Abstract
A rare combination of mutations within mitochondrial DNA subhaplogroup T2e is identified as affiliated with Sephardic Jews, a group that has received relatively little attention. Four investigations were pursued: Search of the motif in 250 000 control region records across 8 databases, comparison of frequencies of T subhaplogroups (T1, T2b, T2c, T2e, T4, T*) across 11 diverse populations, creation of a phylogenic median-joining network from public T2e control region entries, and analysis of one Sephardic mitochondrial full genomic sequence with the motif. It was found that the rare motif belonged only to Sephardic descendents (Turkey, Bulgaria), to inhabitants of North American regions known for secret Spanish–Jewish colonization, or were consistent with Sephardic ancestry. The incidence of subhaplogroup T2e decreased from the Western Arabian Peninsula to Italy to Spain and into Western Europe. The ratio of sister subhaplogroups T2e to T2b was found to vary 40-fold across populations from a low in the British Isles to a high in Saudi Arabia with the ratio in Sephardim more similar to Saudi Arabia, Egypt, and Italy than to hosts Spain and Portugal. Coding region mutations of 2308G and 14499T may locate the Sephardic signature within T2e, but additional samples and reworking of current T2e phylogenetic branch structure is needed. The Sephardic Turkish community has a less pronounced founder effect than some Ashkenazi groups considered singly (eg, Polish), but other comparisons of interest await comparable averaging. Registries of signatures will benefit the study of populations with a large number of smaller-size founders.
Keywords: mitochondrial DNA, sephardim, haplogroup T, founder effect, subhaplogroup T2, Iberia
Introduction
Genetic lines can be very successful. In the founding of some communities, only a handful of individuals were responsible for a large number of the individuals in those communities today. Such a narrow ‘founder effect'1 has been of considerable interest.2, 3 Behar et al4, 5 have investigated the size of the founder effect in 15 Jewish communities with samples of mitochondrial DNA obtained from 1725 individuals. Their impressive undertaking has provided, as they note, ‘… a nearly comprehensive picture of the maternal genetic landscape of the entire Jewish population' (p. 13).
A notable result is that the Ashkenazim, which comprise the largest Jewish population worldwide, shows a prominent founder effect with only 4 mitochondrial haplotypes comprising 40% of the modern population and with the most frequent haplotype found in 19% of individuals.4, 6 Sephardim, descendents from Spanish and Portuguese Jews who were the largest Jewish population until the 18th century, instead show a large variation in maternal founders of the communities of the Ottoman Empire.5 This was especially apparent in their results for Turkey, with the 4 most frequent haplotypes found in only 17% of the present-day descendent population and the most frequent haplotype found in not quite 6% (for Bulgaria, the numbers are 27 and 8.5%, respectively). The Ottoman Empire, including Turkey and Bulgaria, received many of the Jewish people who were exiled from Spain and Portugal in the late 15th century.7 The contrasting findings for the two prominent Jewish populations suggest a greater genetic diversity in mitochondrial DNA in Sephardim than in Ashkenazim.
However, the seemingly higher genetic variability in the present-day Sephardic population, which will be revisited in the ‘Discussion' section, does not preclude uncovering Sephardic signatures. Although the term ‘signatures' has been used in various manners,3, 8 a maternal signature can be considered to be a specific sequence of mutations in mitochondrial DNA (ie, a haplotype) that is substantially and significantly overrepresented in a community of interest compared with the host population or more general comparison group. The issue of signatures is independent of the issue of size of founder effects. Identification of signatures can allow groups of any size in the population to be tracked in their migration through space and time. In addition, whole collections of even infrequent signatures would allow for ethnic origins to be clarified, a practical application especially important for potential descendents of Sephardic Jews. There were many Crypto-Jews who practiced the Sephardic rite in secret, notably in Mexico to escape the long arm of the Spanish Inquisition.9 Finally, maternal signatures can generate interest in less-studied mitochondrial subhaplogroups about which relatively little is known.
Suspicion of a signature in a minority ethnic group can be initiated with as little as a haplotype match in two unrelated individuals from that group. We investigate one such Sephardic signature. The haplotype of a suspected Sephardic origin has mutations 16114T-16126T-16153A-16192T-16294T-16519C in the first control region of mitochondrial DNA. The motif falls within haplogroup T,10, 11, 12 which is present in close to 10% of individuals with European ancestry.3, 13 Criteria for inclusion in T in the first control region are transitions from base ‘C' to base ‘T' at positions 16126 and 16294. The suspected Sephardic haplotype is further identifiable as a branch of T2e,14 formerly T5,2 characterized by the addition of a mutation at 16153 in the first control region2 (and at 150 in the second control region). Reclassification occurred after determination that sequences classified as T5 on the basis of the control region alone were found to have mutations in the coding region that define subhaplogroup T2,15, 16 namely 11812G and 14233G.15 Therefore, former T5 is a sister lineage to other subhaplogroups of T2, such as the more well-known T2b.17 Thus, criteria for the proposed Sephardic signature are the presence of a previously undocumented combination of transitions at control region positions 16114 and 16192 within subhaplogroup T2e. This new cluster will be referred to here as T2e5.
To further investigate the spatial and temporal affiliations of T2e5 specifically and T2e generally, four avenues are pursued: (1) A search is conducted throughout multiple databases of the first control region of mitochondrial DNA for the T2e5 motif to ascertain the prevalence and geographic affiliation of the new haplotype. (2) One T2e5 sample is sequenced for polymorphisms along the entire mitochondrial DNA and compared with T2e sequences to identify any potential coding region mutations that are important for the Sephardic sequence and its relation to other branches. (3) A phylogenetic tree is built from T2e control sequences to provide further information on the relation among lineages including the Sephardic cluster. Although full genomic sequences are usually preferable to avoid misclassifications based on control region information alone, T2e is an ideal subhaplogroup to exploit the more abundant control region data because it is defined by mutations in the control regions alone. Time to the most recent common ancestor is estimated to address questions of when the lineage emerged as well as where. (4) The frequencies of T subhaplogroups are compared across growing published literature of various populations including from Europe, the Americas, and the Near East. Although the geographic distribution of haplogroup T has been investigated, less is known about the different subhaplogroups, especially T2e.
