Abstract
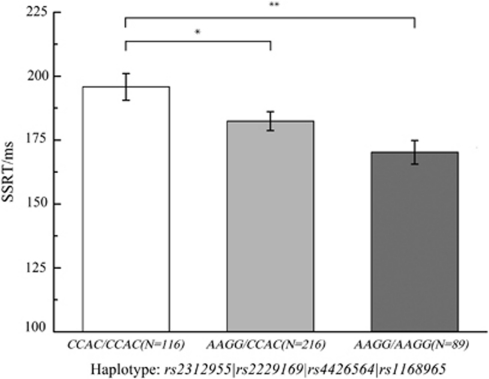
Response inhibition refers to the suppression of inappropriate or irrelevant responses. It has a central role in executive functions, and has been linked to a wide spectrum of prevalent neuropsychiatric disorders. Increasing evidence from neuropharmacological studies has suggested that gene variants in the norepinephrine neurotransmission system make specific contributions to response inhibition. This study genotyped five tag single-nucleotide polymorphisms covering the whole alpha-2B-adrenergic receptor (ADRA2B) gene and investigated their associations with response inhibition in a relatively large healthy Chinese sample (N=421). The results revealed significant genetic effects of the ADRA2B conserved haplotype polymorphisms on response inhibition as measured by stop-signal reaction time (SSRT) (F(2, 418)=5.938, p=0.003). Individuals with the AAGG/AAGG genotype (n=89; mean SSRT=170.2 ms) had significantly shorter SSRTs than did those with either the CCAC/AAGG genotype (n=216; mean SSRT=182.4 ms; uncorrected p=0.03; corrected p=0.09) or the CCAC/CCAC genotype (n=116; mean SSRT=195.8 ms; corrected p<0.002, Cohen's d=0.51). This finding provides the first evidence from association research in support of a critical role of the norepinephrine neurotransmission system in response inhibition. A better understanding of the genetic basis of response inhibition would allow us to develop more effective diagnosis, treatment, and prevention of deficient or underdeveloped response inhibition as well as its related prevalent neuropsychiatric disorders.
Keywords: ADRA2B gene, response inhibition, stop-signal task, haplotype, norepinephrine neurotransmission system
INTRODUCTION
Response inhibition refers to the suppression of actions that are no longer required or that are inappropriate (Aron et al, 2004; Logan and Cowan, 1984; Logan et al, 1997; Verbruggen and Logan, 2008). As a central element of executive functions, response inhibition is posited to be essential for flexible and goal-directed behaviors in ever-changing environments (Chamberlain et al, 2011; Verbruggen and Logan, 2008). Deficient or underdeveloped response inhibition has been linked to a wide spectrum of prevalent neuropsychiatric disorders, such as attention deficit/hyperactivity disorder (Alderson et al, 2007; Chamberlain et al, 2011; Lijffijt et al, 2005), obsessive–compulsive disorder (Chamberlain et al, 2005), and schizophrenia (Lipszyc and Schachar, 2010). Therefore, a better understanding of the neurobiological mechanism of response inhibition, including its genetic basis and underlying neurotransmission systems, could potentially promote more effective diagnosis, treatment, and prevention of deficient or underdeveloped response inhibition as well as its related prevalent neuropsychiatric disorders.
There is accumulating evidence that the brain norepinephrine neurotransmission system has a critical role in response inhibition (Chamberlain and Sahakian, 2007; Pattij and Vanderschuren, 2008). Several prevalent psychostimulants, which increase dopamine and norepinephrine neurotransmission in the prefrontal cortex and thalamic regions, can effectively improve the ability of response inhibition in rodents (Robinson et al, 2008), clinical subjects (Chamberlain et al, 2007), and healthy volunteers (Chamberlain et al, 2006, 2009). These psychostimulants included methylphenidate (Aron et al, 2003), amphetamine and modafinil (Turner et al, 2003, 2004), and atomoxetine (Chamberlain et al, 2006, 2007, 2009). Initially, researchers believed that these psychostimulants solely targeted the dopamine neurotransmission system, but it is now believed that they may also function through the norepinephrine neurotransmission system. In particular, the specific norepinephrine reuptake inhibitor, atomoxetine, shows consistent positive effects on the behavioral performance in the stop-signal task (Chamberlain et al, 2006, 2007; Robinson et al, 2008) and has also been found to modulate the activities of the response-inhibition-related fronto-basal-thalamic loop circuits (Chamberlain et al, 2009). Moreover, researchers have proposed that the norepinephrine neurotransmission system might be specifically related to the maintenance of vigilance, a key component of response inhibition (Aron and Poldrack, 2005).
Human alpha-2B-adrenergic receptor (ADRA2B), also termed the α2Badrenergic receptor, is coded by the human ADRA2B gene located on chromosome 2 (Lomasney et al, 1990). It is a G-protein-coupled presynaptic receptor with high specificity and affinity for norepinephrine in the central nervous system, which has a negative feedback function and modulates norepinephrine neurotransmission (Cousijn et al, 2010). There are two common functional polymorphisms in the ADRA2B gene. One polymorphism consists of 9-nucleotide Ins/Del at the position +901(+901 Ins/Del) in the encoding region, as indicated in Figure 1, leading to an Ins/Del of three glutamic acid residues in the third intracellular loop of the receptor protein (ADRA2B Ins/Del301-303) (Heinonen et al, 1999), which is a very important region for G-protein coupling (Eason et al, 1992). Several previous genetic imaging studies consistently found that the +901 Del was related to enhanced neural activations in the emotion processing regions (the amygdala and the insula) and better behavioral performance (de Quervain et al, 2007; Rasch et al, 2009). Another polymorphism in the ADRA2B gene was recently identified as the 12-nucleotide Ins/Del at the position −4825 (−4825 Del/Ins) in the promoter region, as indicated in Figure 1 (Crassous et al, 2010). The above two common functional Ins/Del polymorphisms are strictly linked to the single-nucleotide polymorphism (SNP) rs2229169 and reside right on the single haplotype block observed in previous studies (Belfer et al, 2005; Cayla et al, 2004; Crassous et al, 2010). Given the critical role of the norepinephrine neurotransmission system in response inhibition, therefore, it is possible that ADRA2B gene variants may also be associated with response inhibition. To our knowledge, no studies have investigated potential associations between ADRA2B gene variants and response inhibition.
