Abstract
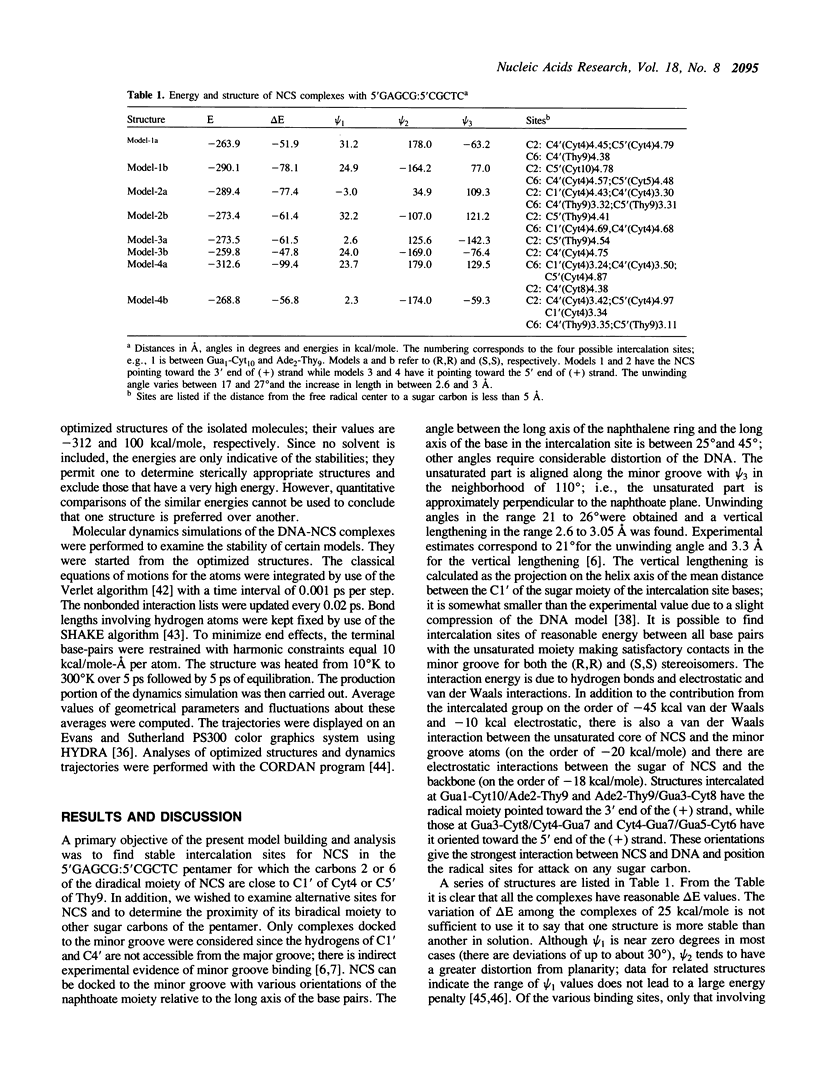
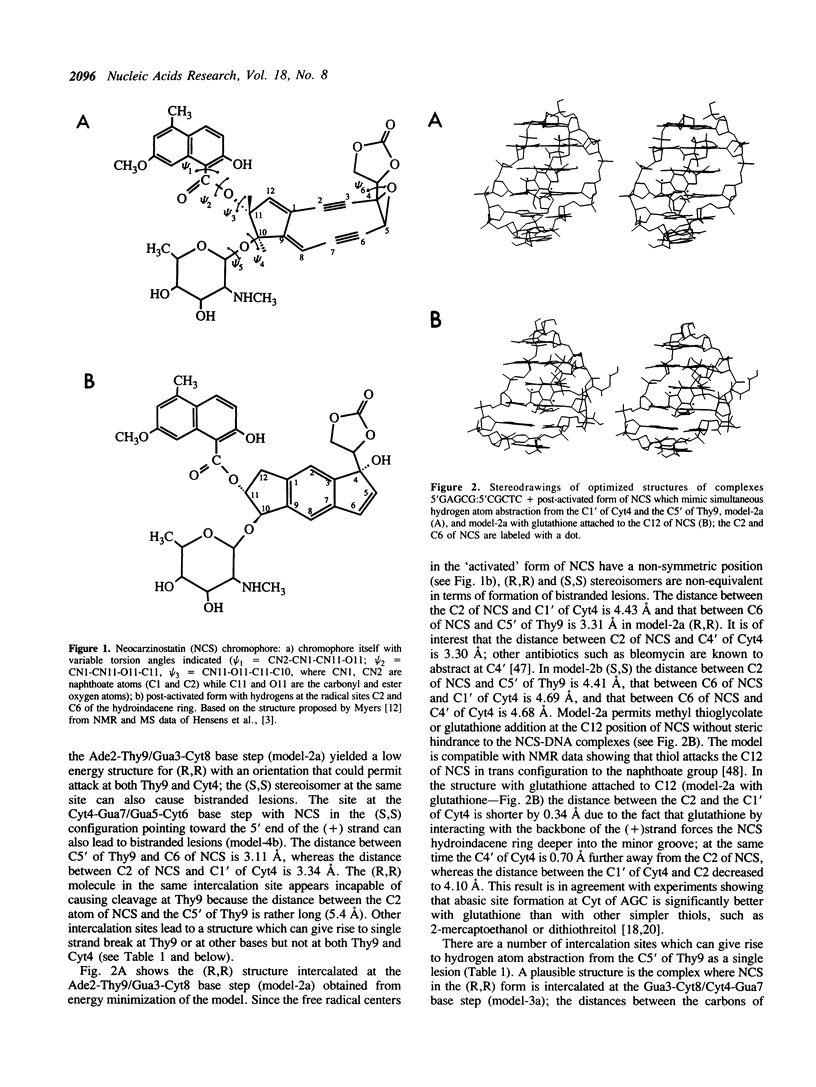
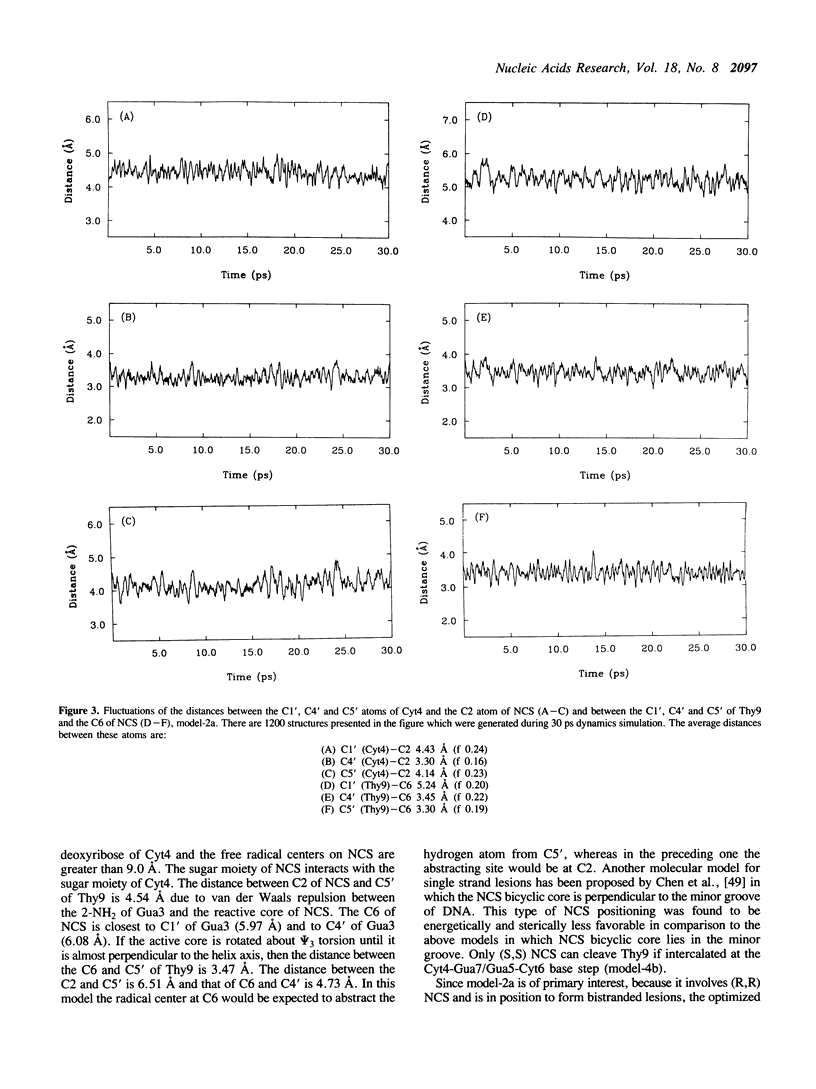
Model building and molecular mechanics and dynamics calculations have been performed on a number of complexes of the post-activated form of the neocarzinostatin chromophore (NCS) with the B-DNA oligomer 5'GAGCG:5'CGCTC. Stable structures with the naphthoic acid moiety intercalated at all base pairs can be constructed. The observed bistranded lesions consisting of an abasic site at the Cyt residue in AGC and a direct break at the Thy residue on the complementary strand can be explained by assuming that NCS in the (R,R) form intercalates between the Ade2-Thy9/Gua3-Cyt8 base step with its 'diradical' core oriented towards the 3'-end of the (+) strand. Sites at C5', C4' and C1' in the minor groove are within a short enough distance from the two radical centers on NCS to permit hydrogen atom abstraction and the formation of the bistranded lesions. Strand cleavage at Thy9 may occur as a single lesion if NCS is intercalated into the Gua3-Cyt8/Cyt4-Gua7 base step with its active core towards the 3'-end of the (-) strand. The results are analyzed, and the utility and limitations of this type of model building are discussed.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arnott S., Hukins D. W. Optimised parameters for A-DNA and B-DNA. Biochem Biophys Res Commun. 1972 Jun 28;47(6):1504–1509. doi: 10.1016/0006-291X(72)90243-4. [DOI] [PubMed] [Google Scholar]