The investigations together should provide a thorough analysis of an infrequently considered mitochondrial subhaplogroup, haplotype, and ethnic population.
Materials and methods
Control region matches
Eight databases of control region 1 were searched for the motif 16114T-16126T-16153A-16192T-16294T-16519C: (1) Mitosearch (n≈34 000, http://www.mitosearch.org), sponsored by Family Tree DNA in the United States; (2) Oxford Genetic Atlas Project (n≈10 000, http://www.bloodoftheisles.net/) from Sykes' study of Britain;18 (3) Sorenson Molecular Genealogy Foundation (n≈73 000, http://www.smgf.org/), which analyzes DNA samples obtained from 173 countries with one-third of the samples from the United States (includes GeneTree customers); (4) FBI database (http://www.fbi.gov/hq/lab/fsc/backissu/april2002/ miller1.htm) with contributing forensic and university labs and pre-2002 sequences from GenBank, European Molecular Biology Laboratory and literature; (5) a worldwide database of published and unpublished sequences (n≈66 000) courtesy of Valery Zaporozhchenko, Research Centre for Medical Genetics, Russian Academy of Medical Sciences; (6) private matches of customers of Family Tree DNA (n≈100 000); (7) public database of Oxford Ancestry customers (n≈2000, http://www.oxfordancestors.com) largely from Britain; (8) EMPOP (n≈10 000, http://www.empop.org), developed by the Institute for Legal Medicine at Innsbruck Medical University and the Institute of Mathematics at University of Innsbruck consisting of forensic data, published literature, and unpublished sequences from a participating lab. The databases together total at least a quarter of a million independent samples.
Full mitochondrial genomic sequence
A T2e5 Sephardic sample with maternal origins descending from Salonica, Turkey, was outsourced to FamilyTreeDNA13 for analysis. The electropherograms were additionally visually inspected and Mutation Surveyor v. 3.30 (SoftGenetics, State College, PA, USA; http://www.softgenetics.com) used to independently verify positions of mutations. T2e sequences for comparison were downloaded from (1) GenBank in FASTA format and assessed for differences from the rCRS with HmtDB (http://www.hmtdb.uniba.it:8080/hmdb/) and (2) from a study19 with sequences not yet deposited with GenBank.
Control region phylogenetic tree
The Mitosearch database was searched for sequences harboring mutations at 16126, 16153, and 16294 (T2e subhaplogroup) under all categories of T listed: T5, T, T*, T2, T3, and T4. ID numbers of matches were checked to ensure an entry was not counted twice. Median-joining networks were created using Network version 4.6.0.0, Fluxus Technology Ltd (Suffolk, England; http://www.fluxus-technology.com/). For estimating time to most recent common ancestor of the purported Sephardic cluster T2e5, coalescence analysis with respect to T2e was performed with the rho statistic and a SD assuming a normal distribution.20, 21 Three different mutations rates were simulated based on estimates in the literature: fast,22 slow,20, 21 and intermediate,23 of one mutation every 1000, 20 180, and 5000 years, respectively. More precise resolutions were deemed unnecessary.
Geographic distribution
All known studies reporting mitochondrial DNA information for Sephardic populations (n=4) were checked for frequency of T2e and other T subhaplogroups. Studies for comparison populations were selected on the basis of large size and availability. If frequencies of subhaplogroups were provided in the text13, 24, 25 or in online supplementary material,26, 27 those assignments were used with the assumption that T3=T2c, T5=T2e, and T*=T other. Otherwise, control region 1 sequences were inspected in the text24, 28 or supplementary materials4, 5, 26, 29, 30 and assigned to subhaplogroups of T. Assignment criteria used were 16163G-16189C (T1), 16304C (T2b), 16292T (T2c), 16153A (T2e), 16324C (T4) within the context of also having 16126C, 16294T (T); remaining motifs were assigned as ‘T other'. Percentage was then calculated from the proportion of each T subhaplogroup to the total number of samples in the study. χ2 Tests for independence were performed on comparisons involving Total T, T2e, and T2b omitting the 70 000 records of National Genographic. They were not performed when the expected frequency was <1 in any cell or <5 in >20% of the cells. Yates' correction, G-test, and Fisher's exact test were considered and used when appropriate; because of the rarity of T2e, certain statistical comparisons with the present sample sizes were not possible. Analyses were performed using Systat software (Systat Software, Inc., Chicago, IL, USA) and with Preacher.31
Results
Control region matches
T2e5 is a rare haplotype.
Exact matches
All known occurrences of the sequence are shown in Table 1. Despite the small size of the cluster, its geographic affiliation is striking. The matches are either definite Sephardic, suggestive of Sephardic ancestry, or readily consistent with this interpretation. One-quarter of the entries is from maternal lineages that are Sephardic through the Ottoman Empire with two from Turkey and one from Bulgaria. Half of the cluster is from Northern Mexico and South Texas, a region with a notable Crypto-Jewish history. These descend from the Mexican states of Nuevo Leon (including its center Monterrey), Tamaulipas, and Coahuila, and in the United States, from the border town of Roma, Texas. Each of these locales has been specifically singled out as harboring Iberian Jewish residents.9, 32, 33, 34 The samples seem to be from unrelated individuals. Information about the final three matches is limited. Their listings of Brazil,24 Portugal,35 and ‘Hispanic' (FBI database) are all consistent with Sephardic ancestry.36
Table 1. Control region haplotypes of the proposed Sephardic signature from a search of published literature and mitochondrial DNA databases.