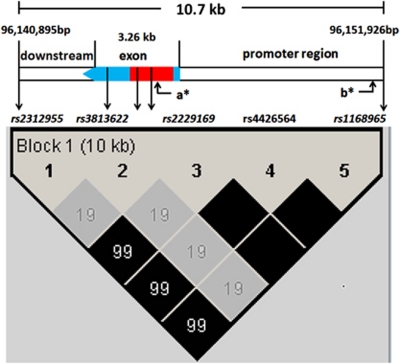
Figure 1.
Schematic representation of the ADRA2B gene and linkage disequilibrium map of selected SNPs (r2). a*, the location of the +901 Insertion/Deletion; b*, the location of −4825 Deletion/Insertion.
In this study, we chose five tag SNPs based on the HapMap data (www.hapmap.org) and previous studies (Belfer et al, 2005; Crassous et al, 2010), including rs2312955 in the downstream region, rs3813662, rs2229169, and rs4426564 in the single exon region (with rs2229169 and rs4426564 in the coding region), and rs1168965 in the promoter region (see Figure 1). These SNPs covered the entire ADRA2B gene plus 4–6 kb upstream and 1–2 kb downstream and captured most variations of the ADRA2B gene because of its single haplotype block structure, including the two common functional +901 Ins/Del and the −4825 Del/Ins polymorphisms (Belfer et al, 2005; Crassous et al, 2010). The rs2229169 was selected as the proxy SNP for the two common functional +901 Ins/Del and the −4825 Del/Ins polymorphisms, with the A allele in strict linkage with+901 Ins and −4825 Del. Therefore, we proposed that the A allele of the rs2229169 polymorphism (same as A/+901Del/−4825 Ins haplotype), which is believed to be linked to enhanced neural activations (de Quervain et al, 2007; Gibbs et al, 2010), would be significantly associated with better response inhibition (ie, shorter SSRT).
PARTICIPANTS AND METHODS
Participants
Participants were 455 healthy undergraduate students (261 females, aged from 19 to 22 years, with a mean of 20.3 years, SD=0.82), which was a subsample who completed the Stop-Signal task (see below) from the larger Gene-Brain-Behavior Project at Beijing Normal University (eg, Chen et al, 2011; He et al, 2010; Li et al, 2011). All subjects were Han Chinese with normal or corrected-to-normal vision and no neurological or psychiatric history based on their self-report. They all signed written informed consent. This study was approved by the Institutional Review Board of the State Key Laboratory of Cognitive Neuroscience and Learning at Beijing Normal University.
Genotyping
A 4 ml venous blood sample was collected from each subject. Genomic DNA was extracted according to the standard method within 2 weeks after the blood sample was collected. All samples were genotyped using the standard Illumina GoldenGate Genotyping protocol (for details, see www.southgene.com.cn).
As described above and in Figure 1 and Table 1, five SNPs in the ADRA2B gene on chromosome 2 were selected, including rs2312955, rs3813662, rs2229169, rs4426564, and rs1168965. One female with a 50% missing call rate was removed, resulting in 454 subjects in subsequent analyses. All five ADRA2B gene SNPs met the criteria of a call rate of >95%, Minor Allele Frequency (MAF) of >0.05, and Hardy–Weinberger equilibrium (HWE) of p>0.05. The allele frequencies in our sample were very similar to those of the Han Chinese in the HapMap data set (see Table 1).
Table 1. Allele Frequencies of Candidate SNPs in the ADRA2B Gene and Their Linkage Shown by Ethnic Groups. Data Were from This Study and the HapMap Website (www.hapmap.org).
| SNP | Base-pair position | Location | Minor/major allelea |
Minor allele frequency |
||||
|---|---|---|---|---|---|---|---|---|
| This study CHB | CHB | JPT | CEU | YRI | ||||
| rs2312955 | 96140895 | Downstream | A/C | 0.47 (N=421) | 0.45 (N=274) | 0.34 (N=224) | 0.33 (N=224) | 0.11 (N=292) |
| rs3813662 | 96143422 | Exon | C/A | 0.14 (N=421) | 0.18 (N=274) | 0.20 (N=226) | 0.004 (N=226) | 0.00 (N=126) |
| rs2229169 | 96144443 | Encoding | A/C | 0.47 (N=421) | 0.48 (N=90) | 0.31 (N=88) | 0.30 (N=130) | 0.25 (N=126) |
| rs4426564 | 96144713 | Encoding | G/A | 0.47 (N=421) | 0.45 (N=270) | 0.33 (N=222) | 0.32 (N=224) | 0.11 (N=294) |
| rs1168965 | 96151626 | Promoter | G/C | 0.47 (N=421) | 0.45 (N=274) | 0.35 (N=226) | 0.32 (N=226) | 0.11 (N=294) |
| Common haplotypes | AAGG/CCAC | 0.47 (N=421) | 0.4 (N=180)b | 0.3 (N=60) | — | |||
| Mean pairwise r2 | — | 0.99 | 0.95 | 1.00 | 0.95 | 0.68 | ||
It should be noted that, on the HapMap website, these specific alleles of SNPs have different labels because of the different coding based on either the forward primer or the reverse primer. For example, the alleles of rs2229169 are A and C in this study (the standard Illumina GoldenGate Genotyping protocol) and in the dbSNP b126, but they are T and G on the HapMap website. In this table, we used the coding of all alleles based on the coding system of the Illumina system.
