Abstract
The pattern of population genetic variation and allele frequencies within a species are unstable and are changing over time according to different evolutionary factors. For humans, it is possible to combine detailed patrilineal genealogical records with deep Y-chromosome (Y-chr) genotyping to disentangle signals of historical population genetic structures because of the exponential increase in genetic genealogical data. To test this approach, we studied the temporal pattern of the ‘autochthonous' micro-geographical genetic structure in the region of Brabant in Belgium and the Netherlands (Northwest Europe). Genealogical data of 881 individuals from Northwest Europe were collected, from which 634 family trees showed a residence within Brabant for at least one generation. The Y-chr genetic variation of the 634 participants was investigated using 110 Y-SNPs and 38 Y-STRs and linked to particular locations within Brabant on specific time periods based on genealogical records. Significant temporal variation in the Y-chr distribution was detected through a north–south gradient in the frequencies distribution of sub-haplogroup R1b1b2a1 (R-U106), next to an opposite trend for R1b1b2a2g (R-U152). The gradient on R-U106 faded in time and even became totally invisible during the Industrial Revolution in the first half of the nineteenth century. Therefore, genealogical data for at least 200 years are required to study small-scale ‘autochthonous' population structure in Western Europe.
Keywords: Brabant, genetic genealogy, historical migration, human population structure, Y-chromosome
Introduction
Since the beginning of genetic research, scientists have been interested in how evolution and history have influenced the population structure of organisms. Patterns of genetic differentiation among populations within species are unstable in time according to the different evolutionary factors influencing allele frequencies (eg, genetic drift, gene flow, selection).1 It is quite difficult to understand how the pattern of genetic variation of a species changed over generations based on the data of modern, living populations, although most phylogeographic studies try to do so.2 Through methods based on coalescence of haploid markers, it is possible to get information about the effect of (pre)-historical events on a species.3 In humans, hereditary surnames and genealogical records may assist these methods by eliminating recent demographic and migration events from the ‘autochthonous' population pattern. This is especially interesting for Y-chromosomal analysis because as the patrilineal hereditary family names are co-inherited with the Y-chromosomes (Y-chr), a surname should, within a genealogy, correlate with a particular Y-chr variant.4
The human Y-chr has proven to be a good detector of historic migration events because of its mostly non-recombining inheritance and its small effective population size.5 Moreover, it is an excellent marker to study population differentiation on a regional scale because it was shown that patrilineal markers exhibit a larger geographic specificity than matrilineal or autosomal ones, because of the plausible reduced mobility of males compared with females.5 By using the link between genealogical records and Y-chr, the effect of migrations on population structures in the last centuries may be identified and temporally distinguished from each other. Today, genetic genealogy has mostly been used to test for relatedness,6, 7 to estimate non-paternity in a population8 and to measure mutation rates of Y-STRs9 and Y-SNPs.10 As genetic genealogical databases are increasing exponentially,4 it becomes possible to combine detailed genealogical records with Y-chr profiles to disentangle the effect of historic events on human population genetics. Y-SNP mutation rates are so low within the time-depth of genealogical records, that the Y-SNPs are not expected to undergo mutations and are ideal for analyses of temporal population differentiation.1 Nevertheless, there may always be inconsistencies between written records and genetic results by events that unlink the connection between Y-chr and genealogy (eg, non-paternity, adoption).7 Moreover, linking Y-chrs to genealogies does not reveal the genetic variation at a past time window but will only give indications about the effects of past migrations based on the genetic variation that is transmitted to the contemporary population.11 Therefore, the value of the genetic-genealogical approach needs to be confirmed based on a known geographically small-scaled population genetic structure.
The Duchy of Brabant in Central–Western Europe was a historical region in the Low Countries containing three contemporary Belgian provinces (Antwerp, Flemish and Walloon Brabant) and one Dutch province (North Brabant). Significant micro-geographical differentiation within this region was detected based on the differences in sub-haplogroup frequencies of the Y-chr.12 To find a signature for population differentiation in Brabant, the donors in this study were assigned to an area within Brabant according to the residence of their oldest reported parental ancestor (ORPA).12 On the basis of this approach, it was assumed to observe a more ‘indigenous' population pattern, which is not blurred by the huge recent migration events of the last decades.13 However, the time period wherein the ORPA lived was different for each individual and varied between the fourteenth till nineteenth century. Moreover, because only participants with an ORPA living in Brabant were selected, an assessment of the effect of immigration on the stability of the population genetic structure was impossible. In this study, we therefore optimized the sampling procedure in order to analyze to which degree the genetic pattern on the Y-chr changed during the last 400 years according to the well-known history and demography of the region.14 We discuss and evaluate the genetic-genealogical approach to analyze temporal genetic differentiation in a particular region.