| HVSI | HVSII | Geographic/ethnic origin | Source |
|---|---|---|---|
| 16114 16126 16153 16192 16221 16294 16519 | 73 150 263 309.1C 315.1C | Sephardic; Salonica, Turkey | Previously unpublished |
| 16114 16126 16153 16192 16294 (up to 16400) | 73 150 263 309.1C 315.1C | Central Portugal | Periera et al29 |
| 16114 16126 16153 16192 16294 16519 | 73 150 263 309.1C 309.2C 315.1C | ‘Hispanic' | FBI database |
| 16067 16114 16126 16153 16192 16294 (up to 16362) | not tested | Northern Brazil | Alves-Silva et al24 |
| 16114 16126 16153 16192 16294 16519 | not tested | Roma, Texas, USA | ahttp://www.mitosearch.org |
| 16114 16126 16153 16192 16294 16519 | not tested | Comales, Tamaulipas Mexico | bhttp://www.mitosearch.org |
| 16114 16126 16153 16192 16291 16294 16519 | 73 150 263 (up to 300) | Sephardic; Bulgaria | Behar et al4 |
| 16114 16126 16153 16192 16294 16519 | 73 150 263 (up to 300) | Sephardic; Turkey | Behar et al4 |
| 16114 16126 16153 16192 16294 16519 | not tested | Monterrey Mexico | Family Tree DNA |
| 16114 16126 16153 16192 16294 16519 | 73 150 263 309.1C 315.1C | General Teran, Nuevo Leon Mexico | Sorenson Foundation |
| 16114 16126 16153 16192 16294 16519 | 73 150 263 309.1C 315.1C | Linares, Nuevo Leon Mexico | Sorenson Foundation |
| 16114 16126 16153 16192 16294 16519 | 73 150 263 309.1C 315.1C | Ramos Arizpe, Coahuila Mexico | Sorenson Foundation |
Abbreviations: HVSI, hypervariable segment 1; HSV2, hypervariable segment 2.
Record retrieved in 2007.
Record retrieved in 2009.
The small T2e5 cluster satisfies criteria for being a signature. Although it is premature to set specific thresholds of a signature, a sample of 25% known Sephardic and 50% suspicion of Sephardic origin is overwhelmingly above what would be expected for a general European haplogroup. The numbers of Sephardim worldwide are very small, and therefore the expected percentage of Sephardim in any haplotype cluster is vanishingly small. The combined databases do not appear to have any biases for Iberia, Mexico, or Sephardim. Moreover, the cluster also does not appear to be merely Iberian because of the relatively small proportion of (presumed non-Jewish) matches from Spain and Portugal than was found. Instead, the data point to a Sephardic signature.
Near matches
Neither of the two mutations that define T2e5 (16114T-16192T) were found without the other in any sequence of T2e. Within those T haplogroup sequences that are not T2e (ie, no mutation at nucleotide 16153), the transition at 16192 was found not infrequently from various regions. A single instance with the transition at 16114 but not at 16192 occurred in a sample from Wales (Oxford Atlas Project, 16114T-16126C-16187-16294T-16296-16324C). Both of these events in haplogroup T are most likely independent of the origins of 16192T and 16114T in T2e. Thus far, T2e5 appears to be a small isolated sequence marked by transitions at both 16114 and 16192 within T2e.
Mitochondrial full genomic sequence
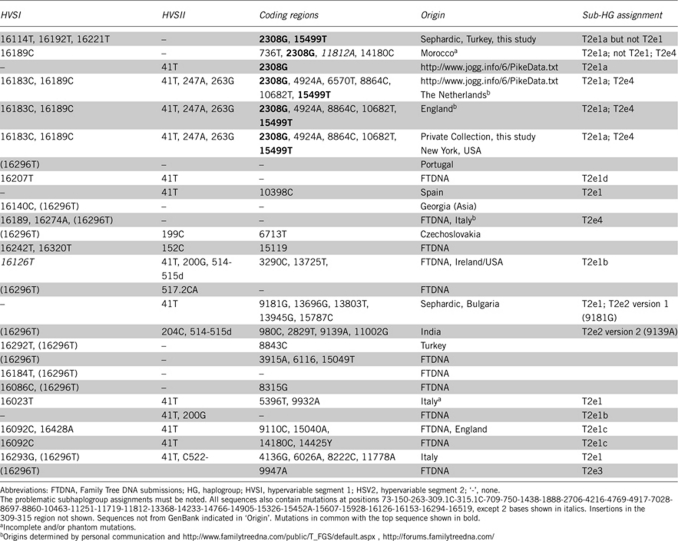
The T2e5 Salonican sample was found to have, as expected, the two T2-defining mutations at 11812 and 14233, as well all of the mutations that are found in the superordinate groups. Two additional coding region mutations were found: 15499T, a synonymous mutation in cytochrome B, and 2308G located in 16S ribosomal RNA.
A total of 23 complete sequences of haplogroup T2e were currently present on GenBank. None of them have either of the control region mutations from the Sephardic signature. Overlap occurred with the coding region and is shown in Table 2. One of the overlapping sequences37 is reported to be missing mutations,16, 38 which makes comparison difficult. Nonetheless, the existence of both 2308G and 15499T in existing T2e sequences suggests that neither of these coding region mutations in the Sephardic signature sample is a defining mutation. The current branch structure of T2e leads to incoherent assignments involving the relevant mutations: 3 of the sequences with 2308G would get assignments in multiple branches (both T2e1a, criterion of 2308G, and T2e4, criterion of 16189C) and 2 would meet the criterion for one branch (T2e1a), yet not meet the criterion for the branch from which it derives (T2e1, 41T). 15499T is currently not a named branch. Several phylogenetic trees that connect the Sephardic signature are possible and await additional samples and reworking of T2e14.
Table 2. Mutations for the new Sephardic sequence (top row) and all T2e sequences on GenBank.
Control region phylogenetic tree
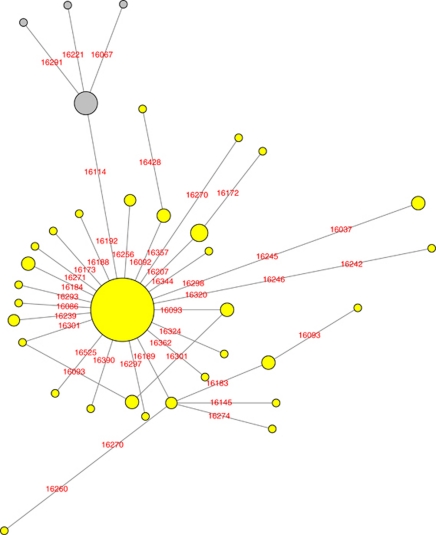
A total of 86 entries were found for subhaplogroup T2e criteria under the T5 category in mitosearch, 2 of which are already part of the Sephardic cluster. Additionally, 33 entries meeting the criteria of T2e were found in categories T and T2. None were listed in T*, T3, or T4. The median-joining network from the 119 samples along with the 10 remaining purported Sephardic T2e5 samples is shown in Figure 1. Nucleotide position 16296 was excluded in the final network because of its instability within Haplogroup T,2 which was verified in a pre-analysis of the data. In addition, 16519 was also downweighted to 0 because data were occasionally absent at that position, although all but 2 reporting the position had 16519C.