In the HapMap website, CHB and JPT are combined into one Asian group for this haplotype.
The Stop-Signal Task
The response inhibition task was the manual response version of the task used in the study by Xue et al (2008). A ‘tracking' paradigm was used to allow for a more precise and sensitive measure of SSRT (Aron et al, 2003; Aron and Poldrack, 2006). This paradigm has been successfully applied under a wide range of experimental conditions and with subjects from different populations (Aron et al, 2007; Band et al, 2003; Verbruggen and Logan, 2008). In addition, a highly accurate reaction time tool—the RTbox (Li et al, 2010)—was used to record and collect subjects' responses. Subjects' performance was indexed by the stop-signal reaction time (SSRT), namely, the amount of time the subjects required to intentionally suppress a response based on the horse race model (Band et al, 2003; Verbruggen and Logan, 2008).
The ‘tracking' stop-signal paradigm was programmed in a custom Matlab code (The MathWorks) (for details, see Xue et al, 2008). On each trial, letter ‘T' or ‘D' was displayed on the computer screen, with the ‘T' requesting a response with the left-hand and the ‘D' requesting a response with the right-hand. Subjects were asked to respond as quickly and accurately as possible to these letters using the index finger of their left or right hand (the Go trials). For 25% (randomly selected) of the trials (the Stop trials), subjects heard a beep after the Go stimulus had appeared, and they should suppress the ongoing response. Each subject performed four blocks of 64 trials per block (16 Stop trials and 48 Go trials per block), totaling 256 trials (64 Stop trials and 192 Go trials). The stop-signal delay (SSD) changed dynamically in a 50-ms increment throughout the experiment depending on the subject's performance. A successful response inhibition led to an increase in the SSD by 50 ms on the next Stop trial to make it more difficult, whereas a failed trial of response inhibition led to a decrease in the SSD by 50 ms on the next Stop trial to make it easier. The tracking procedure ensured that over time, the probability of successful inhibition would ideally stabilize around 0.5. There were four staircases of SSD starting at 140, 180, 220, and 260 ms, respectively. The staircases were independent and randomly interleaved. In each staircase, the last 6 converged Stop trials and the 18 corresponding Go trials were used to calculate the SSRT. Four staircases were visually inspected for convergence and consistency. The observed SSRT was estimated by subtracting the given SSD after successful inhibition from the corresponding Go reaction time (Aron et al 2003; Aron and Poldrack, 2006; Band et al, 2003). In sum, the five variables from the ‘tracking' stop-signal paradigm were the correct rate of Go trials (Go Rate), the successful stop rate (Stop Rate), SSD, SSRT, and Go RT.
Data Analyses
Analyses were conducted using Plink v1.07 (Purcell et al, 2007) and SPSS for Windows (release 16.0).
In the total sample of 454 subjects with both genetic and behavioral data, 8 subjects (5 males) with the <50% correct rate of Go trials and 14 subjects (6 males) with no response inhibition were removed. In a recent review of studies using the stop-signal task (n=126), the range of SSRTs in the young healthy adults was 75.76∼344.86 ms (Congdon et al, 2010). On the basis of this report, 11 subjects with shorter than 60 ms SSRT were excluded because such SSRT seemed too short for response inhibition. Therefore, to examine potential biases resulting from the data cleaning, we conducted several preliminary analyses. First, for the deleted 33 subjects, we tested the HWE for all five SNPs and there were no significant results (p>0.32), indicating that the exclusion of these subjects was not due to the individuals' genotype. Second, we examined the mean age and intelligence scores (WAIS-RC) (Gong, 1992) of this excluded sample as compared with the final sample and found no significant differences, confirming that the exclusion of these subjects did not bias the sampling of subjects in terms age and IQ. The exclusion of these 33 subjects because of their questionable SSRT data did not bias the genetic association results. The final sample for the genetic association analysis was 421 subjects (242 females/179 males, mean age=20.3 years, SD=0.8). Finally, to avoid the effects of outliers, an isolated cluster of 7 subjects with longer than 400 ms SSRT (outside of the mean ±3 SD) were rescaled to the max SSRT (370 ms) of the remaining sample. It is worth noting that a separate analysis excluding these seven subjects showed very similar results (the haplotype effect on SSRT, F(2, 411)=−4.979, p=0.007).
Quantitative trait genetic association analysis was carried out by using Plink. In order to test the group differences between different genotypes, ANOVA and the Fisher's least significant difference post hoc tests (t-tests) were performed in SPSS. As the results of linkage disequilibrium analysis showed that four SNPs (rs2312955, rs2229169, rs4426564, and rs1168965) in the ADRA2B gene were in strong linkage with one another and constructed a conserved haplotype block (mean pairwise r2=0.999), we conducted the association analysis with the haplotype as well as the fifth SNP rs3813662. The SNP rs3813662 and the haplotype block were coded as minor allele/haplotype dosage (homozygote of major allele=0, heterozygote=1, homozygote of minor allele=2). There were two common haplotypes AAGG and CCAC (rs2312955∣rs2229169∣rs4426564∣rs1168965) with respective frequencies of 0.47 and 0.53. There was a rare haplotype CAGG in only one subject. Phase analysis showed this subject with the CAGG/CCAC genotype was similar to the ones with the AAGG/CCAC genotype. This subject was thus assigned to the AAGG/CCAC heterozygote group. As there were only 10 C/C homozygotes of minor allele of rs3813662, we merged them with the heterozygotes C/A (n=108) with the code value of 1 and coded the A/A homozygotes (n=303) with the code value of 0. Bonferroni correction for multiple comparisons was used in this analysis.