Materials and methods
Sampling and Y-chr genotyping
Samples were selected from a list of participants collected via genealogical societies in Belgium, the Netherlands, Grand Duchy of Luxembourg and Northern France. As well as a DNA-sample, the requirement for participation was the availability of patrilineal genealogical data with the ORPA born before 1800 and patrilineal presence in Western Europe for at least two generations (to exclude immigrant waves of the last decades). After receiving all genealogical data, only participants currently living in Brabant or from which at least one paternal ancestor of their paternal line was born in Brabant (Figure 1), were selected for this study. To have a representative sample, this requirement was not communicated to the audience. A fraction of the participants were already partly genotyped for the Y-chr.12
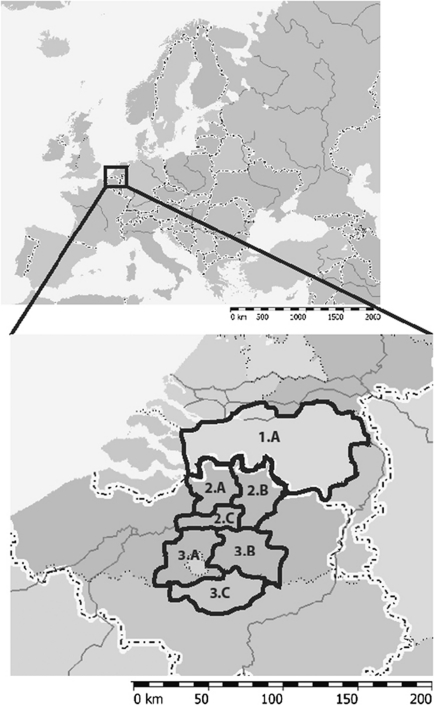
Figure 1.
Geographical location of Brabant within Western Europe and the seven defined areas within Brabant. 1.A: Province North Brabant (The Netherlands); 2.A: Arrondissement (Arro) Antwerp (Belgium, BE); 2.B: Arro Turnhout (BE); 2.C: Arro Mechelen (BE); 3.A: Arro Brussels–Halle–Vilvoorde (BHV; BE); 3.B: Arro Leuven (BE); 3.C: Arro Nijvel (BE, parallel to the Belgian province Walloon Brabant). The number of each area refers to one of the three main regions it belongs to in the population genetic analysis.
A buccal swab sample from each selected participant was collected for DNA-extraction by using the Maxwell 16 System (Promega, Madison, WI, USA) followed by real-time PCR quantification (Quantifiler Human DNA kit, Applied Biosystems, Foster City, CA, USA). In total, 38 STR loci were genotyped as described in previous studies,12, 15 with the addition of Y-STR DYS635. DYS635 was additionally genotyped for all individuals who were already genotyped for 37 Y-STRs in Larmuseau et al.12 All haplotypes were submitted to Whit Atheys' Haplogroup Predictor16 to obtain probabilities for the inferred haplogroups. On the basis of these results, the samples were assigned to specific SNP assays to confirm the haplogroup and to assign the sub-haplogroup to the lowest possible level of the latest Y-chr tree reported by Karafet et al.17 and according to the update on the Y Chromosome Consortium web page (http://ycc.biosci.arizona.edu/nomenclature_system/index.html), with the exception of the substructuring within haplogroup A, R1b1b2a1 (R-U106) and R1b1b2a2g (R-U152). Also a set of recently characterized Y-SNPs, which improved resolution of the haplogroup G phylogeny was included.18 All haplogroup G samples which were already SNP-genotyped in Larmuseau et al.,12 were additionally characterized with this new set. Sixteen multiplex systems with 110 Y-SNPs were developed using SNaPshot mini-sequencing assays (Applied Biosystems) and analyzed on an ABI3130XL Genetic Analyzer (Applied Biosystems) according to previously published protocols.19 All primer sequences and concentrations for the analysis of the 110 Y-SNPs are available from the authors on request.
Genealogical data sets
The genealogical data from each participant underwent a high-quality control through the demonstration of their research with official documents. Pairs with a common official ancestor in paternal lineage but with a different Y-chr sub-haplogroup or Y STR-haplotypes with >6 differences (out of 38 Y-STRs) were excluded from all data sets. On the basis of the general Y-STR mutation rate, >6 mutations out of 38 Y-STRs is not likely to occur between recent genealogical relationship.20 Furthermore, one individual from each pair which (i) showed no difference in surname (or close variant of surname), (ii) belonged to the same Y-chr sub-haplogroup with a related Y-haplotype (≤6 mutations in the 38 genotyped Y-STRs) and (iii) had identical residence regions for all a priori defined time frames (see further), was excluded from the analysis. This may exclude the possibility of a family bias when different members of one family have subscribed to this project.
The assignment of all Y-chr sub-haplogroups to residence regions for different time periods is based on the genealogical records of date and place of baptism and the date of death of the paternal ancestors of the participants. These records are most reliable and available in Brabant.14 For each selected participant, we noted the place of baptism of the oldest patrilineal person (because we assume that there was more than one living patrilineal ancestor at a given moment) living in the years 1600, 1625, 1650, 1675, 1700, 1725, 1750, 1775, 1800, 1825, 1850, 1875, 1900, 1925, 1950, 1975, 2000 and 2010. All places of baptism are then assigned to one of the residence regions, based on contemporary administrative borders; North Brabant, arrondissement Antwerp, Turnhout, Mechelen, Leuven, Brussels–Halle–Vilvoorde (BHV) and Nijvel (Figure 1). These present administrative units are not based on physical borders that might represent barriers to migration but are established based on the range of influence of a certain city (eg, Antwerp, Mechelen and Leuven) and the highly complicated history of Brabant.21 Individuals with a residence region outside Brabant for a specific time period are excluded from the data set of that particular period. In total, we obtained 18 different data sets, further referred to as, for example, the ‘1600 data set'. The data set of all participants with a present residence (PR) within Brabant is further called the ‘PR' data set.