Figure 1.
Phylogenetic network of hypervariable region 1 of all 119 entries of mitochondrial DNA subhaplogroup T2e (T5) from the public database mitosearch and the sequences of the Sephardic signature. The size of the circle is proportional to the number of entries, and the distance between connected nodes reflects the number of mutations that separate them. Nodes repositioned slightly to allow space for the labels. Sephardic signature circles shown filled in dark gray. Created with Network version 4.6.0.0, Fluxus Technology Ltd.
The Sephardic T2e5 cluster can be seen above the central node where it is apparent that it reflects fit data. There are no reticulations of the mutations in which the same mutation appears in different branches and it appears to be one offshoot of T2e. It is a nearly orphaned taxon separated by two mutations from its nearest neighbor of ancestral T2e. This reflects, as noted earlier, that thus far no instances of one mutation without the other (16114T, 16192T) within T2e has been identified.
Elsewhere within T2e, there were a total of 35 distinct haplotypes, which constitutes 29.4% of the 119 mitosearch sequences, similar to the percentage of distinct control region haplotypes (28.5%) that have been reported for all of T.39 T2e is widespread with reported maternal ancestry in order of frequency: United States, Unknown, Germany, Ireland, Scotland, England, France, Italy, Spain, Poland, Romania, Canada, USSR, and Mexico, and with 1 report each, Argentina, Australia, Austria, Azores, Belarus, Czech Republic, Greece, Iran, Latvia, The Netherlands, Norway, Slovakia, Sweden, and Switzerland. As the most frequent European ancestry for Americans is German followed by Irish (http://www.census.gov), this likely is a strong contributor to large representations of these locations in the American-centered data set. Despite bias of the public database, it is nonetheless apparent that this infrequent subhaplogroup of T is geographically diverse, like haplogroup T overall.
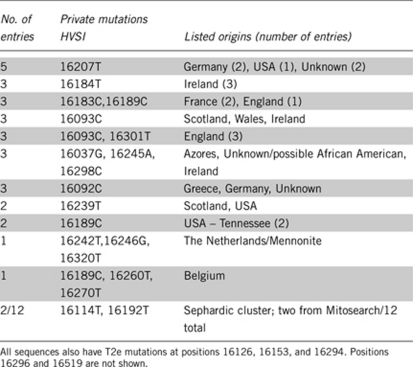
A total of 43 distinct mutations were found, with 16189C the most frequent occurring in 9 of the 119 individuals, or ∼7.5%. This is a common mutation that has independently arisen in many backgrounds.13 The phylogeny of T2e was found to be star-like with the most frequent haplotype the ancestral sequence (16126T-16153A-16294T, not considering positions 16296, 16519, or control region 2), with numerous much smaller clusters descended from that central node. In a previous study of haplogroup T control region,39 T2e (‘T5') was not star-like, presumably because there were only 15 T2e samples at the time. The ancestral T2e sequence was found in 68 samples or more than half (57.1%) of the present data set. The next most frequent haplotype was found in 5 individuals, followed by 6 haplotypes of 3 individuals each and 3 haplotypes of 2 individuals. These amounted to 29 individuals or approximately another quarter (24.4%) of the mitosearch data set. All haplotypes with two or more individuals are shown in Table 3. The remaining 25 haplotypes were unique and comprised less than a fifth (18.5%) of the data set. There is one reticulation that can be clearly seen in the network (square below central node) involving 16093 (lines 4 and 5 in Table 3), a position at which a relatively large amount of heteroplasmy has been reported40. There are also 2 orphan nodes separated by 3 mutations from the closest neighbor (lines 6 and 10, Table 3) and 2 nearly orphan nodes (lines 11 and 12, Table 3), including the Sephardic signature. The latter may be due to the very low numbers of Sephardim from which to draw samples or the possibility that sequences with only16114T or only 16192T within T2e do not reflect a surviving lineage.
Table 3. List of first control region (HVS1) private mutations within subhaplogroup T2e (former T5) from Mitosearch that have two or more entries with the same haplotype and/or reflect orphan or nearly orphan motifs differing by three or two mutations from the nearest match.
Coalescence analysis
Time estimates to the most recent common ancestor of the Sephardic signature T2e5 ranged all the way from after the expulsion – clearly impossible – to >15 000 years before present (YBP) (Fast: 338 YBP, 95% confidence interval (95% CI)=present to 763 YBP; Intermediate: 688 YBP, 95% CI=present−3820 YBP; slow: 6811 YBP, CI=present to 15 245). Given mutations rates that vary by two orders of magnitude,22 as well as other issues with mutation rates and the rho statistic,23, 41 at present coalescence analysis cannot be used to distinguish between different plausible timelines for the proposed Sephardic cluster.