RESULTS
As shown in Table 2, the mean Go RT, SSD, and SSRT were 616.1 ms (SD=145.2 ms), 430.6 ms (SD=147.6 ms), and 183.5 ms (SD=53.7 ms), respectively. As mentioned earlier, SSRTs were within the 62.2∼370 ms range. The mean Go Rate was 0.93 (SD=0.06) and the mean Stop Rate 0.62 (SD=0.13). These numbers were consistent with results from previous studies as summarized in a recent review (Congdon et al, 2010) as well as with the previous study using the same version of the task (Xue et al, 2008). Although the group-averaged Stop Rate (0.62) in this study was a little higher than the theoretical 0.5, the range of Stop Rate in this study was consistent with previous researches (Congdon et al, 2010) and in the theoretically acceptable range (Band et al, 2003; Logan and Cowan, 1984).
Table 2. Behavioral Data on the Signal Stop Task and Differences Based on Haplotype.
| Haplotypea | Total | Major haplotype | Heterozygote | Minor haplotype | ANOVA F | Contrast test |
|---|---|---|---|---|---|---|
| N | 421 | 116 | 216 | 89 | ||
| SSRT/ms | 183.5 (53.6) | 195.8 (55.5) | 182.4 (55.1) | 170.2 (43.8) | 5.938b | Major>minorc heterzygote>minord |
| GO RT/ms | 616.1 (145.2) | 618.3 (133.3) | 617.0 (151.4) | 610.8 (146.1) | 0.076 | — |
| SSD/ms | 430.6 (147.6) | 419.0 (140.0) | 432.7 (156.3) | 440.6 (135.8) | 0.583 | — |
| Go rate | 0.93 (0.10) | 0.94 (0.08) | 0.92 (0.10) | 0.93 (0.11) | 1.27 | — |
| Stop rate | 0.62 (0.13) | 0.62 (0.14) | 0.61 (0.13) | 0.63 (0.11) | 0.79 | — |
The haplotype was constructed from four SNPs: rs2312955∣rs2229129∣rs4426564∣rs116896; rs2312955, rs2229129, rs4426564, and rs1168965 are in strictly linkage and each of them could tag the haplotype.
Note: numbers represent the mean (SD).
p<0.01.
Adjusted p<0.01, Cohen's d=0.51.
Unadjusted p<0.05.
There were no significant differences in Stop Rates among the three genetic groups, which excluded a potential confounding effect of the Stop Rate on genetic differences in SSRT. There were also no significant effects of age and intelligence on Go RT and SSRT.
As shown in Table 2, ANOVAs and post hoc tests showed that minor homozygotes and heterozygotes of the haplotype block had significantly shorter SSRT and better response inhibition as compared with major homozygotes (F(2, 418)=5.938, p=0.003). As shown in Figure 2, individuals with the AAGG/AAGG genotype showed significantly shorter SSRT than those with the CCAC/CCAC genotype (t(418)=−3.42, corrected p<0.01, Cohen's d=0.51) and the CCAC/AAGG genotype (t(418)=−2.19, uncorrected p=0.03), but the difference between CCAC/CCAC and CCAC/AAGG individuals was not significant. There were no significant results of rs3813662. There were also no genetic effects on Go RT, Go Rate, and Stop Rate.
Figure 2.
Association between haplotype and SSRT. SSRT was compared across different genotypes by one-way ANOVA followed by Fisher's least significant difference (LSD) post hoc test. *Unadjusted p<0.05. **Adjusted p<0.001. Error bars are SEM.
DISCUSSION
In this study, we chose five tag SNPs that covered the whole ADRA2B gene to investigate this gene's effects on response inhibition in normal Chinese adults. Results showed that a single conserved haplotype block structure spanning the whole ADRA2B gene was significantly linked to response inhibition. Individuals with the AAGG/AAGG haplotype had significantly shorter SSRT (ie, better response inhibition) compared with those with the CCAC/AAGG or CCAC/CCAC haplotype. These results further confirmed the critical role of the norepinephrine neurotransmission system in response inhibition as shown in previous neuropharmacological studies (Chamberlain et al, 2006, 2007, 2009).
These findings can be integrated with multiple lines of human and animal research (including biochemical, pharmacological, genetic, and brain imaging studies) on the ADRA2B gene and its protein product (the ADRA2B) as well as the norepinephrine neurotransmission system. First, there is convergent evidence of anatomical overlapping between the expression distribution of the ADRA2B gene and the neural substrates of response inhibition. Messenger RNA of the ADRA2B gene has been found to be expressed in the subcortical nucleus and extensive cortical regions, such as the hypothalamus, thalamus, and right inferior frontal cortex (Scheinin et al, 1994; Tavares et al, 1996). Brain imaging studies have shown that these same brain regions have a central role in response inhibition (Aron et al, 2007; Aron and Poldrack, 2006; Xue et al, 2008). Furthermore, variants of the ADRB2B gene have been associated with activations in these brain regions, particularly the right inferior frontal gyrus, the insula, and the basal ganglia system. For example, the +901 Del/Ins variant of the ADRA2B gene modulates the neural activity of the amygdala/insula/inferior frontal gyrus (de Quervain et al, 2007; Rasch et al, 2009; Cousijn et al, 2010; Urner et al, 2011).
