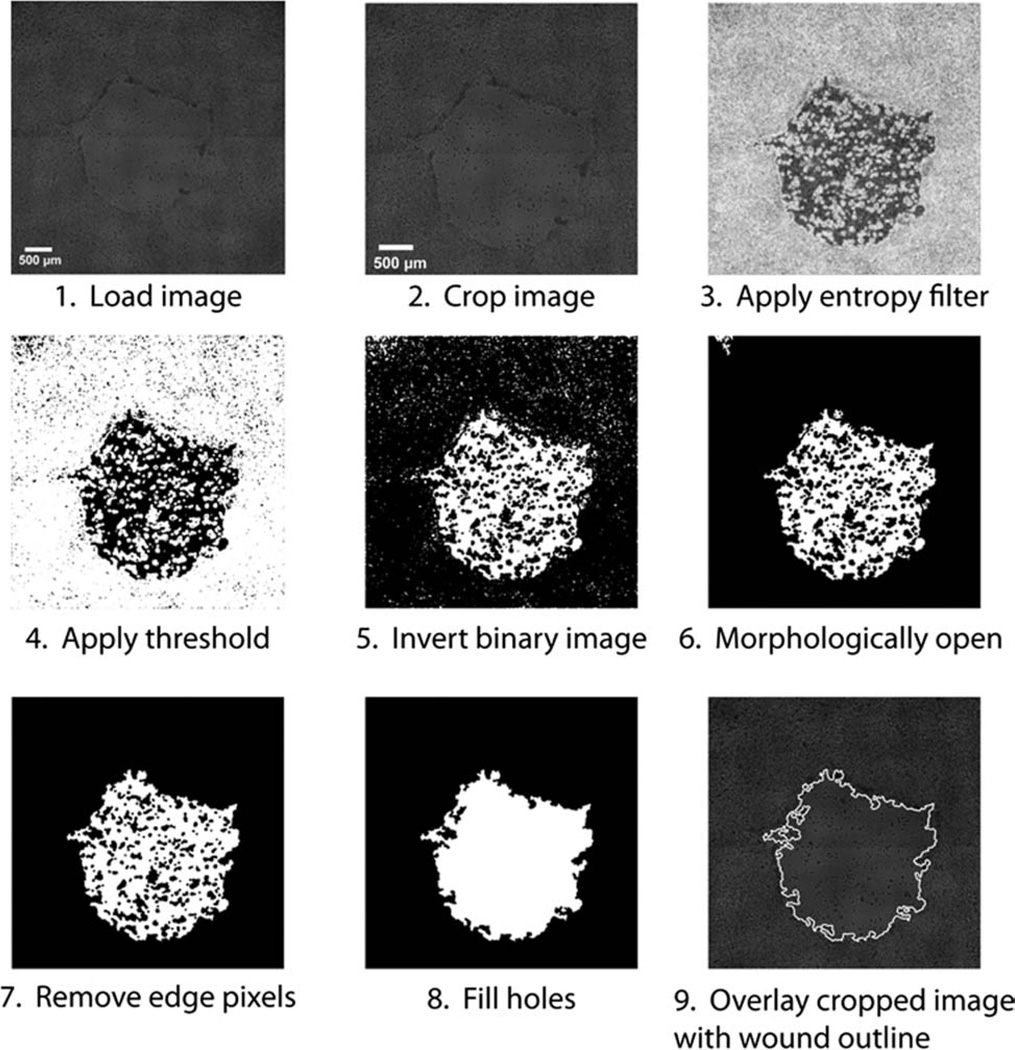
Figure 2.
The wound healing image analysis algorithm. First, the image is loaded (1) and cropped (2). An entropy filter is then applied to generate a texture image (3). The texture image is thresholded to give a binary image (4), which is then inverted (5). Small objects are then removed by morphologically opening the binary image (6). Objects touching the edge of the image are removed (7). The holes are then filled to create the wound mask (8). The wound area is calculated and then an outline of the wound is superimposed on the cropped image for quality control and analysis (9).