Geographic distribution
Haplogroup T, comparison populations
The total T lineage occurs in many regions (Table 4). It also differs in its incidence across the populations (α2=95.10, P<0.001, df=9) ranging from 4.3% in an Ashkenazi group to 24.4% in Jewish Near Eastern communities of Iran and Iraq. There is also a high prevalence of total T in Italy, as expected, and in parts of Saudi Arabia. The large data sets from mitosearch and Genographic are in close accordance with the incidence of Haplogroup T in the smaller British and Irish Isles study (Scotland, Wales, England, Cornwall, Ireland) of ∼9% and likely reflect the large number of Americans with Western European ancestry that comprise these data sets.13
Table 4. Prevalence of subhaplogroups of T across different studies as percentage of all sequences found in each study.
| HG | OTT SEPH | OTT SEPH | SPA nw | POR | BRAZILa | IR J | ASH | ASH | GBI | ITA nc | SAU w | Mitb | NG |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | 194 | 191 | 995 | 549 | 96/247 | 217 | 565 | 762 | 1,610 | 395 | 72 | 34 000 | 76 638 |
| Ref | 5 | 25 | 27 | 29 | 24 | 5 | 5 | 25 | 27 | 30 | 26 | 13 | |
| T1 | 1.51% | – | 0.8 | 2.19 | 2.08/0.81 | 5.06 | 0.88 | – | 2.1 | 3.80 | 6.9 | 1.81 | 1.82 |
| T2e | 2.52 | – | 0.9 | 0.73 | 3.12/1.20 | 0 | 0.53 | – | 0.2 | 2.02 | 2.8 | 0.35 | 0.32 |
| T2b | 2.02 | – | 1.9 | 4.37 | 5.21/2.02 | 0.46 | 1.42 | – | 4.2 | 3.54 | 1.4 | 4.06 | 3.91 |
| T2c | 1.01 | – | 1.1 | 0.55 | 1.04/0.40 | 13.82 | 0.18 | – | 0.4 | 1.77 | 1.4 | 0.30 | 0.43 |
| (T2 total | 5.55 | – | 3.9 | 5.65 | 9.37/3.62 | 14.28 | 2.13 | – | 4.8 | 7.34 | 5.6 | 4.71 | 4.67 |
| T4 | 0 | – | 0.2 | 0.18 | 0/0 | 0 | 0.71 | – | 0.2 | 0.25 | 0 | 0.20 | 0.24 |
| T other | 5.04 | – | 2.0 | 1.09 | 2.08/0.81 | 5.06 | 1.06 | – | 2.0 | 2.02 | 0 | 2.38 | 1.93 |
| T tot | 11.11 | 14.00 | 6.9 | 9.21 | 13.58/5.24 | 24.40 | 4.76 | 4.3 | 9.1 | 13.67 | 12.5 | 9.07 | 8.66 |
Abbreviations: ASH, Ashkenazi; GBI, Great Britain and Ireland; HG, haplogroup; IRJ, Iraqi and Iranian Jewish; ITAnc, North Central Italy; Mit, Mitosearch; n, number of subjects; NG, National Genographic; OTT SEPH, Ottoman Sephardic; POR, Portugal; Ref, reference; SAUw, Western Saudi Arabia; SPA nw, Northwest Spain; T tot, Total % of all Ts; -, unknown.
The greater prevalence of T2b than T2e except in Sephardim and Saudi Arabia must be noted.
First number is % of European haplogroups only; second number is % of entire sample.
T tot includes correction of .10 for double counting 35 samples in both T2e and its listed category.
Haplogroup T, Sephardic population
The four studies involving Sephardic populations have incidences of T of 4.9%, not reported, 14 and 11.1%. The first study28 was based on a limited number of samples (n=41, 2 of haplogroup T), and in the second study,42 Morocco, with its large pre-existing Jewish populations in the North African region before the Iberian influx, was the only representation of Sephardim. The remaining two estimates are shown in Table 4. The reported 14% incidence25 may be an inflated estimate because it includes non-Sephardic native near Eastern Jews from Israel that consider themselves to be Sephardic (remaining subjects descend from Cyprus, Bulgaria, Greece, Spain, Italy, and Turkey). The final estimate5 with >100 subjects from Turkey and Bulgaria is likely currently the most accurate representation of the Iberian Sephardic population, at least for Ottoman groups. Comparison with the other geographic regions finds this incidence of total T to be larger than the host population of Northern Spain (α2=4.46, P<0.05, df=1), and may be slightly larger than the host population of Portugal, although it is not found to be significantly different (P>0.1). The incidence of all of haplogroup T in Sephardim is comparable to many regions, including countries of Western Europe combined, a world-wide database and Italy.
Subhaplogroup T2e and T2b in Sephardic and comparison populations
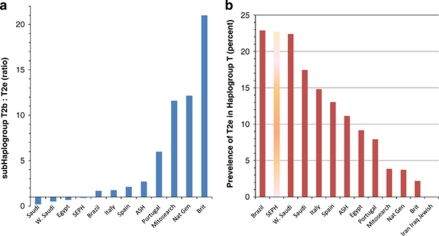
More illuminating is the geographic distribution of T2e compared with other subhaplogroups of T, which was found to be notably different across the populations. Although T2e is found widely, it is less frequent than the sister lineage T2b in all the populations shown in Table 4 except two: Sephardic Ottoman and Western Saudi Arabia (likely Egypt as well, see Abu-Amero et al26, supplementary materials.) This suggests that despite the special problems inherent in comparing Sephardic Ottoman with its previous Iberian geographic hosts – that is, any similarity could merely indicate that the comparison host group contained a substantial presence of once Sephardic Conversos – Sephardic subhaplogroups can be distinguished as different from those host populations. The proportion of T2e to T2b in Ottoman Sephardim is closer to Western Saudi Arabia than to Spain or Portugal. The frequency of T2e vs T2b is significantly different between Sephardim and Spain (Fisher's exact test, P<0.025, although due to the rarity of T2e, comparisons should be repeated with larger samples). The distribution of the T haplogroup within Spain and Portugal is more similar to that found throughout Europe, with a high relative incidence of T2b instead. The overall incidence of T2e in the current comparison groups is the highest in Western Saudi Arabia (2.8% of all haplogroup sequences), Sephardim (2.5%), and Italy (2.0%) where it is appears 10 times more frequent than in Britain (0.2%). The ratio of subhaplogroup T2e to T2b and the incidence of T2e within T overall are shown in Figure 2.
Figure 2.
Prevalence of subhaplogroup T2e in different regional data samples. (a) The proportion of T2b to T2e in each population. A ratio of 1 (horizontal axis) reflects equal incidence of T2e and T2b, values <1 indicate greater incidence of T2e, and values >1 indicate greater incidence of T2b than T2e. (b) The incidence of T2e as a percentage of all haplogroup T present in the study. Abbreviations: Saudi=Saudi Arabia, W Saudi=West Saudi Arabia, SEPH (shown as lighter shaded bars)=Ottoman Sephardic, ASH=Ashkenazi, Nat Gen=National Genographic Project, Brit=British and Irish Isles, J=Jewish.