Second, cumulating evidence from clinical and molecular neuropharmacological studies on animals and humans supports the critical role of the norepinephrine neurotransmission system in response inhibition (Aston-Jones and Gold, 2009; Chamberlain et al, 2006, 2007, 2009). Psychostimulants that enhance the brain noradrenergic neurotransmission can improve response inhibition in clinical samples with attention-deficit/hyperactivity disorders (Aron et al, 2003; Turner et al, 2004) and normal controls (Turner et al, 2003). Atomoxetine, a specific norepinephrine reuptake inhibitor, was found to increase the noradrenergic neurotransmission in the brain (Bymaster et al, 2002) and to modulate the activity of the response-inhibition-related cortico-basal thalamic circuits at rest state in rats (Easton et al, 2007). Atomoxetine has been found to enhance activation in the right inferior frontal gyrus during the stop-signal task in healthy subjects (Chamberlain et al, 2009), and to improve the ability of response inhibition in both clinical and healthy subjects (Chamberlain et al, 2006, 2007, 2009). These studies established relations among the brain noradrenergic neurotransmission, the neural activity of the right inferior frontal gyrus involved in the task, and the behavioral ability of response inhibition.
Third, the ADRA2B gene has a large conserved haplotype block linking the common tag SNPs (such as those typed in this study) and the common functional +901 Del/Ins and –4825 Ins/Del variants. Cayla et al (2004) first reported that the polymorphism at position +1182 relative to the start codon (rs2229169) was in strict linkage with the +901 Del/Ins variant, which was then confirmed and extended by Belfer et al (2005) and most recently by Crassous et al (2010) to include other SNPs. Our data further confirmed this block with more SNPs and a large Chinese sample.
With this strict linkage, the direction of genetic effects of the ADRA2B gene on response inhibition observed in this study can be interpreted based on previous biochemical research on the +901 Del/Ins. The +901 Del deletes three glutamic acid residues in the third intracellular loop of the receptor protein (also termed ADRA2B Ins/Del301-303) (Heinonen et al, 1999), which is a very important region for G-protein coupling (Eason et al, 1992). The ADRA2B receptors with deletion of these three glutamic acid residues display a decreased agonist-promoted phosphorylation and consequently a complete loss of desensitization, which in turn could lead to a continuingly enhanced noradrenergic neurotransmission (Small et al, 2001). Therefore, carriers of the AAGG/AAGG genotype (same as +901 Del/Del carriers, who showed enhanced noradrenergic neurotransmission) were better at response inhibition indexed by the shorter SSRT than were their counterparts with the CCAC/CCAC genotype (same as +901 Ins/Ins) or AAGG/CCAC genotype (same as +901 Del/Ins).
Taking together our results and evidence from previous brain imaging and neuropharmacological studies, we propose that ADRA2B gene variants influence response inhibition by regulating noradrenergic neurotransmission in the brain and modulating the neural activation associated with some important components in the stop-signal task, such as vigilance, detection of temporal unpredictable stop signal, and attention shift with task switching (Aston-Jones and Cohen, 2005; Aron and Poldrack, 2005).
Three main limitations of this study need to be mentioned. First, this was an association study. The biochemical and physiological functions of the haplotype polymorphisms were not directly explored in this study. Although the linked +901 Del/Ins and −4825 Ins/Del polymorphisms could be the potential cause based on evidence from previous studies (Belfer et al, 2005; Crassous et al, 2010), biochemical research is needed to investigate the role of the polymorphisms in the intron and noncoding regions because they can also exert transcriptional suppression of the genes, influence mRNA secondary structure (Nackley et al, 2006), and regulate gene expression and splicing (Zhang et al, 2007). Second, because this was a college sample, we used only self-report to screen for neurological and psychiatric disorders. It is plausible to have some potential confounding effects. Future research should employ an objective systematic tool for that purpose. (Although we did not administer a systematic battery of objective assessments for neurological diseases and mental disorders, the larger project included measures of two main mental health issues relevant to college students: depression and anxiety. They were measured by Beck's depression (BDI-II) and anxiety inventories (BAI) (Beck, 1990; Beck et al, 1996). According to the cutoff scores and interpretive labels provided by Beck (1990) and Beck et al (1996), three subjects scored in the ‘severe' range (29–63) of BDI-II and 22 subjects scored in the ‘severe' range (26–63) of BAI. As depression and anxiety were not related to response inhibition, we did not exclude these subjects from our analyses.) Third, this study was conducted in one ethnic group (ie, Han Chinese college students) to avoid ethnic stratification problems. These SNPs and the haplotype block constructed from SNPs have different MAF in different ethnic populations based on the HapMap Data (www.hapmap.org, see Table 1). Thus, these genetic effects on SSRT found in this study need to be replicated in other ethnic populations.
In conclusion, this study found significant effects of ADRA2B haplotype polymorphisms on response inhibition in a large Chinese sample, with the minor AAGG haplotype associated with shorter SSRT (ie, better response inhibition) compared with the major CCAC haplotype. These effects were independent of subjects' intelligence, age, and gender. These findings provided new genetic associative evidence for the critical role of the norepinephrine neurotransmission system in response inhibition and a new insight into the function of the ADRA2B receptor in the central nervous system. Future studies need to combine genetic, behavioral, and functional imaging techniques to investigate the neural mechanisms underlying the genetic effects on response inhibition. Future studies should also examine the gene × drug interactions on treatment of disorders with deficient response inhibition, because several studies of ADHD have been conducted on a related gene—the ADRA2A gene (the alpha-2A-adrenergic receptor gene in the same family as the ADRA2B gene) (Cheon et al, 2009; da Silva et al, 2008; Polanczyk et al, 2007). Results from such studies will not only improve our understanding the genetic basis and the neural mechanism of response inhibition, but also potentially contribute to the diagnosis and treatment of specific psychiatric disorders associated with deficient or underdeveloped response inhibition, such as ADHD, OCD, and addictions.
