Abstract
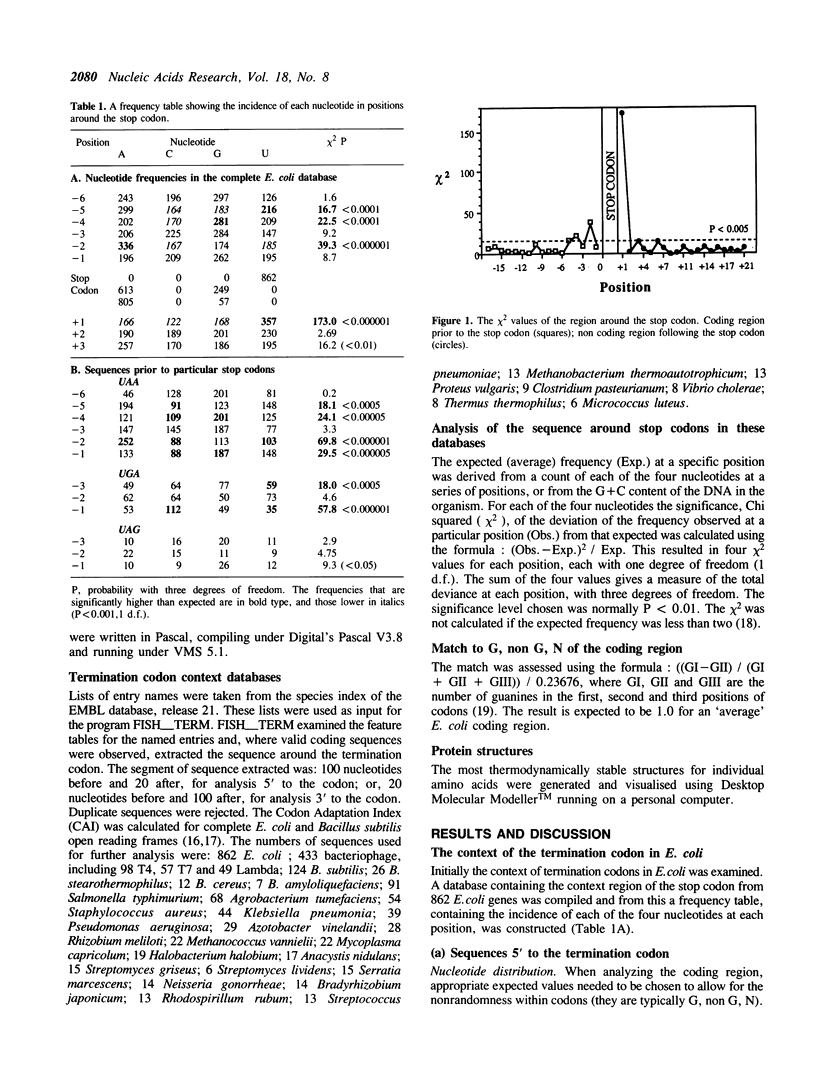
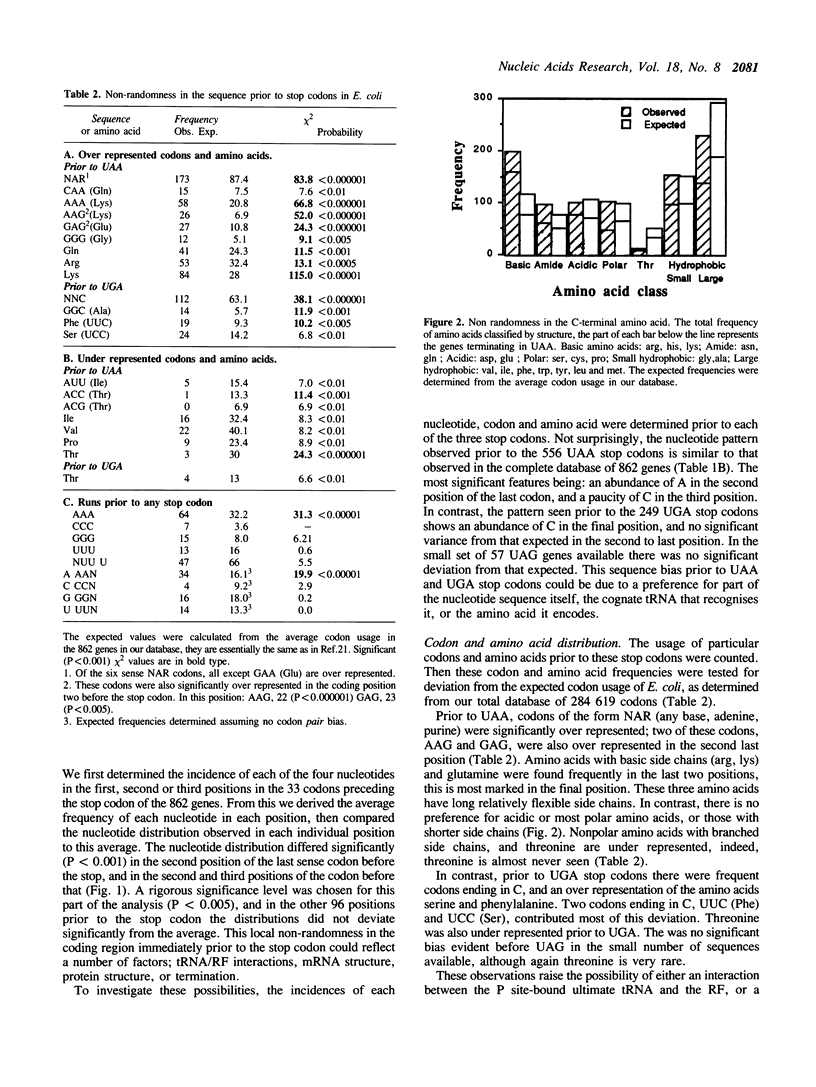
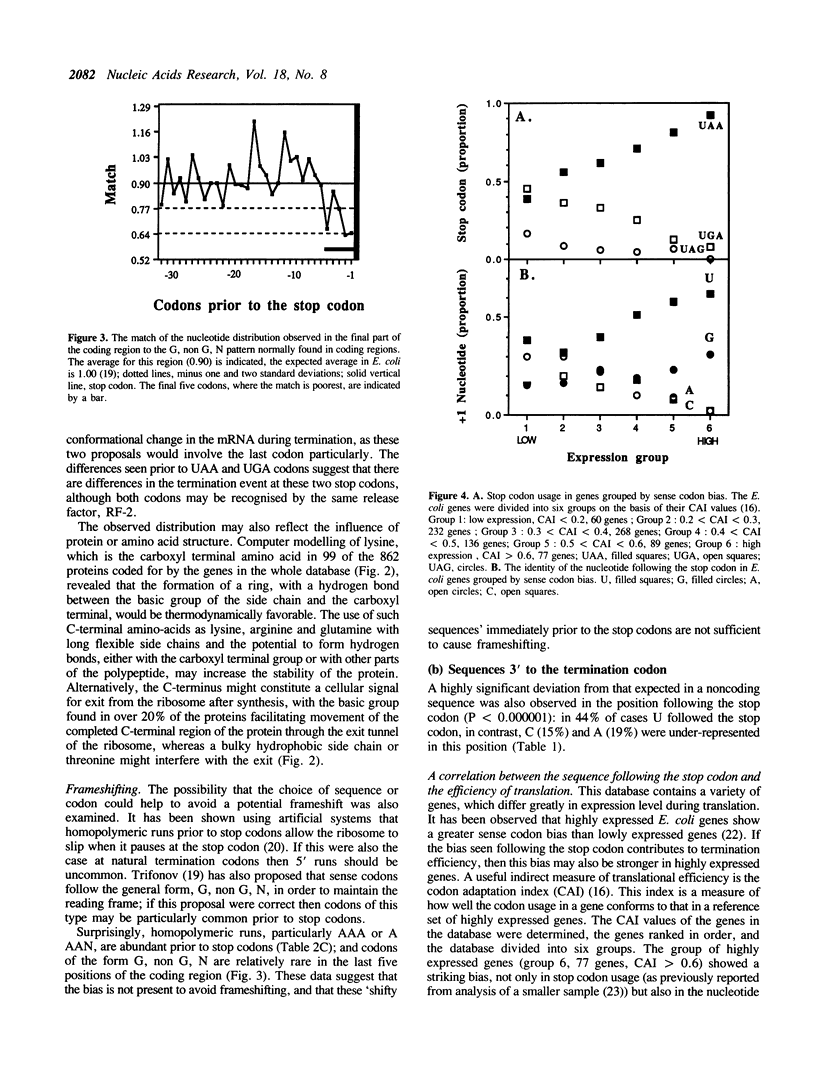
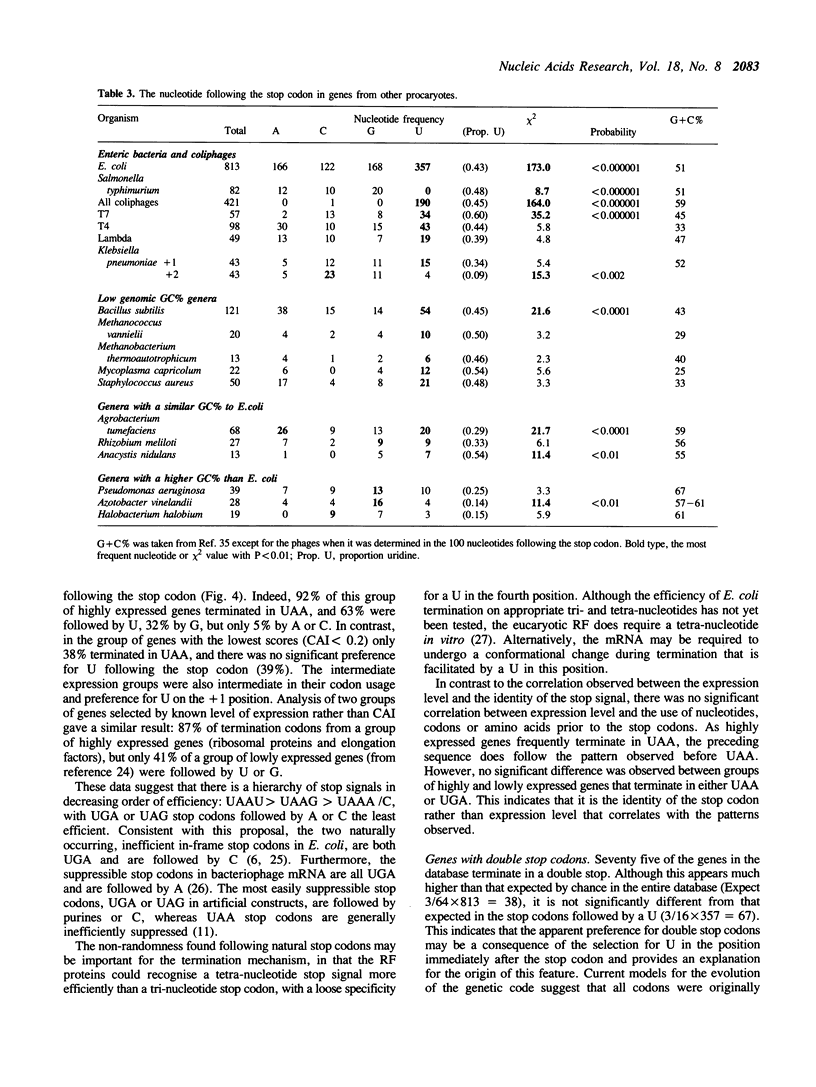
The sequences around the stop codons of 862 Escherichia coli genes have been analysed to identify any additional features which contribute to the signal for the termination of protein synthesis. Highly significant deviations from the expected nucleotide distribution were observed, both before and after the stop codon. Immediately prior to UAA stop codons in E. coli there is a preference for codons of the form NAR (any base, adenine, purine), and in particular those that code for glutamine or the basic amino acids. In contrast, codons for threonine or branched nonpolar amino acids were under-represented. Uridine was over-represented in the nucleotide position immediately following all three stop codons, whereas adenine and cytosine were under-represented. This pattern is accentuated in highly expressed genes, but is not as marked in either lowly expressed genes or those that terminate in UAG, the codon specifically recognised by polypeptide chain release factor-1. These observations suggest that for the efficient termination of protein synthesis in E. coli, the 'stop signal' may be a tetranucleotide, rather than simply a tri-nucleotide codon, and that polypeptide chain release factor-2 recognises this extended signal. The sequence following stop codons was analysed in genes from several other procaryotes and bacteriophages. Salmonella typhimurium, Bacillus subtilis, bacteriophages and the methanogenic archaebacteria showed a similar bias to E. coli.