Discussion
A small Sephardic signature (‘T2e5') has been identified within subhaplogroup T2e mitochondrial DNA. Its exclusive presence in individuals of Iberian descent points to origins in Iberia, which hosted Sephardim for 1500 years. However, it cannot be ruled out that the distinguishing mutations originated elsewhere, with migration to Iberia the only surviving lineage.
We found T2e to be widespread geographically, despite the infrequency of this subhaplogroup. In addition, the incidence decreases from the Western Arabian Peninsula to Italy to Iberia and into Western Europe. Saudi Arabia may be a recipient of migration rather than the center of expansion for T2e because ancestral sequences are not found there,26 just as Abu-Amero et al26 conclude for the highly prevalent non-ancestral J haplogroups in the region. The ratio of T2e to sister subhaplogroup T2b was found to vary 40-fold, with caveats about dividing by small numbers. This study is to our knowledge the first to report the greatly changing presence of T2e across geographic regions.
In Ottoman Sephardic Jews, the incidence of T2e was greater than hosts Spain or Portugal and more similar to Northern and Western Saudi Arabia, North Central Italy, and Egypt. The frequency of T2e was higher than T2b, like Saudi Arabia and Egypt, and unlike the remaining comparison populations. Interestingly, T2e does not appear more prevalent than T2b in any of the Near Eastern region of Iran, Palestine, Jordan, or Turkey.26 The greater genetic similarity of Sephardim to parts of Saudi Arabia, Italy, and Egypt than to Spain or Portugal suggests a maternal migration path from the Middle East with one route going back to Western Africa and another route to Iberia, with a stop in Italy (ignoring back migrations from Iberia). This not-unexpected migration route is consistent with many scenarios of interest for origins of the Sephardic cluster in Iberia. These include Jewish settlers seeking asylum after destruction of temples in Jerusalem by Romans and Babylonians 2000–2500 years ago, slightly earlier Jewish settlers in Iberia,7, 43 non-Jewish Muslims in the dispersal of Islam 1000+ years ago, non-Jewish Iberian peopling 2500+ years ago that predates all Jewish influx,44 and settlers in Iberia (or Italy) >5000 years ago that entirely predate the existence of Jewish groups. Thus, what is arguably the most contentious issue of whether there is genetic evidence of original Jewish DNA for the Sephardic line cannot be resolved.
The relatively high presence of T2e in Sephardim suggests that the subhaplogroup occurred relatively early in the Sephardic population because if it appeared instead at the end of the community's isolation in Iberia, there would be insufficient time for its spread in the population. Why then is the frequency of T2e matches in Spain and Portugal currently so low? Similarly, fewer Sephardic signature T2e5 matches were found in Iberia than in Northern Mexico and Southwest United States. Given once-Jewish Conversos are present in Iberia and not only in the New World, why is the frequency of T2e5 matches in Spain and Portugal currently so low? Infrequent T2e and T2e5 would have constituted a greater proportion of those that left compared with those that stayed behind. Less competition implies a greater opportunity to flourish. Consequences of competition may be especially notable in less successful subhaplogroups that start out with low frequencies in the host population and may grow to greater numbers only in the exile populations.
Is it meaningful to state that there is a less pronounced founder effect in Sephardic than in Ashkenazi communities? At least 200 000 of Spain and Portugal's Jewish exilers settled in the Ottoman Empire,7 bringing with them their genetic diversity. Ashkenazim during the same time period had a smaller population, which can contribute to less genetic variability and therefore a more prominent founder effect of larger, but fewer, successful founding lineages. However, despite the seeming consistency of history, the comparative claim still requires scrutiny. Behar et al5 noted that an earlier study by Thomas et al45 reach the opposite conclusion of non-Ashkenazi Jewish groups having the more narrow founder effect. An important difference comes from the Ashkenazi data: whereas Behar et al reported that one haplogroup (K) is found in about a third of the Ashkenazi maternal lineage, the top five haplotypes found in the study by Thomas et al were not from that haplogroup. Haplogroup K manifests in close to 40% in the Polish Jewish community, but much less so in other Ashkenazi groups (Behar et al,4 supplementary materials; also see Feder et al42). Testing from Ashkenazi backgrounds having little representation of the haplogroup (not reported in the study by Thomas et al) would account for not finding the strong founder effect and instead reaching a different conclusion. The difference between the conclusions highlights the fact that different averages leads to different interpretations. A conclusion that the Ashkenazi maternal founder effect is more pronounced (ie, less genetic variability) than in many Sephardic communities is based on pooling all of the Ashkenazi communities but keeping the Sephardic communities separate. At present, we know that the Sephardic Turkish population has a less narrow founder effect than does the Ashkenazi Polish population. Further comparisons of interest among Jewish populations await comparable averaging.
In conclusion, the findings in the present study suggest that within the genetic diversity of Sephardim, subhaplogroup T2e5 is a small signature. Clearly, the majority of Sephardim will not have this haplotype. Nonetheless, populations that have large numbers of small-sized founders without prominent founder effects will benefit from registries of signatures. Haplogroup T, an ancient lineage from the Near East, may have a notable presence in Sephardim carried through for centuries, bringing with it the possible consequences of its unusual mutations that affect mitochondrial ATP production, such as increased coronary artery disease,46 cardiomyopathy,47 macular degeneration,48 and unlikely endurance athletes.49 In any event, one woman from Iberia who lived between 500 and likely 2000 years ago has modern-day decedents who remained in Portugal, migrated to Turkey, Bulgaria, the United States, Mexico, and Brazil.
Acknowledgments
I thank Family Tree DNA, especially, Bennett Greenspan and Thomas Krahn, for sequencing the Salonican sample, supplying the pherograms, and providing the total number of records in Mitosearch; Valery Zaporozhchenko for searching database no. 5; Antonia Picornell for pherograms of the Turkish samples from Picornell et al; and Sergio Pena for discussion of sample Br 164 from Alves-Silva et al.