Acknowledgments
This study was supported by the 111 Project of the Ministry of Education of China (B07008). We thank all graduate research assistants who helped us with data collection.
The authors declare no conflict of interest.
References
- Alderson RM, Rapport MD, Kofler MJ. Attention-deficit/hyperactivity disorder and behavioral inhibition: a meta-analytic review of the stop-signal paradigm. J Abnorm Child Psychol. 2007;35:745–758. doi: 10.1007/s10802-007-9131-6. [DOI] [PubMed] [Google Scholar]
- Aron AR, Dowson JH, Sahakian BJ, Robbins TW. Methylphenidate improves response inhibition in adults with attention-deficit/hyperactivity disorder. Biol Psychiatry. 2003;54:1465–1468. doi: 10.1016/s0006-3223(03)00609-7. [DOI] [PubMed] [Google Scholar]
- Aron AR, Durston S, Eagle DM, Logan GD, Stinear CM, Stuphorn V. Converging evidence for a fronto-basal-ganglia network for inhibitory control of action and cognition. J Neurosci. 2007;27:11860–11864. doi: 10.1523/JNEUROSCI.3644-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aron AR, Poldrack RA. The cognitive neuroscience of response inhibition: relevance for genetic research in attention-deficit/hyperactivity disorder. Biol Psychiatry. 2005;57:1285–1292. doi: 10.1016/j.biopsych.2004.10.026. [DOI] [PubMed] [Google Scholar]
- Aron AR, Poldrack RA. Cortical and subcortical contributions to Stop signal response inhibition: role of the subthalamic nucleus. J Neurosci. 2006;26:2424–2433. doi: 10.1523/JNEUROSCI.4682-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aron AR, Robbins TW, Poldrack RA. Inhibition and the right inferior frontal cortex. Trends Cognitive Sci. 2004;8:170–177. doi: 10.1016/j.tics.2004.02.010. [DOI] [PubMed] [Google Scholar]
- Aston-Jones G, Cohen JD. An integrative theory of locus coeruleus-norepinephrine function: adaptive gain and optimal performance. Ann Rev Neurosci. 2005;28:403–450. doi: 10.1146/annurev.neuro.28.061604.135709. [DOI] [PubMed] [Google Scholar]
- Aston-Jones G, Gold JI. How we say no: norepinephrine, inferior frontal gyrus, and response inhibition. Biol Psychiatry. 2009;65:548–549. doi: 10.1016/j.biopsych.2009.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Band GP, van der Molen MW, Logan GD. Horse-race model simulations of the stop-signal procedure. Acta Psychologica. 2003;112:105–142. doi: 10.1016/s0001-6918(02)00079-3. [DOI] [PubMed] [Google Scholar]
- Beck AT. Beck Anxiety Inventory Manual. Psychological Corporation: San Antonio, TX; 1990. [Google Scholar]
- Beck AT, Steer RA, Ball R, Ranieri W. Comparison of Beck depression inventories -IA and -II in psychiatric outpatients. J Personality Assess. 1996;67:588–597. doi: 10.1207/s15327752jpa6703_13. [DOI] [PubMed] [Google Scholar]
- Belfer I, Buzas B, Hipp H, Phillips G, Taubman J, Lorincz I, et al. Haplotype-based analysis of alpha 2A, 2B, and 2C adrenergic receptor genes captures information on common functional loci at each gene. J Hum Genet. 2005;50:12–20. doi: 10.1007/s10038-004-0211-y. [DOI] [PubMed] [Google Scholar]
- Bymaster FP, Katner JS, Nelson DL, Hemrick-Luecke SK, Threlkeld PG, Heiligenstein JH, et al. Atomoxetine increases extracellular levels of norepinephrine and dopamine in prefrontal cortex of rat: a potential mechanism for efficacy in attention deficit/hyperactivity disorder. Neuropsychopharmacology. 2002;27:699–711. doi: 10.1016/S0893-133X(02)00346-9. [DOI] [PubMed] [Google Scholar]
- Cayla C, Heinonen P, Viikari L, Schaak S, Snapir A, Bouloumie A, et al. Cloning, characterisation and identification of several polymorphisms in the promoter region of the human alpha2B-adrenergic receptor gene. Biochem Pharmacol. 2004;67:469–478. doi: 10.1016/j.bcp.2003.09.029. [DOI] [PubMed] [Google Scholar]
- Chamberlain SR, Blackwell AD, Fineberg NA, Robbins TW, Sahakian BJ. The neuropsychology of obsessive compulsive disorder: the importance of failures in cognitive and behavioural inhibition as candidate endophenotypic markers. Neurosci Biobehav Rev. 2005;29:399–419. doi: 10.1016/j.neubiorev.2004.11.006. [DOI] [PubMed] [Google Scholar]
- Chamberlain SR, Del Campo N, Dowson J, Muller U, Clark L, Robbins TW, et al. Atomoxetine improved response inhibition in adults with attention deficit/hyperactivity disorder. Biol Psychiatry. 2007;62:977–984. doi: 10.1016/j.biopsych.2007.03.003. [DOI] [PubMed] [Google Scholar]
- Chamberlain SR, Hampshire A, Muller U, Rubia K, Del Campo N, Craig K, et al. Atomoxetine modulates right inferior frontal activation during inhibitory control: a pharmacological functional magnetic resonance imaging study. Biol Psychiatry. 2009;65:550–555. doi: 10.1016/j.biopsych.2008.10.014. [DOI] [PubMed] [Google Scholar]