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aota S., Gojobori T., Ishibashi F., Maruyama T., Ikemura T. Codon usage tabulated from the GenBank Genetic Sequence Data. Nucleic Acids Res. 1988;16 (Suppl):r315–r402. doi: 10.1093/nar/16.suppl.r315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudet A. L., Caskey C. T. Mammalian peptide chain termination. II. Codon specificity and GTPase activity of release factor. Proc Natl Acad Sci U S A. 1971 Mar;68(3):619–624. doi: 10.1073/pnas.68.3.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benkovic S. J., Fierke C. A., Naylor A. M. Insights into enzyme function from studies on mutants of dihydrofolate reductase. Science. 1988 Mar 4;239(4844):1105–1110. doi: 10.1126/science.3125607. [DOI] [PubMed] [Google Scholar]
- Bossi L. Context effects: translation of UAG codon by suppressor tRNA is affected by the sequence following UAG in the message. J Mol Biol. 1983 Feb 15;164(1):73–87. doi: 10.1016/0022-2836(83)90088-8. [DOI] [PubMed] [Google Scholar]
- Bossi L., Ruth J. R. The influence of codon context on genetic code translation. Nature. 1980 Jul 10;286(5769):123–127. doi: 10.1038/286123a0. [DOI] [PubMed] [Google Scholar]
- Craigen W. J., Caskey C. T. Translational frameshifting: where will it stop? Cell. 1987 Jul 3;50(1):1–2. doi: 10.1016/0092-8674(87)90652-0. [DOI] [PubMed] [Google Scholar]
- Craigen W. J., Cook R. G., Tate W. P., Caskey C. T. Bacterial peptide chain release factors: conserved primary structure and possible frameshift regulation of release factor 2. Proc Natl Acad Sci U S A. 1985 Jun;82(11):3616–3620. doi: 10.1073/pnas.82.11.3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggertsson G., Söll D. Transfer ribonucleic acid-mediated suppression of termination codons in Escherichia coli. Microbiol Rev. 1988 Sep;52(3):354–374. doi: 10.1128/mr.52.3.354-374.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelberg-Kulka H., Schoulaker-Schwarz R. A flexible genetic code, or why does selenocysteine have no unique codon? Trends Biochem Sci. 1988 Nov;13(11):419–421. doi: 10.1016/0968-0004(88)90209-5. [DOI] [PubMed] [Google Scholar]