Afterward, each Y-chr was also assigned to a region within Brabant on the basis of the place of baptism of the ORPA. This data set is further referred to as the ‘genealogical residence' (GR) data set. The GR data set is additionally filtered out based on extra genealogical data and the anthroponymy of the surname, namely the ‘purified GR' (PGR) data set. First, all participants who are known descendants of a foundling or a child with an unknown biological father were excluded from this data set. Next, participants were excluded based on the anthroponymical analysis of the surname of their ORPA. The language (inclusive dialect) and the etymology of the surnames, and the archive data with earliest appearance of each surname in the Low Countries were scientifically examined, as defined by standard sources22 and based on the databank of the State Archive of Belgium and the Meertens Institute (Royal Netherlands Academy of Arts and Sciences; www.meertens.knaw.nl). All surnames with an indication for a toponym, which is not located in the ‘GR' region of the participant were excluded, as well as foreign surnames or surnames with a non-Brabant dialect, which may indicate previous migrations. Moreover, also each participant with a surname which is not found in the national archives before 1500, were excluded from the ‘PGR' data set because this can be an indication of a non-autochthonous surname. Finally, we excluded furthermore from the latter data set all participants without an ORPA born before 1750 in the data set named ‘PGR <1750' (PGRb). The full approach is schematically illustrated in supplementary figures (Supplementary Figures S1 and S2).
Genetic and demographical analysis
Estimations of FST-values were calculated based on Y-SNP sub-haplogroup frequencies to determine the genetic relationship between all regions, between the three main groups of regions (namely North Brabant, Antwerp–Turnhout–Mechelen and Leuven–BHV–Nijvel) and between all Dutch versus Belgian individuals. All values were estimated using ARLEQUIN v.3.123 and tested for statistical significance by means of random permutation of samples in 10 000 replicates. For the pairwise FST-values, the sequential Bonferroni correction was applied to correct significance levels for multiple testing.24 The frequencies of the main observed sub-haplogroups were compared between the three main groups of regions based on a three-sample test of equality of proportions without continuity correction. These tests together with the SD of the frequencies for the main observed sub-haplogroups were calculated using the software R v.2.13.0.25
Census population data from Belgium and the Netherlands for the period 1600–2010 were collected from literature14, 26 and official instances, namely the Directorate General Statistics and Economic Information (Belgian government) and Statistics Netherlands (Dutch government).
Results
In total, 881 Western-European males sent their genealogical data. Only 20% of the participants could present reliable genealogical records before 1600. From 1650 onward, this number increased above 50% and even more than 90% could give high-quality data from 1750 till today. However, 10% of the participants did not want to distribute the genealogical data of their last two generations because of privacy reasons. Of the 881 participants in total, 247 males were excluded for further analysis because there was no known paternal ancestor born in Brabant or their PR is not in this region. All individuals were correctly assigned to the main haplogroups using the Whit Athey's Haplogroup Predictor. The single exception was a Y-chr belonging to haplogroup A, which is not included in the Predictor. However, according to a recent study on the root of the human Y-chromosomal phylogenetic tree by Cruciani et al.,27 haplogroup A is not monophyletic and therefore this concrete Y-chr is further referred as belonging to paragroup Y*(xBT). In total, nine main haplogroups were observed with almost ca 85% of all samples belonging to haplogroup R (65%) and I (20%). At the lowest observed level of the phylogenetic tree, 40 different sub-haplogroups were observed in the data set (including the Y-chr assigned to haplogroup Y*(xBT)). Nearly 70% of all samples belonged to only four sub-haplogroups: R1b1b2a1 (R-U106; 27.6%), R1b1b2a2* (R-P312*, 20.1%), I1* (I-M253*, 11.7%) and R1b1b2a2g (R-U152, 9.6%).
The total sampling sizes for each residence region in each data set is given in supplementary materials (Supplementary Table S1). It is clear that the sampling size for Nijvel is too low (<15 individuals) to analyze it separately. Moreover, the sampling sizes for several residence regions are also too low for data sets 1600, 1625 and 1650. The distribution tables for all other data sets are given in supplementary materials (Supplementary Tables S2–S20). The PGR data set has in total 61 participants less than the GR data set. The reason for the exclusion was a toponym outside the residence region within the surname (24 individuals), a surname of a foreign language or a non-Brabant dialect (exclusively a French surname within the traditionally French-speaking part of Brabant, Nijvel) (12 individuals), no archive data found for the surname before 1500 (17 individuals) or the descendant of a foundling or child of unknown father (8 individuals).