- Bose K. K., Tatsumi K., Strauss B. S. Apurinic/apyrimidinic endonuclease sensitive sites as intermediates in the in vitro degradation of deoxyribonucleic acid by neocarzinostatin. Biochemistry. 1980 Oct 14;19(21):4761–4766. doi: 10.1021/bi00562a007. [DOI] [PubMed] [Google Scholar]
- Boye E., Köhnlein W., Skarstad K. Characterization of intracellular DNA strand breaks induced by neocarzinostatin in Escherichia coli cells. Nucleic Acids Res. 1984 Nov 12;12(21):8281–8291. doi: 10.1093/nar/12.21.8281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K. X., Gresh N., Pullman B. A tentative model of the intercalative binding of the neocarzinostatin chromophore to double-stranded tetranucleotides. Nucleic Acids Res. 1987 Mar 11;15(5):2175–2189. doi: 10.1093/nar/15.5.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin D. H., Zeng C. H., Costello C. E., Goldberg I. H. Sites in the diyne-ene bicyclic core of neocarzinostatin chromophore responsible for hydrogen abstraction from DNA. Biochemistry. 1988 Oct 18;27(21):8106–8114. doi: 10.1021/bi00421a020. [DOI] [PubMed] [Google Scholar]
- Dasgupta D., Goldberg I. H. Mode of reversible binding of neocarzinostatin chromophore to DNA: evidence for binding via the minor groove. Biochemistry. 1985 Nov 19;24(24):6913–6920. doi: 10.1021/bi00345a025. [DOI] [PubMed] [Google Scholar]
- Dickerson R. E. Base sequence and helix structure variation in B and A DNA. J Mol Biol. 1983 May 25;166(3):419–441. doi: 10.1016/s0022-2836(83)80093-x. [DOI] [PubMed] [Google Scholar]
- Edo K., Akiyama Y., Saito K., Mizugaki M., Koide Y., Ishida N. Absolute configuration of the amino sugar moiety of the neocarzinostatin chromophore. J Antibiot (Tokyo) 1986 Nov;39(11):1615–1619. doi: 10.7164/antibiotics.39.1615. [DOI] [PubMed] [Google Scholar]