Validation Study: Measuring the Healing Response Due to a Known Growth Factor
To test the automated wound healing assay, we measured the healing response to the known growth factor neuregulin-2β (15,16). MCF7 cells were seeded in 96-well plates as described previously. The wounds were created in each well by laser ablation. The wells were washed and the medium was replaced by basal medium supplemented with 0, 3, 30 or 100 nM neuregulin-2β. Neuregulin-2β was purified using the protocol developed by the Riese laboratory (15,16). The plate was then imaged in bright field at 0, 24, 48, 72, and 96 h. The wound area was measured using the texture segmentation algorithm for each well at each time point. The percent healing of each well was determined by dividing the wound area at each time point by the initial wound area and multiplying by 100. The cells were serum starved before the wounds were created to decrease the basal levels of cell migration. Additionally, the cells were grown without serum after the wounds in order to study cell motility caused directly by neuregulin 2β as opposed to growth factors present in serum.
Results
Reproducible Wound Formation Using LEAP™
The LEAP™ instrument can consistently produce circular wounds in a confluent monolayer of cells using laser ablation. The wounds had a consistent geometry. This is demonstrated in Figure 3, which shows the individual wounds produced in each well of a 96-well plate. The initial wound area was measured using our texture segmentation method. The initial wounds have a standard deviation of 9.16% across the whole plate. The throughput of this automated wound healing assay is very rapid. It takes less than 30 min to create the wounds in every well of a 96-well plate, and less than 20 min to image a whole plate at 3× magnification.
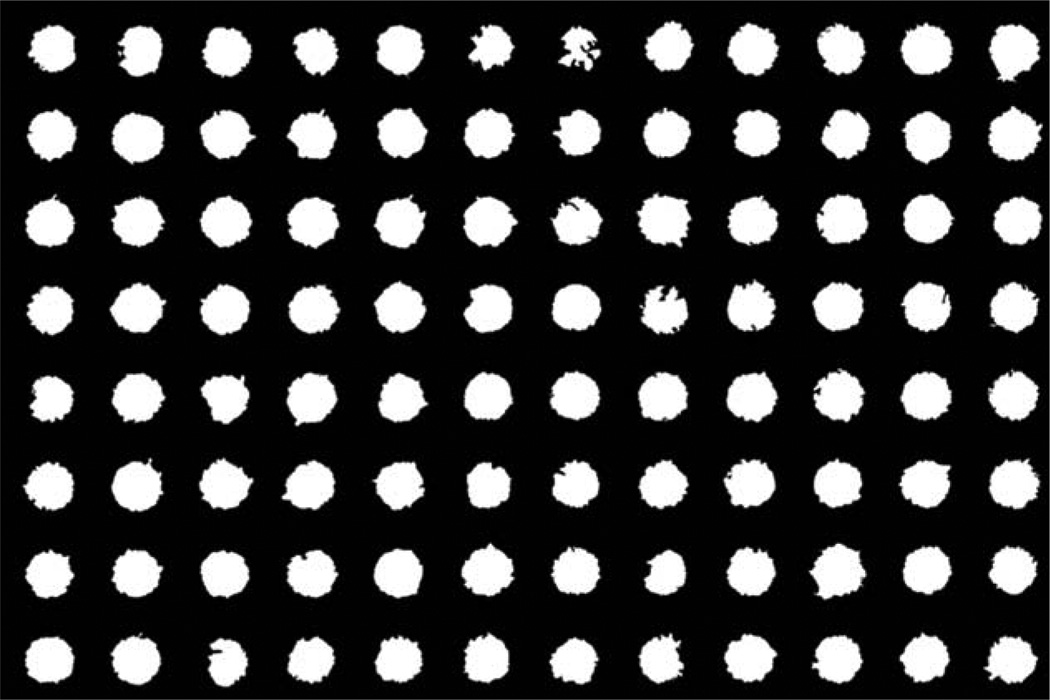
Figure 3.
Masks of the created wounds in a 96-well plate. The wounds have a consistent shape. The size of the created wounds had a standard deviation of 9.16%.
Accuracy of Texture Segmentation Algorithm to Determine Wound Area
The image analysis performed by our texture segmentation algorithm accurately identifies the bare wound area from the confluent cell monolayer. The wound area for many different wells and time points is shown in Figure 4. This indicates that there is accurate segmentation regardless of the size or shape of the wound. The segmentation is robust and is able to accurately identify the wound area regardless of errors in background illumination and focus. The MATLAB based algorithm requires 11 s to process each whole well image.