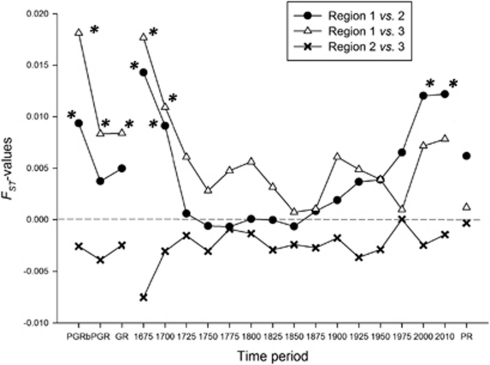
The genetic relationship between all defined regions, between the three main groups of regions – namely North Brabant (region 1), Antwerp-Turnhout-Mechelen (region 2) and Leuven–BHV–Nijvel (region 3) – and between Dutch versus Belgian individuals were assessed by means of FST based on the Y-SNP sub-haplogroup frequencies. Between the three main groups, seven values were significant after sequential Bonferroni correction, namely for the GR data sets and the two earliest time periods (1675 and 1700) between regions 1 and 2 and regions 1 and 3 (Figure 2). Moreover, also the FST-values between regions 1 and 2 were significant for the 2000 and 2010 data sets. Till 1875, there is a clear trend for isolation-by-distance because the FST-values were larger between regions 1 and 3 than between regions 1 and 2. The values between regions 2 and 3 were always estimated to be negative and therefore considered to be zero. This is also visible based on the two-sample tests for equality of Y-sub-haplogroup proportions with continuity correction between Dutch and Belgian individuals, which were significant for all data sets except for the periods between 1725 and 1900 (Supplementary Table S21).
Figure 2.
Relative changes of the pairwise FST-values between the three main regions within Brabant during time. *significant value; region 1, North Brabant (The Netherlands); region 2, Antwerp–Turnhout–Mechelen; region 3, BHV–Leuven–Nijvel. Abbreviations: GR, genealogical residence; PGRb, purified genealogical residence <1750; PGR, purified genealogical residence; PR, present residence.
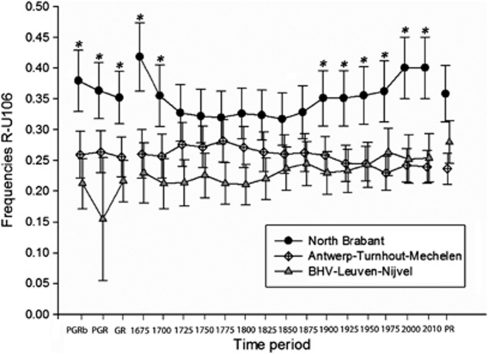
The SD for the frequencies of R1b1b2a1 (R-U106) within Brabant were large because of relatively low sample sizes, however, the three-sample test for equality of R-U106 proportions revealed significant differentiation for the earliest time periods (1675 and 1700) and for the three GR data sets (Figure 3). Furthermore, significant differentiation was also found between the three main regions in Brabant for the latest time periods (1975, 2000 and 2010). Next to R-U106, a (nonsignificant) trend for differentiation was observed as well for R1b1b2a2g (R-U152); the frequency of R-U152 was lower in North Brabant (8% in the PGRb) versus regions 2 and 3 (12% and 11% in the PGRb, respectively). No trend of differentiation between the main regions was found based on other sub-haplogroups.
Figure 3.
Relative changes in frequency of R1b1b2a1 (R-U106) within the three main defined regions within Brabant during time. The error bars represent standard errors; an asterisk (*) represents a significant value between North Brabant and both Belgian regions. The abbreviations for particular time periods are similar to Figure 2.
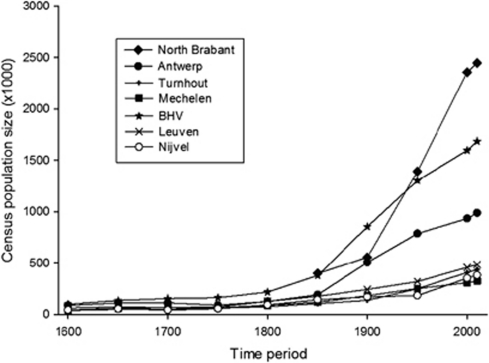
Census population sizes in all regions of Brabant are given in Figure 4. No reliable data were found for North Brabant before 1850, however, this does not influence the observed pattern. Between 1600 and 1850, the population sizes are quite stable and similar to all regions. From 1800 to 1850, there is an increasing growth of the population size and even exponentially in North Brabant, BHV and Antwerp.
Figure 4.
The historical demography for the seven areas within Brabant.