- Engelberg-Kulka H. UGA suppression by normal tRNA Trp in Escherichia coli: codon context effects. Nucleic Acids Res. 1981 Feb 25;9(4):983–991. doi: 10.1093/nar/9.4.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folley L. S., Yarus M. Codon contexts from weakly expressed genes reduce expression in vivo. J Mol Biol. 1989 Oct 5;209(3):359–378. doi: 10.1016/0022-2836(89)90003-x. [DOI] [PubMed] [Google Scholar]
- Gouy M., Gautier C. Codon usage in bacteria: correlation with gene expressivity. Nucleic Acids Res. 1982 Nov 25;10(22):7055–7074. doi: 10.1093/nar/10.22.7055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutman G. A., Hatfield G. W. Nonrandom utilization of codon pairs in Escherichia coli. Proc Natl Acad Sci U S A. 1989 May;86(10):3699–3703. doi: 10.1073/pnas.86.10.3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagervall T. G., Björk G. R. Undermodification in the first position of the anticodon of supG-tRNA reduces translational efficiency. Mol Gen Genet. 1984;196(2):194–200. doi: 10.1007/BF00328050. [DOI] [PubMed] [Google Scholar]
- Kohli J., Grosjean H. Usage of the three termination codons: compilation and analysis of the known eukaryotic and prokaryotic translation termination sequences. Mol Gen Genet. 1981;182(3):430–439. doi: 10.1007/BF00293932. [DOI] [PubMed] [Google Scholar]
- Lee C. C., Timms K. M., Trotman C. N., Tate W. P. Isolation of a rat mitochondrial release factor. Accommodation of the changed genetic code for termination. J Biol Chem. 1987 Mar 15;262(8):3548–3552. [PubMed] [Google Scholar]