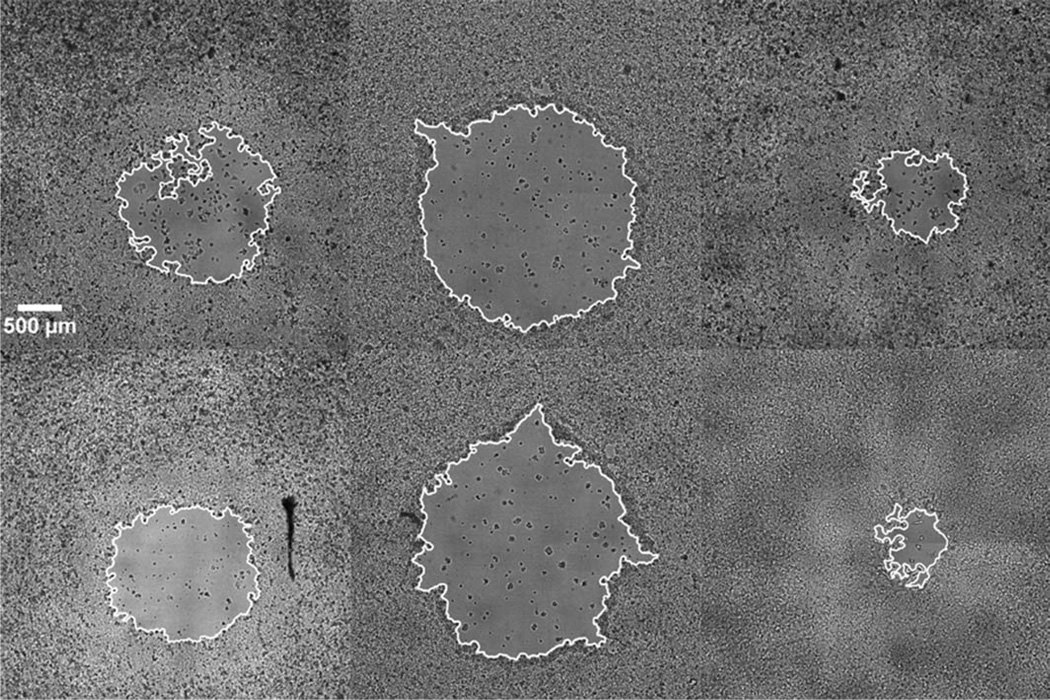
Figure 4.
The measured wound area (outlined) as measured by our texture segmentation algorithm for several different wounds. The images were collected and processed in an automated fashion, and the robust segmentation successfully identifies the wound area. The segmentation is successful for wounds of various sizes and shapes.
Tracking Growth Factor Mediated Wound Healing Over Time
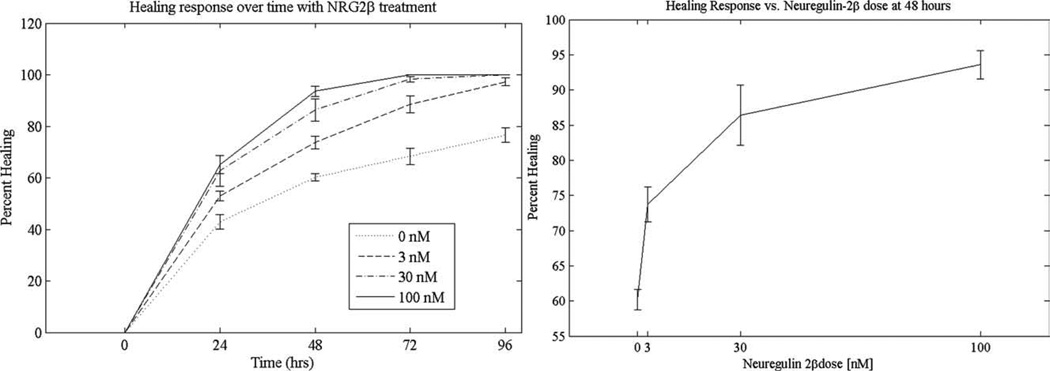
In order to validate this new automated wound healing assay, MCF7 cells were treated with different doses of the known growth factor neuregulin-2β and the subsequent wound healing was measured every 24 h for 4 days. The healing progression of a single wound over 96 h is shown in Figure 5. The wounds healed in a dose dependant manner as shown in Figure 6. All of the wounds treated with neuregulin-2β healed significantly faster than the untreated control, with complete healing of the wounds by 72 h for wells treated with 30 or 100 nM neuregulin-2β. The Z′-factor of this assay was 0.685, indicating that it is a high quality assay for screening wound healing response.