Discussion
Authentic north–south clinal variation within Brabant
Spatial differentiation was found within Brabant, especially for the oldest time periods investigated (1675 and 1700) and the ‘GR' data sets. Within these data sets the three main regions – North Brabant (region 1), Antwerp–Turnhout–Mechelen (region 2) and BHV–Leuven–Nijvel (region 3) – significantly differed from each other. The principle of isolation-by-distance was valid next to a trend of population differentiation at the level of sub-haplogroup frequencies. A north–south cline was observed for the most frequent sub-haplogroup, R1b1b2a1 (R-U106), with the highest frequency in region North Brabant (37% in the PGRb data set), an intermediate frequency in region 2 (26%) and the lowest frequency in region 3 (21%) (Figure 3). Next to R-U152, a nonsignificant and opposite north–south trend was found for the frequencies of R1b1b2a2g (R-U152) with a lower frequency in North Brabant (8% in PGRb) than in the Belgian regions 2 and 3 (12% and 11%, respectively).
Recent studies on a European scale reported a frequency peak of R-U106 in Northern–Central Europe with a steep frequency fall to the south.28, 29 The strong observed decreasing north–south gradient for R1b1b2a1 (R-U106) in Brabant (from 37% in North Brabant to 21% in BHV–Leuven–Nijvel) is therefore an authentic signal and only a subset of the entire gradient ranging between the Netherlands (37.2%) and France (7.1%).30 For R-U152, a similar but reversed pattern was observed in Europe with the highest occurrence in France/Northern Italy and a frequency fall to the north.28, 29 Although the trend of a north–south gradient for R-U152 within Brabant (8% in North Brabant and 11% in BHV–Leuven–Nijvel) turned out to be nonsignificant, it might belong to the macro-scale gradient from France to Northern Europe.
The significant gradient of R-U106 and the trend for R-U152 may most likely be the cause of a previously assumed genetic barrier between the Netherlands and France.31 Even the latest YHRD-data from these two countries revealed a barrier zone of two clusters of Y-chr haplotypes based on a geostatistical approach.32 Our sampling on a micro-geographical scale shows that there is most likely no sudden strong decline of the R-U106 frequency somewhere between the Netherlands and France because of a physical or cultural border for the ‘GR' and earliest (eg, 1675, 1700) data sets. The pattern of differences in the R-U106 frequencies seems to occur in a geographically large stepwise cline. The geographical range of a cline is of course difficult to observe based on forensic data of present populations. On the basis of our study, the population genetic pattern in Western Europe is therefore best explained by a painting of Leonardo Da Vinci based on the sfumato technique; on a macro-scale there is a good picture/structure visible with clear color/genetic differences but on a micro-scale you see a large cline and not an abrupt line/barrier.
Temporal differentiation within Brabant
Clinal variation in human haplogroups might be subjected to entropy because of continuous migration of families and therefore it will fade in time. The strongest clinal variation between the analyzed regions within Brabant was indeed observed for the data sets before 1750 and for the ‘GR data sets' reflecting the most authentic population structure in Brabant. In our genealogical data set, (interregional) migrations are observed continuously in time mostly because of partner choice, as was also observed in historic demographical studies of Brabant.14 Moreover, based on all genealogical records no single individual in our data set could illustrate that his entire patrilineal line stayed in one single community/parish in the period between 1675 and 2010. On the other hand, families from the Low Countries are quite sedentary and they even returned to their birth region after large-scale migration events, for example, the well-known migration of West-Flemish families during World War I after which most of the families returned back to their previous hometown even if their whole property and community was erased by war.33
As 1750, the FST-values and R-U106 frequencies show a decreasing genetic differentiation between the Netherlands and the Belgian regions. The decrease may most likely be amplified by the huge well-known migration events because of religious and economical motives from Flanders to the Netherlands in the sixteenth and seventeenth century.33 On the basis of our approach, the effect of real migrations will be visible later in time because the place of baptism of the oldest living patrilineal person of the family was the chosen parameter. The date and place of baptism together with the date of the funeral, are the best documented and most reliable records in pre-Napoleonic time, in contrast to the place of funeral and residence place(s).14
From 1850 onward, the gradient of genetic differences becomes undetectable, especially for the small R-U106 trend within Belgium. Most likely, this must be linked to the demographical growth in all regions within Brabant started at the beginning of the nineteenth century, especially for North Brabant, BHV and Antwerp (Figure 4). In this period, the increasing population size was mainly a result of massive immigration, especially in the industrialized cities in Brabant.34 Owing to the Industrial Revolution, the transport capacity increased and many families moved to the city where they found more opportunities for employment in new factories. Nevertheless, after 1900, the genetic differences increased again between the Dutch and Belgian regions mainly based on the frequencies of R-U106 with a higher frequency in North Brabant and a lower one in Belgium (Figure 3; Supplementary Table S21). The FST-values between North Brabant and Antwerp–Turnhout–Mechelen also became significant for the data sets of 2000 and 2010 (Figure 2). The genetic difference between the Netherlands and Belgium thus increased after 1900 and may be explained by the breakup of the United Kingdom of the Netherlands in these two countries in 1830. As it is observed between Germany and Poland based on Y-chr,35 administrative borders may affect the current population genetic structure because of the fact that migration occurs mainly within a country. Once there was a notable administrative border between the Netherlands and Belgium, North Brabant received for its textile industry many immigrants from the North where the frequency of R-U106 is relatively high, and the region of Antwerp received more immigrants from the South where this frequency is much lower.36
Validation of the genetic-genealogical approach
A detailed view of the surnames and genealogical data of the selected individuals showed that all highly frequent surnames and all main historical events in Brabant are representatively covered in the data sets, guaranteeing a good representation of the indigenous population. Nevertheless, unreligious, Jewish and gypsy families will not be covered in our data set because genealogical data mostly rely on Christian church records in the pre-Napoleonic period (<1800). These groups were, however, marginal and strongly isolated in Brabant37 and will not influence the overall population analysis of this region. Moreover, today a lot of families with roots outside Western Europe (the so-called gastarbeiders or migrant workers from Italy and North Africa since 1960–1970, recent immigrants for the European Union administration, refugee families, and so on) are living in Brabant and are not included in this temporal analysis. Therefore, the pattern we observed is only based on families living already for more than two generations in Western Europe.