- Galat A., Goldberg I. H. Analysis of microdensitometric data in terms of probability of cleavage of DNA. Comput Appl Biosci. 1987 Nov;3(4):333–338. doi: 10.1093/bioinformatics/3.4.333. [DOI] [PubMed] [Google Scholar]
- Goldberg I. H. Free radical mechanisms in neocarzinostatin-induced DNA damage. Free Radic Biol Med. 1987;3(1):41–54. doi: 10.1016/0891-5849(87)90038-4. [DOI] [PubMed] [Google Scholar]
- Hatayama T., Goldberg I. H. DNA damage and repair in relation to cell killing in neocarzinostatin-treated HeLa cells. Biochim Biophys Acta. 1979 Jun 20;563(1):59–71. doi: 10.1016/0005-2787(79)90007-8. [DOI] [PubMed] [Google Scholar]
- Hawley R. C., Kiessling L. L., Schreiber S. L. Model of the interactions of calichemicin gamma 1 with a DNA fragment from pBR322. Proc Natl Acad Sci U S A. 1989 Feb;86(4):1105–1109. doi: 10.1073/pnas.86.4.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensens O. D., Dewey R. S., Liesch J. M., Napier M. A., Reamer R. A., Smith J. L., Albers-Schönberg G., Goldberg I. H. Neocarzinostatin chromophore: presence of a highly strained ether ring and its reaction with mercaptan and sodium borohydride. Biochem Biophys Res Commun. 1983 Jun 15;113(2):538–547. doi: 10.1016/0006-291x(83)91759-x. [DOI] [PubMed] [Google Scholar]
- Hensens O. D., Goldberg I. H. Mechanism of activation of the antitumor antibiotic neocarzinostatin by mercaptan and sodium borohydride. J Antibiot (Tokyo) 1989 May;42(5):761–768. doi: 10.7164/antibiotics.42.761. [DOI] [PubMed] [Google Scholar]