- Martin R., Weiner M., Gallant J. Effects of release factor context at UAA codons in Escherichia coli. J Bacteriol. 1988 Oct;170(10):4714–4717. doi: 10.1128/jb.170.10.4714-4717.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. H., Albertini A. M. Effects of surrounding sequence on the suppression of nonsense codons. J Mol Biol. 1983 Feb 15;164(1):59–71. doi: 10.1016/0022-2836(83)90087-6. [DOI] [PubMed] [Google Scholar]
- Osawa S., Jukes T. H. Codon reassignment (codon capture) in evolution. J Mol Evol. 1989 Apr;28(4):271–278. doi: 10.1007/BF02103422. [DOI] [PubMed] [Google Scholar]
- Osawa S., Jukes T. H. Evolution of the genetic code as affected by anticodon content. Trends Genet. 1988 Jul;4(7):191–198. doi: 10.1016/0168-9525(88)90075-3. [DOI] [PubMed] [Google Scholar]
- Rydén S. M., Isaksson L. A. A temperature-sensitive mutant of Escherichia coli that shows enhanced misreading of UAG/A and increased efficiency for some tRNA nonsense suppressors. Mol Gen Genet. 1984;193(1):38–45. doi: 10.1007/BF00327411. [DOI] [PubMed] [Google Scholar]
- Salser W. The influence of the reading context upon the suppression of nonsense codons. Mol Gen Genet. 1969 Oct 13;105(2):125–130. doi: 10.1007/BF00445682. [DOI] [PubMed] [Google Scholar]
- Sharp P. M., Bulmer M. Selective differences among translation termination codons. Gene. 1988;63(1):141–145. doi: 10.1016/0378-1119(88)90553-7. [DOI] [PubMed] [Google Scholar]
- Sharp P. M., Li W. H. The codon Adaptation Index--a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987 Feb 11;15(3):1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. M., Rogers M. S., McConnell D. J. Selection pressures on codon usage in the complete genome of bacteriophage T7. J Mol Evol. 1984;21(2):150–160. doi: 10.1007/BF02100089. [DOI] [PubMed] [Google Scholar]