Using the genetic genealogical approach to determine temporal differentiation in a population also required a correct link between Y-chr and the genealogical records. As it was observed for two pairs of individuals in our original data set, non-paternity or unknown adoption may break the connection between genetics and genealogy. Therefore, we can assume that the more we look back in time, the more we have to be aware of the degree of incorrect assignment of Y-chr to a certain region. Nevertheless, in our initial data set, almost all participants with an overlapping part in their genealogy showed the same Y-chr variation based on their sub-haplogroup and haplotype. Accordingly, there is agreement in the literature that past rates of non-paternity are <5% per generation and in some populations even <1%, mainly calculated based on the differences between expected and observed genetic diversity within common and rare surnames.8, 38 This is also consistent with contemporary estimates when there is no previous suspicion of non-paternity.4 But the most important notion on the genetic genealogical approach is that this method does not observe the genetic variation in a population in the past. The temporal analysis is based on the genetic variation that is transmitted to the contemporary population. Therefore, the reconstruction is based on a part of the past variation as a lot of the Y-chr variation will be lost by genetic drift (or the so-called ‘daughtering out'). With our approach the signal of migration on macro-scale as well within Brabant became visible, providing useful insight into the genetic effect of past migration events. The fact that the genetic-genealogical approach showed an authentic signal of a cline conforming the continental genetic pattern26 and the temporal differences of the found cline can be associated with the historical context of the region, guarantees the usefulness of our approach.
Finally, next to the analysis of the genetic variation during several time periods, three different population analyses were done based on the place of baptism of the ORPA, the so-called ‘GR' data sets. The rationale behind these analyses is that in this way, the confounding effects of large and regional migrations during the last decades (and centuries) are minimized, so that it gives access to a sample, which infers a more reliable picture of the ancient population structure at the time of the beginning of genealogical records. All three data sets showed a quite similar pattern based on FST-values and main sub-haplogroup frequencies and may therefore be useful to detect the ‘indigenous' population structure before the Industrial Revolution. Future studies need to give insights into whether surname-selection may still give an extra possibility to provide an even older sample for the population genetic analysis as patrilineal surnames are already present in Western Europe earlier than the seventeenth century.22, 39
Conclusion
By linking a huge number of Y-chr sub-haplogroups to a certain location at a specific time period, it became possible to study the effects of past migrations on the micro-geographic population genetic pattern within the Western-European region of Brabant. Significant genetic differentiation was observed in the oldest studied time periods (1675–1700). However, the differentiation faded over time and the north–south trend for sub-haplogroup R-U106 almost disappeared during the Industrial Revolution in the first half of the nineteenth century. On the basis of this temporal analysis, it is clear that categorizing donors into local sub-populations on the basis of at least two or three generations of residence (as commonly applied in human population genetic/genomic studies),40, 41 may not enable the identification of small-scale ‘autochthonous' population structures in Western Europe. Our approach will be useful to prepare future studies comparing ancient and modern DNA variability in human populations. Therefore, this study exemplifies the usefulness of a strong collaboration between genetic researchers and the genetic genealogy community.