Figure 5.
Tracking wound healing of one sample over time. The top row consists of an outline of the wound mask superimposed on top of the initial bright field image at 0 (left), 48 (middle), and 96 h (right) after wound formation. The bottom row shows the binary wound mask of the same wound at 0 (left), 48 (middle), and 96 h (right) after wound formation.
Figure 6.
The results of a preliminary wound-healing assay that evaluates the effect of neuregulin-2β on the healing of wounds in a culture of MCF7 cells. On the left is a healing curve of four different concentrations of neuregulin-2β showing that the treated cells healed faster than did the untreated cells. On the right, there is a dose response curve of neuregulin-2β on healing at 48 h after wound creation and concurrent growth factor administration. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Discussion
In this report, we have described the development of a novel high-throughput wound healing assay using the LEAP™ instrument. The LEAP™ instrument is used to first create reproducible wounds in each well of a multiwell plate, and then perform bright field imaging of each well over time. A custom texture segmentation image analysis algorithm was developed to identify and measure the wound area in these bright field images. We then demonstrated an application of the technique by measuring the dose dependent healing response of the known growth factor neuregulin-2β.
This new technique provides several improvements to the current methods of performing the wound healing assay. First, consistent wounds are created in each well by laser ablation. This is advantageous to the traditional mechanical “scratch” because the wound is created in a complete sterile environment, and size and shape of the wounds are more reproducible. This method uses completely automated wound creation and image analysis, so human error is reduced. In traditional wound healing analysis, the wound areas are measured manually in a very laborious manner. Additionally the manual analysis of images makes assumptions about the shape of the wound, typically fitting a rough wound to a rectangular geometry. The automated bright field image analysis in this new technique is much faster and calculates a wound area that is significantly more accurate than what a human can measure. Image sets that previously took weeks to analyze can now be analyzed in a few hours.
The bright-field wound healing assay has great potential for use as an assay platform. The image analysis is performed in a marker-less fashion. Fluorescence imaging can still be performed to study the biology of the healing response, such as actin staining or labeling a protein with GFP, without needing to use a fluorescent label to identify the wound area. While the proof-of-principle assay used serum starvation and 96-h time points, the assay can be easily adapted to a variety of culture conditions, and different cells can be used. Additionally, the laser-induced wounds can be smaller, which will result in shorter healing times.
This method also has several advantages over fluorescence based high throughput wound healing assays. The main advantage is that this method uses bright field imaging, so the cells do not have to be fixed and stained for analysis. This means that the healing of the wound can be calculated by comparing it with the initial size of the wound. The fluorescence-based wound healing assays typically only measure the wound at a single time point, and it is assumed that all wells had identical wounds. Also, each well can be imaged at multiple time points to monitor the healing progression. Implementation of the LEAP™ high-throughput wound-healing assay holds great promise for use in screening compounds that affect cell motility and migration.
Acknowledgments
The authors acknowledge the support of Dr. Fred Koller of Cyntellect and Dr. Michelle Zatcoff formerly of Cyntellect for their help in adapting the LEAP™ to perform wound creation. The LEAP instrument was purchased with support from the NIH Shared Instrument Grant Program, grant 1S10 RR023651-01A2 under the ARRA program. This report was reported as a poster at the CYTO 2010 International Congress of ISAC in Seattle, Washington on May 8–12, 2010.