- Shields D. C., Sharp P. M. Synonymous codon usage in Bacillus subtilis reflects both translational selection and mutational biases. Nucleic Acids Res. 1987 Oct 12;15(19):8023–8040. doi: 10.1093/nar/15.19.8023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D., Yarus M. tRNA-tRNA interactions within cellular ribosomes. Proc Natl Acad Sci U S A. 1989 Jun;86(12):4397–4401. doi: 10.1073/pnas.86.12.4397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stormo G. D., Schneider T. D., Gold L. Quantitative analysis of the relationship between nucleotide sequence and functional activity. Nucleic Acids Res. 1986 Aug 26;14(16):6661–6679. doi: 10.1093/nar/14.16.6661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trifonov E. N. Translation framing code and frame-monitoring mechanism as suggested by the analysis of mRNA and 16 S rRNA nucleotide sequences. J Mol Biol. 1987 Apr 20;194(4):643–652. doi: 10.1016/0022-2836(87)90241-5. [DOI] [PubMed] [Google Scholar]
- Valle R. P., Morch M. D. Stop making sense: or Regulation at the level of termination in eukaryotic protein synthesis. FEBS Lett. 1988 Aug 1;235(1-2):1–15. doi: 10.1016/0014-5793(88)81225-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss R., Gallant J. Mechanism of ribosome frameshifting during translation of the genetic code. 1983 Mar 31-Apr 6Nature. 302(5907):389–393. doi: 10.1038/302389a0. [DOI] [PubMed] [Google Scholar]
- Zinoni F., Birkmann A., Stadtman T. C., Böck A. Nucleotide sequence and expression of the selenocysteine-containing polypeptide of formate dehydrogenase (formate-hydrogen-lyase-linked) from Escherichia coli. Proc Natl Acad Sci U S A. 1986 Jul;83(13):4650–4654. doi: 10.1073/pnas.83.13.4650. [DOI] [PMC free article] [PubMed] [Google Scholar]