Acknowledgments
We thank all the volunteers who donated DNA samples and provided genealogical data used in this study. They acknowledge the Flemish Society for Genealogical Research, Familiekunde Vlaanderen, which was involved in the collection of the samples and data. They are grateful to Anja Termote and Michel Willems of the Directorate General Statistics and Economic Information (Belgian Government) for providing demographic data. Many thanks also to Pieter Van Camp, Koenraad Matthys, Jean-Jacques Cassiman, Marc Van Den Cloot, Inge Neyens and Lucrece Lernout for useful assistance and discussions. We also want to thank Antonette Anandarajah and three anonymous referees for their useful corrections on an earlier version of this paper. Maarten HD Larmuseau is postdoctoral fellow of the FWO-Vlaanderen (Research Foundation-Flanders). This study was funded by the Flemish Society for Genealogical Research ‘Familiekunde Vlaanderen' (Antwerp), the Flanders Ministry of Culture and the KU Leuven BOF-Centre of Excellence Financing on ‘Eco- and socio-evolutionary dynamics' (Project number PF/2010/07).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Jobling MA, Hurles ME, Tyler-Smith C. Human Evolutionary Genetics: Origins, Peoples and Disease. London/New York: Garland Science Publishing; 2004. [Google Scholar]
- Avise JC. Phylogeography: The History and Formation of Species. Cambridge, MA: Harvard University Press; 2000. [Google Scholar]
- Larmuseau MHD, Van Houdt JKJ, Guelinckx J, Hellemans B, Volckaert FAM. Distributional and demographic consequences of Pleistocene climate fluctuations for a marine demersal fish in the north-eastern Atlantic. J Biogeography. 2009;36:1138–1151. [Google Scholar]
- King TE, Jobling MA. What's in a name? Y chromosomes, surnames and the genetic genealogy revolution. Trends Genet. 2009;25:351–360. doi: 10.1016/j.tig.2009.06.003. [DOI] [PubMed] [Google Scholar]
- Underhill PA, Kivisild T. Use of Y chromosome and mitochondrial DNA population structure in tracing human migrations. Ann Rev Genet. 2007;41:539–564. doi: 10.1146/annurev.genet.41.110306.130407. [DOI] [PubMed] [Google Scholar]
- Kayser M, Vermeulen M, Knoblauch H, Schuster H, Krawczak M, Roewer L. Relating two deep-rooted pedigrees from Central Germany by high-resolution Y-STR haplotyping. Forensic Sci Int Genet. 2007;1:125–128. doi: 10.1016/j.fsigen.2007.02.004. [DOI] [PubMed] [Google Scholar]
- Soodyall H, Nebel A, Morar B, Jenkins T. Genealogy and genes: tracing the founding fathers of Tristan da Cunha. Eur J Human Genet. 2003;11:705–709. doi: 10.1038/sj.ejhg.5201022. [DOI] [PubMed] [Google Scholar]
- King TE, Jobling MA. Founders, drift, and infidelity: the relationship between Y chromosome diversity and patrilineal surnames. Mol Biol Evol. 2009;26:1093–1102. doi: 10.1093/molbev/msp022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyer E, Puymirat J, Dieltjes P, Bakker E, deKnijff P. Estimating Y chromosome specific microsatellite mutation frequencies using deep rooting pedigrees. Human Mol Genet. 1997;6:799–803. doi: 10.1093/hmg/6.5.799. [DOI] [PubMed] [Google Scholar]
- Xue YL, Wang QJ, Long Q, et al. Human Y chromosome base-substitution mutation rate measured by direct sequencing in a deep-rooting pedigree. Curr Biol. 2009;19:1453–1457. doi: 10.1016/j.cub.2009.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helgason A, Hrafnkelsson B, Gulcher JR, Ward R, Stefansson K. A populationwide coalescent analysis of Icelandic matrilineal and patrilineal genealogies: evidence for a faster evolutionary rate of mtDNA lineages than Y chromosomes. Am J Human Genet. 2003;72:1370–1388. doi: 10.1086/375453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larmuseau MHD, Vanderheyden N, Jacobs M, Coomans M, Larno L, Decorte R. Micro-geographic distribution of Y-chromosomal variation in the central-western European region Brabant. Forensic Sci Int Genet. 2011;5:95–99. doi: 10.1016/j.fsigen.2010.08.020. [DOI] [PubMed] [Google Scholar]
- Hill EW, Jobling MA, Bradley DG. Y-chromosome variation and Irish origins. Nature. 2000;404:351–352. doi: 10.1038/35006158. [DOI] [PubMed] [Google Scholar]
- Klep PMM. Bevolking en arbeid in transformatie - een onderzoek naar de ontwikkelingen in Brabant, 1700-1900. Nijmegen: Socialistiese Uitgeverij Nijmegen; 1981. [Google Scholar]
- Jacobs M, Janssen L, Vanderheyden N, Bekaert B, Van de Voorde W, Decorte R. Development and evaluation of multiplex Y-STR assays for application in molecular genealogy. Forensic Sci Int Genet Suppl Ser. 2009;2:57–59. [Google Scholar]
- Athey WT. Haplogroup prediction from Y-STR values using a Bayesian-allele-frequency approach. J Genet Geneal. 2006;2:34–39. [Google Scholar]