- Chamberlain SR, Muller U, Blackwell AD, Clark L, Robbins TW, Sahakian BJ. Neurochemical modulation of response inhibition and probabilistic learning in humans. Science. 2006;311:861–863. doi: 10.1126/science.1121218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlain SR, Robbins TW, Winder-Rhodes S, Muller U, Sahakian BJ, Blackwell AD, et al. Translational approaches to frontostriatal dysfunction in attention-deficit/hyperactivity disorder using a computerized neuropsychological battery. Biol Psychiatry. 2011;69:1192–1203. doi: 10.1016/j.biopsych.2010.08.019. [DOI] [PubMed] [Google Scholar]
- Chamberlain SR, Sahakian BJ. The neuropsychiatry of impulsivity. Curr Opin Psychiatry. 2007;20:255–261. doi: 10.1097/YCO.0b013e3280ba4989. [DOI] [PubMed] [Google Scholar]
- Chen C, Chen C, Moyzis R, Dong Q, He Q, Zhu B, et al. Sex modulates the associations between the COMT gene and personality traits. Neuropsychopharmacology. 2011;36:1593–1598. doi: 10.1038/npp.2011.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheon KA, Cho DY, Koo MS, Song DH, Namkoong K. Association between homozygosity of a G allele of the alpha-2a-adrenergic receptor gene and methylphenidate response in Korean children and adolescents with attention-deficit/hyperactivity disorder. Biol Psychiatry. 2009;65:564–570. doi: 10.1016/j.biopsych.2008.12.003. [DOI] [PubMed] [Google Scholar]
- Congdon E, Mumford JA, Cohen JR, Galvan A, Aron AR, Xue G, et al. Engagement of large-scale networks is related to individual differences in inhibitory control. Neuroimage. 2010;53:653–663. doi: 10.1016/j.neuroimage.2010.06.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cousijn H, Rijpkema M, Qin S, van Marle HJ, Franke B, Hermans EJ, et al. Acute stress modulates genotype effects on amygdala processing in humans. Proc Natl Acad Sci USA. 2010;107:9867–9872. doi: 10.1073/pnas.1003514107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crassous PA, Blaise R, Marquette A, Snapir A, Scheinin M, Paris H, et al. Identification of a novel 12-nucleotide insertion polymorphism in the promoter region of ADRA2B: full linkage with the 9-nucleotide deletion in the coding region and influence on transcriptional activity. Biochem Pharmacol. 2010;79:407–412. doi: 10.1016/j.bcp.2009.08.024. [DOI] [PubMed] [Google Scholar]
- da Silva TL, Pianca TG, Roman T, Hutz MH, Faraone SV, Schmitz M, et al. Adrenergic alpha2A receptor gene and response to methylphenidate in attention-deficit/hyperactivity disorder-predominantly inattentive type. J Neural Transm. 2008;115:341–345. doi: 10.1007/s00702-007-0835-0. [DOI] [PubMed] [Google Scholar]
- de Quervain DJ, Kolassa IT, Ertl V, Onyut PL, Neuner F, Elbert T, et al. A deletion variant of the alpha2b-adrenoceptor is related to emotional memory in Europeans and Africans. Nat Neurosci. 2007;10:1137–1139. doi: 10.1038/nn1945. [DOI] [PubMed] [Google Scholar]
- Eason MG, Kurose H, Holt BD, Raymond JR, Liggett SB. Simultaneous coupling of alpha 2-adrenergic receptors to two G-proteins with opposing effects. Subtype-selective coupling of alpha 2C10, alpha 2C4, and alpha 2C2 adrenergic receptors to Gi and Gs. J Biol Chem. 1992;267:15795–15801. [PubMed] [Google Scholar]
- Easton N, Marshall F, Fone K, Marsden C. Atomoxetine produces changes in cortico-basal thalamic loop circuits: assessed by phMRI BOLD contrast. Neuropharmacology. 2007;52:812–826. doi: 10.1016/j.neuropharm.2006.09.024. [DOI] [PubMed] [Google Scholar]
- Gibbs AA, Naudts KH, Azevedo RT, David AS. Deletion variant of alpha2b-adrenergic receptor gene moderates the effect of COMT val(158)met polymorphism on episodic memory performance. Eur Neuropsychopharmacol. 2010;20:272–275. doi: 10.1016/j.euroneuro.2009.12.007. [DOI] [PubMed] [Google Scholar]
- Gong Y. The Manual of Wechsler Adult Intelligence Scale Revised in China. Hunan Medical University Press: Changsh Hunan, China; 1992. [Google Scholar]
- He Q, Xue G, Chen C, Lu Z, Dong Q, Lei X, et al. Serotonin transporter gene-linked polymorphic region (5-HTTLPR) influences decision making under ambiguity and risk in a large Chinese sample. Neuropharmacology. 2010;59:518–526. doi: 10.1016/j.neuropharm.2010.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinonen P, Koulu M, Pesonen U, Karvonen MK, Rissanen A, Laakso M, et al. Identification of a three-amino acid deletion in the alpha2B-adrenergic receptor that is associated with reduced basal metabolic rate in obese subjects. J Clin Endocrinol Metabol. 1999;84:2429–2433. doi: 10.1210/jcem.84.7.5818. [DOI] [PubMed] [Google Scholar]
- Li J, Chen C, He Q, Li H, Moyzis RK, Xue G, et al. Neurotensin receptor 1 gene (NTSR1) polymorphism is associated with working memory. PLoS One. 2011;6:e17365. doi: 10.1371/journal.pone.0017365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Liang Z, Kleiner M, Lu ZL. RTbox: a device for highly accurate response time measurements. Behav Res Methods. 2010;42:212–225. doi: 10.3758/BRM.42.1.212. [DOI] [PubMed] [Google Scholar]
- Lijffijt M, Kenemans JL, Verbaten MN, van Engeland H. A meta-analytic review of stopping performance in attention-deficit/hyperactivity disorder: deficient inhibitory motor control. J Abnormal Psychol. 2005;114:216–222. doi: 10.1037/0021-843X.114.2.216. [DOI] [PubMed] [Google Scholar]