The author declares no conflict of interest.
Footnotes
Note
Accession numbers for sequences referred to are: JN819272 (this study), AF381985, non-GenBank, non-GenBank, JN030346, JN828512 (this study), EF177410.1, EU258890.1, FJ178379.1, AF346982.1, EU703624.1, EU597536.1, FJ238094.1, GU944474.1, GU565218.1, EF556188.1, AY714029, HM852862. JF831421, JF921152, JF937679, EF661001, JF893457, JF831146, JF903810, EF060363, and JF96544
References
- Mayr E. Animal Species and Evolution. Cambridge: Belknap Press of Harvard University Press; 1963. [Google Scholar]
- Richards M, Macaulay V, Hickey E, et al. Tracing European founder lineages in the Near Eastern mtDNA pool. Am J Hum Genet. 2000;67:1251–1276. [PMC free article] [PubMed] [Google Scholar]
- Richards MB, Macaulay VA, Bandelt H-J, Sykes BC. Phylogeography of mitochondrial DNA in Western Europe. Ann Hum Genet. 1998;62:241–260. doi: 10.1046/j.1469-1809.1998.6230241.x. [DOI] [PubMed] [Google Scholar]
- Behar DM, Metspalu E, Kivisild T, et al. Counting the founders: the matrilineal genetic ancestry of the Jewish Diaspora. PLoS ONE. 2008;3:e2062. doi: 10.1371/journal.pone.0002062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behar DM, Metspalu E, Kivisild T, et al. The matrilineal ancestry of Ashkenazi Jewry: portrait of a recent founder event. Am J Hum Genet. 2006;78:487–497. doi: 10.1086/500307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behar DM, Hammer MF, Garrigan D, et al. MtDNA evidence for a genetic bottleneck in the early history of the Ashkenazi Jewish population. Eur J Hum Genet. 2004;12:355–364. doi: 10.1038/sj.ejhg.5201156. [DOI] [PubMed] [Google Scholar]
- Gerber J.The Jews of Spain: A History of the Sephardic Experience1st ed.New York: Free Press; 1994 [Google Scholar]
- Campbell A, Mrázek J, Karlin S. Genome signature comparisons among prokaryote, plasmid, and mitochondrial DNA. Proc Natl Acad Sci USA. 1999;96:9184–9189. doi: 10.1073/pnas.96.16.9184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebman SB. The Jews in New Spain: Faith, Flame, and the Inquisition. Coral Gables, FL: Univ of Miami Pr; 1970. [Google Scholar]
- Richards M, Côrte-Real H, Forster P, et al. Paleolithic and neolithic lineages in the European mitochondrial gene pool. Am J Hum Genet. 1996;59:185–203. [PMC free article] [PubMed] [Google Scholar]
- Macaulay V, Richards M, Hickey E, et al. The emerging tree of West Eurasian mtDNAs: a synthesis of control-region sequences and RFLPs. Am J Hum Genet. 1999;64:232–249. doi: 10.1086/302204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torroni A, Huoponen K, Francalacci P, et al. Classification of European mtDNAs from an analysis of three European populations. Genetics. 1996;144:1835–1850. doi: 10.1093/genetics/144.4.1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behar DM, Rosset S, Blue-Smith J, et al. The Genographic Project public participation mitochondrial DNA database. PLoS Genet. 2007;3:e104. doi: 10.1371/journal.pgen.0030104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Oven M, Kayser M. Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Hum Mutat. 2009;30:E386–E394. doi: 10.1002/humu.20921. [DOI] [PubMed] [Google Scholar]
- Ingman M, Kaessmann H, Pääbo S, Gyllensten U. Mitochondrial genome variation and the origin of modern humans. Nature. 2000;408:708–713. doi: 10.1038/35047064. [DOI] [PubMed] [Google Scholar]
- Palanichamy MG, Sun C, Agrawal S, et al. Phylogeny of mitochondrial DNA macrohaplogroup N in India, based on complete sequencing: implications for the peopling of South Asia. Am J Hum Genet. 2004;75:966–978. doi: 10.1086/425871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrnstadt C, Elson JL, Fahy E, et al. Reduced-median-network analysis of complete mitochondrial DNA coding-region sequences for the major African, Asian, and European haplogroups. Am J Hum Genet. 2002;70:1152–1171. doi: 10.1086/339933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sykes B. The Blood of the Isles. London: Bantam Press; 2006. [Google Scholar]
- Pike DA, Barton TJ, Bauer SL, Kipp EB. mtDNA haplogroup T phylogeny based on full mitochondrial sequences. J Genet Geneal. 2010;6:1–24. [Google Scholar]
- Forster P, Harding R, Torroni A, Bandelt HJ. Origin and evolution of Native American mtDNA variation: a reappraisal. Am J Hum Genet. 1996;59:935–945. [PMC free article] [PubMed] [Google Scholar]
- Saillard J, Forster P, Lynnerup N, Bandelt HJ, Nørby S. mtDNA variation among Greenland Eskimos: the edge of the Beringian expansion. Am J Hum Genet. 2000;67:718–726. doi: 10.1086/303038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons TJ, Muniec DS, Sullivan K, et al. A high observed substitution rate in the human mitochondrial DNA control region. Nat Genet. 1997;15:363–368. doi: 10.1038/ng0497-363. [DOI] [PubMed] [Google Scholar]
- Santos C, Montiel R, Sierra B, et al. Understanding differences between phylogenetic and pedigree-derived mtDNA mutation rate: a model using families from the Azores Islands (Portugal) Mol Biol Evol. 2005;22:1490–1505. doi: 10.1093/molbev/msi141. [DOI] [PubMed] [Google Scholar]