- Karafet TM, Mendez FL, Meilerman MB, Underhill PA, Zegura SL, Hammer MF. New binary polymorphisms reshape and increase resolution of the human Y chromosomal haplogroup tree. Genome Res. 2008;18:830–838. doi: 10.1101/gr.7172008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims LM, Garvey D, Ballantyne J. Improved resolution haplogroup G phylogeny in the Y chromosome, revealed by a set of newly characterized SNPs. Plos One. 2009;4:e5792. doi: 10.1371/journal.pone.0005792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caratti S, Gino S, Torre C, Robino C. Subtyping of Y-chromosomal haplogroup E-M78 (E1b1b1a) by SNP assay and its forensic application. Int J Legal Med. 2009;123:357–360. doi: 10.1007/s00414-009-0350-y. [DOI] [PubMed] [Google Scholar]
- Walsh B. Estimating the time to the most recent common ancestor for the Y chromosome or mitochondrial DNA for a pair of individuals. Genetics. 2001;158:897–912. doi: 10.1093/genetics/158.2.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Uytven R, Bruneel C, Koldeweij AM, van de Sande AWFM, van Oudheusden JAFM. Geschiedenis van Brabant van het hertogdom tot heden. Zwolle: Uitgeverij Waanders; 2011. [Google Scholar]
- Debrabandere F. Woordenboek van de familienamen in België en Noord-Frankrijk. Amsterdam/Antwerpen: LJ Veen/Het Taalfonds; 2003. [Google Scholar]
- Excoffier L, Laval G, Schneider S. ARLEQUIN ver.3.0: an integrated software package for population genetics data analysis. Evol Bioinform Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Rice WR. Analyzing tables of statistical tests. Evolution. 1989;43:223–225. doi: 10.1111/j.1558-5646.1989.tb04220.x. [DOI] [PubMed] [Google Scholar]
- The R Foundation for Statistical Computing R version 2.13.0 (2011
- Vrielinck S. De territoriale indeling van België (1795-1963) - bestuurgeografisch en statistisch repertorium van de gemeenten en de supra-communale eenheden (administratief en gerechtelijk). Met de officiële uitslagen van de volkstellingen. Leuven: Universitaire Pers Leuven; 2000. [Google Scholar]
- Cruciani F, Trombetta B, Massaia A, Destro-Bisol G, Sellitto D, Scozzari R. A revised root for the human Y chromosomal phylogenetic tree: the origin of patrilineal diversity in Africa. Am J Human Genet. 2011;88:814–818. doi: 10.1016/j.ajhg.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruciani F, Trombetta B, Antonelli C, et al. Strong intra- and inter-continental differentiation revealed by Y chromosome SNPs M269, U106 and U152. Forensic Sci Int Genet. 2011;5:E49–E52. doi: 10.1016/j.fsigen.2010.07.006. [DOI] [PubMed] [Google Scholar]
- Busby GBJ, Brisighelli F, Sánchez-Diz P, et al. The peopling of Europe and the cautionary tale of Y chromosome lineage R-M269 Proc R Soc B(in press). [DOI] [PMC free article] [PubMed]
- Myres NM, Ritchie KH, Lin AA, Hughes RH, Woodward SR, Underhill PA. Y-chromosome short tandem repeat intermediate variant alleles DYS392.2, DYS449.2, and DYS385.2 delineate new phylogenetic substructure in human Y-chromosome haplogroup tree. Croatian Med J. 2009;50:239–249. doi: 10.3325/cmj.2009.50.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosser ZH, Zerjal T, Hurles ME, et al. Y-chromosomal diversity in Europe is clinal and influenced primarily by geography, rather than by language. Am J Human Genet. 2000;67:1526–1543. doi: 10.1086/316890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Lacava A, Walier M, Willuweit S, et al. Geostatistical inference of main Y-STR-haplotype groups in Europe. Forensic Sci Int Genet. 2011;5:91–94. doi: 10.1016/j.fsigen.2010.09.010. [DOI] [PubMed] [Google Scholar]
- Winkler J, Twilhaar JN. Achternamen in Nederland & Vlaanderen: Oorsprong, Geschiedenis en Betekenis. Den Haag: SDU Uitgevers; 2006. [Google Scholar]
- Winter A. Migrants and Urbans Change: Newcomers to Antwerp, 1760-1860. London: Pickering and Chatto; 2009. [Google Scholar]
- Kayser M, Lao O, Anslinger K, et al. Significant genetic differentiation between Poland and Germany follows present-day political borders, as revealed by Y-chromosome analysis. Human Genet. 2005;117:428–443. doi: 10.1007/s00439-005-1333-9. [DOI] [PubMed] [Google Scholar]
- Nederlands Interdisciplinair Demografisch Instituut (Compiler): Bevolkingsatlas Van Nederland: Demografische Ontwikkeling Van 1850 Tot Heden. Rijswijk: Elmar BV; 2003. [Google Scholar]
- Vanhemelryck F. Marginalen in de Geschiedenis. Leuven: Davidsfonds; 2004. [Google Scholar]
- McEvoy B, Bradley DG. Y-chromosomes and the extent of patrilineal ancestry in Irish surnames. Human Genet. 2006;119:212–219. doi: 10.1007/s00439-005-0131-8. [DOI] [PubMed] [Google Scholar]
- Bowden GR, Balaresque P, King TE, et al. Excavating past population structures by surname-based sampling: the genetic legacy of the Vikings in northwest England. Mol Biol Evol. 2008;25:301–309. doi: 10.1093/molbev/msm255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalloua PA, Platt DE, El Sibai M, et al. Identifying genetic traces of historical expansions: phoenician footprints in the Mediterranean. Am J Human Genet. 2008;83:633–642. doi: 10.1016/j.ajhg.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King RJ, DiCristofaro J, Kouvatsi A, et al. The coming of the Greeks to Provence and Corsica: Y-chromosome models of archaic Greek colonization of the western Mediterranean. BMC Evol Biol. 2011;11:69. doi: 10.1186/1471-2148-11-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.