- Lipszyc J, Schachar R. Inhibitory control and psychopathology: a meta-analysis of studies using the stop signal task. J Int Neuropsychol Soc. 2010;16:1064–1076. doi: 10.1017/S1355617710000895. [DOI] [PubMed] [Google Scholar]
- Logan GD, Cowan WB. On the ability to inhibit thought and action: a theory of an act of control. Psychol Rev. 1984;91:295–327. doi: 10.1037/a0035230. [DOI] [PubMed] [Google Scholar]
- Logan GD, Schachar RJ, Tannock R. Impulsivity and inhibitory control. Psychol Sci. 1997;8:60–64. [Google Scholar]
- Lomasney JW, Lorenz W, Allen LF, King K, Regan JW, Yang-Feng TL, et al. Expansion of the alpha 2-adrenergic receptor family: cloning and characterization of a human alpha 2-adrenergic receptor subtype, the gene for which is located on chromosome 2. Proc Natl Acad Sci USA. 1990;87:5094–5098. doi: 10.1073/pnas.87.13.5094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nackley AG, Shabalina SA, Tchivileva IE, Satterfield K, Korchynskyi O, Makarov SS, et al. Human catechol-O-methyltransferase haplotypes modulate protein expression by altering mRNA secondary structure. Science. 2006;314:1930–1933. doi: 10.1126/science.1131262. [DOI] [PubMed] [Google Scholar]
- Pattij T, Vanderschuren LJ. The neuropharmacology of impulsive behaviour. Trends Pharmacol Sci. 2008;29:192–199. doi: 10.1016/j.tips.2008.01.002. [DOI] [PubMed] [Google Scholar]
- Polanczyk G, Zeni C, Genro JP, Guimaraes AP, Roman T, Hutz MH, et al. Association of the adrenergic alpha2A receptor gene with methylphenidate improvement of inattentive symptoms in children and adolescents with attention-deficit/hyperactivity disorder. Arch Gen Psychiatry. 2007;64:218–224. doi: 10.1001/archpsyc.64.2.218. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Human Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasch B, Spalek K, Buholzer S, Luechinger R, Boesiger P, Papassotiropoulos A, et al. A genetic variation of the noradrenergic system is related to differential amygdala activation during encoding of emotional memories. Proc Natl Acad Sci USA. 2009;106:19191–19196. doi: 10.1073/pnas.0907425106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson ES, Eagle DM, Mar AC, Bari A, Banerjee G, Jiang X, et al. Similar effects of the selective noradrenaline reuptake inhibitor atomoxetine on three distinct forms of impulsivity in the rat. Neuropsychopharmacology. 2008;33:1028–1037. doi: 10.1038/sj.npp.1301487. [DOI] [PubMed] [Google Scholar]
- Scheinin M, Lomasney JW, Hayden-Hixson DM, Schambra UB, Caron MG, Lefkowitz RJ, et al. Distribution of alpha 2-adrenergic receptor subtype gene expression in rat brain. Brain research. Mol Brain Res. 1994;21:133–149. doi: 10.1016/0169-328x(94)90386-7. [DOI] [PubMed] [Google Scholar]
- Small KM, Brown KM, Forbes SL, Liggett SB. Polymorphic deletion of three intracellular acidic residues of the alpha 2B-adrenergic receptor decreases G protein-coupled receptor kinase-mediated phosphorylation and desensitization. J Biol Chem. 2001;276:4917–4922. doi: 10.1074/jbc.M008118200. [DOI] [PubMed] [Google Scholar]
- Tavares A, Handy DE, Bogdanova NN, Rosene DL, Gavras H. Localization of alpha 2A- and alpha 2B-adrenergic receptor subtypes in brain. Hypertension. 1996;27 (3 Part 1:449–455. doi: 10.1161/01.hyp.27.3.449. [DOI] [PubMed] [Google Scholar]
- Turner DC, Clark L, Dowson J, Robbins TW, Sahakian BJ. Modafinil improves cognition and response inhibition in adult attention-deficit/hyperactivity disorder. Biol Psychiatry. 2004;55:1031–1040. doi: 10.1016/j.biopsych.2004.02.008. [DOI] [PubMed] [Google Scholar]
- Turner DC, Robbins TW, Clark L, Aron AR, Dowson J, Sahakian BJ. Cognitive enhancing effects of modafinil in healthy volunteers. Psychopharmacology. 2003;165:260–269. doi: 10.1007/s00213-002-1250-8. [DOI] [PubMed] [Google Scholar]
- Urner M, van Wingen G, Franke B, Rijpkema M, Fernandez G, Tendolkar I. Genetic variation of the alpha2b-adrenoceptor affects neural correlates of successful emotional memory formation. Hum Brain Mapp. 2011;21:21171. doi: 10.1002/hbm.21171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbruggen F, Logan GD. Response inhibition in the stop-signal paradigm. Trends Cogn Sci. 2008;12:418–424. doi: 10.1016/j.tics.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue G, Aron AR, Poldrack RA. Common neural substrates for inhibition of spoken and manual responses. Cerebral Cortex. 2008;18:1923–1932. doi: 10.1093/cercor/bhm220. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Bertolino A, Fazio L, Blasi G, Rampino A, Romano R, et al. Polymorphisms in human dopamine D2 receptor gene affect gene expression, splicing, and neuronal activity during working memory. Proc Natl Acad Sci USA. 2007;104:20552–20557. doi: 10.1073/pnas.0707106104. [DOI] [PMC free article] [PubMed] [Google Scholar]