- Alves-Silva J, da Silva Santos M, Guimarães PE, Ferreira AC, Bandelt HJ, Pena SD, Prado VF. The ancestry of Brazilian mtDNA lineages. Am J Hum Genet. 2000;67:444–461. doi: 10.1086/303004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feder J, Blech I, Ovadia O, et al. Differences in mtDNA haplogroup distribution among 3 Jewish populations alter susceptibility to T2DM complications. BMC Genomics. 2008;9:198. doi: 10.1186/1471-2164-9-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abu-Amero KK, Larruga JM, Cabrera VM, González AM. Mitochondrial DNA structure in the Arabian Peninsula. BMC Evol Biol. 2008;8:45. doi: 10.1186/1471-2148-8-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García O, Fregel R, Larruga JM, Alvarez V, Yurrebaso I, Cabrera VM, González AM. Using mitochondrial DNA to test the hypothesis of a European post-glacial human recolonization from the Franco-Cantabrian refuge [Internet] Heredity. 2010;106:37–45. doi: 10.1038/hdy.2010.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picornell A, Giménez P, Castro JA, Ramon MM. Mitochondrial DNA sequence variation in Jewish populations. Int J Legal Med. 2006;120:271–281. doi: 10.1007/s00414-006-0083-0. [DOI] [PubMed] [Google Scholar]
- Pereira L, Cunha C, Amorim A. Predicting sampling saturation of mtDNA haplotypes: an application to an enlarged Portuguese database. Int J Legal Med. 2004;118:132–136. doi: 10.1007/s00414-003-0424-1. [DOI] [PubMed] [Google Scholar]
- Turchi C, Buscemi L, Previderè C, et al. Italian mitochondrial DNA database: results of a collaborative exercise and proficiency testing. Int J Legal Med. 2008;122:199–204. doi: 10.1007/s00414-007-0207-1. [DOI] [PubMed] [Google Scholar]
- Preacher KJ.Calculation for the chi-square test: an interactive calculation tool for chi-square tests of goodness of fit and independence [Computer software] http://www.quantpsy.org , 2001
- Raphael DT. Conquistadores and Crypto-Jews of Monterrey. Valley Village, Calif: Carmi House; 2001. [Google Scholar]
- K AMG. Diccionario Porrua De Historia, Biografia Y Geografia De Mexico. Cuarta Edicion. Mexico: Editorial Porrua S.A;1976
- deSola CardozaA.Texas Mexican Secret Spanish Jews Today [Internet] Los Muestros; 1997. 11Available from: http://www.sefarad.org/publication/lm/011/texas.html . [Google Scholar]
- Pereira L, Prata MJ, Amorim A. Diversity of mtDNA lineages in Portugal: not a genetic edge of European variation. Ann Hum Genet. 2000;64 (Pt 6:491–506. doi: 10.1046/j.1469-1809.2000.6460491.x. [DOI] [PubMed] [Google Scholar]
- Lavender A. Brazil's Secret Jews and New Christians: scholarly disagreements about numbers, identities, and the future. J Spanish Portuguese Italian Crypto Jews. 2010;2:77–109. [Google Scholar]
- Maca-Meyer N, González AM, Larruga JM, Flores C, Cabrera VM. Major genomic mitochondrial lineages delineate early human expansions. BMC Genet. 2001;2:13. doi: 10.1186/1471-2156-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y-G, Salas A, Logan I, Bandelt H-J.mtDNA data mining in GenBank needs surveying Am J Hum Genet 200985929–933.author reply 933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pike DA. Phylogenetic networks for the human mtDNA haplogroup T. J Genet Geneal. 2006;2:1–11. [Google Scholar]
- Brandstätter A, Parson W. Mitochondrial DNA heteroplasmy or artefacts–a matter of the amplification strategy. Int J Legal Med. 2003;117:180–184. doi: 10.1007/s00414-002-0350-7. [DOI] [PubMed] [Google Scholar]
- Cox MP. Accuracy of molecular dating with the rho statistic: deviations from coalescent expectations under a range of demographic models. Hum Biol. 2008;80:335–357. doi: 10.3378/1534-6617-80.4.335. [DOI] [PubMed] [Google Scholar]
- Feder J, Ovadia O, Glaser B, Mishmar D. Ashkenazi Jewish mtDNA haplogroup distribution varies among distinct subpopulations: lessons of population substructure in a closed group. Eur J Hum Genet. 2007;15:498–500. doi: 10.1038/sj.ejhg.5201764. [DOI] [PubMed] [Google Scholar]
- Malka ES. Sephardi Jews A Pageant of Spanish Portuguese and Oriental Judaism between the Cross and the Crescent. Trenton, NJ: Edmond s. Malka; 1979. [Google Scholar]
- Sampietro ML, Caramelli D, Lao O, et al. The genetics of the pre-Roman Iberian Peninsula: a mtDNA study of ancient Iberians. Ann Hum Genet. 2005;69 (Pt 5:535–548. doi: 10.1111/j.1529-8817.2005.00194.x. [DOI] [PubMed] [Google Scholar]
- Thomas MG, Weale ME, Jones AL, et al. Founding mothers of Jewish communities: geographically separated Jewish groups were independently founded by very few female ancestors. Am J Hum Genet. 2002;70:1411–1420. doi: 10.1086/340609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofler B, Mueller EE, Eder W, et al. Mitochondrial DNA haplogroup T is associated with coronary artery disease and diabetic retinopathy: a case control study. BMC Med Genet. 2009;10:35. doi: 10.1186/1471-2350-10-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro MG, Huerta C, Reguero JR, et al. Mitochondrial DNA haplogroups in Spanish patients with hypertrophic cardiomyopathy. Int J Cardiol. 2006;112:202–206. doi: 10.1016/j.ijcard.2005.09.008. [DOI] [PubMed] [Google Scholar]
- SanGiovanni JP, Arking DE, Iyengar SK, et al. Mitochondrial DNA variants of respiratory complex I that uniquely characterize haplogroup T2 are associated with increased risk of age-related macular degeneration. PLoS ONE. 2009;4:e5508. doi: 10.1371/journal.pone.0005508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro MG, Terrados N, Reguero JR, Alvarez V, Coto E. Mitochondrial haplogroup T is negatively associated with the status of elite endurance athlete. Mitochondrion. 2007;7:354–357. doi: 10.1016/j.mito.2007.06.002. [DOI] [PubMed] [Google Scholar]