Literature Cited
- 1.Coomber B, Gotlieb A. In vitro endothelial wound repair. Interaction of cell migration and oughproliferation. Arterioscler Thromb Vasc Biol. 1990;10:215–222. doi: 10.1161/01.atv.10.2.215. [DOI] [PubMed] [Google Scholar]
- 2.Todaro GJ, Lazar GK, Green H. The initiation of cell division in a contact-inhibited mammalian cell line. J Cell Comp Physiol. 1965;66:325–333. doi: 10.1002/jcp.1030660310. [DOI] [PubMed] [Google Scholar]
- 3.Wong MK, Gotlieb AI. The reorganization of microfilaments, centrosomes, and microtubules during in vitro small wound reendothelialization. J Cell Biol. 1988;107:1777–1783. doi: 10.1083/jcb.107.5.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fronza M, Heinzmann B, Hamburger M, Laufer S, Merfort I. Determination of the wound healing effect of Calendula extracts using the scratch assay with 3T3 fibroblasts. J Ethnopharmacol. 2009;126:463–467. doi: 10.1016/j.jep.2009.09.014. [DOI] [PubMed] [Google Scholar]
- 5.Ronot X, Doisy A, Tracqui P. Quantitative study of dynamic behavior of cell monolayers during in vitro wound healing by optical flow analysis. Cytometry. 2000;41:19–30. [PubMed] [Google Scholar]
- 6.Keese CR, Wegener J, Walker SR, Giaever I. Electrical wound-healing assay for cells in vitro. Proc Natl Acad Sci USA. 2004;101:1554–1559. doi: 10.1073/pnas.0307588100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yarrow JC, Perlman ZE, Westwood NJ, Mitchison TJ. A high-throughput cell migration assay using scratch wound healing, a comparison of image-based readout methods. BMC Biotechnol. 2004;4:21. doi: 10.1186/1472-6750-4-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Horst D, Scheel SK, Liebmann S, Neumann J, Maatz S, Kirchner T, Jung A. The cancer stem cell marker CD133 has high prognostic impact but unknown functional relevance for the metastasis of human colon cancer. J Pathol. 2009;219:427–434. doi: 10.1002/path.2597. [DOI] [PubMed] [Google Scholar]
- 9.van Horssen R, ten Hagen TLM. Crossing barriers: The new dimension of 2D cell migration assays. J Cell Physiol. 2011;226:288–290. doi: 10.1002/jcp.22330. [DOI] [PubMed] [Google Scholar]
- 10.Geback T, Koumoutsakos P. Edge detection in microscopy images using curvelets. BMC Bioinformatics. 2009;10 doi: 10.1186/1471-2105-10-75. article 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geback T, Schulz MM, Koumoutsakos P, Detmar M. TScratch: A novel and simple software tool for automated analysis of monolayer wound healing assays. Biotechniques. 2009;46:265–274. doi: 10.2144/000113083. [DOI] [PubMed] [Google Scholar]
- 12.Koller MR, Palsson BO, Eisfeld TM. Method for inducing a response in one or more targeted cells. 6,642,018. Patent. 2003
- 13.Koller MR, Hanania EG, Stevens J, Eisfeld TM, Sasaki GC, Fieck A, Palsson BO. High-throughput laser-mediated in situ cell purification with high purity and yield. Cytometry A. 2004;61:153–161. doi: 10.1002/cyto.a.20079. [DOI] [PubMed] [Google Scholar]
- 14.Zordan MD, Leary JF. Image and data analysis for wound healing assay. 65613.P1.US. US Patent. 2010
- 15.Hobbs SS, Coffing SL, Le AT, Cameron EM, Williams EE, Andrew M, Blommel EN, Hammer RP, Chang H, Riese DJ., II Neuregulin isoforms exhibit distinct patterns of ErbB family receptor activation. Oncogene. 2002;21:8442–8452. doi: 10.1038/sj.onc.1205960. [DOI] [PubMed] [Google Scholar]
- 16.Wilson KJ, Mill CP, Cameron EM, Hobbs SS, Hammer RP, Riese DJ., II Inter-conversion of neuregulin2 full and partial agonists for ErbB4. Biochem Biophys Res Commun. 2007;364:351–357. doi: 10.1016/j.bbrc.2007.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]