- Kappen L. S., Chen C. Q., Goldberg I. H. Atypical abasic sites generated by neocarzinostatin at sequence-specific cytidylate residues in oligodeoxynucleotides. Biochemistry. 1988 Jun 14;27(12):4331–4340. doi: 10.1021/bi00412a021. [DOI] [PubMed] [Google Scholar]
- Kappen L. S., Goldberg I. H. Activation of neocarzinostatin chromophore and formation of nascent DNA damage do not require molecular oxygen. Nucleic Acids Res. 1985 Mar 11;13(5):1637–1648. doi: 10.1093/nar/13.5.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappen L. S., Goldberg I. H. Deoxyribonucleic acid damage by neocarzinostatin chromophore: strand breaks generated by selective oxidation of C-5' of deoxyribose. Biochemistry. 1983 Oct 11;22(21):4872–4878. doi: 10.1021/bi00290a002. [DOI] [PubMed] [Google Scholar]
- Kappen L. S., Goldberg I. H. Identification of 2-deoxyribonolactone at the site of neocarzinostatin-induced cytosine release in the sequence d(AGC). Biochemistry. 1989 Feb 7;28(3):1027–1032. doi: 10.1021/bi00429a016. [DOI] [PubMed] [Google Scholar]
- Napier M. A., Goldberg I. H. Neocarzinostatin chromophore. Assignment of spectral properties and structural requirements for binding to DNA. Mol Pharmacol. 1983 Mar;23(2):500–510. [PubMed] [Google Scholar]
- Napier M. A., Holmquist B., Strydom D. J., Goldberg I. H. Neocarzinostatin: spectral characterization and separation of a non-protein chromophore. Biochem Biophys Res Commun. 1979 Jul 27;89(2):635–642. doi: 10.1016/0006-291x(79)90677-6. [DOI] [PubMed] [Google Scholar]
- Platzer K. E., Momany F. A., Scheraga H. A. Conformational energy calculations of enzyme-substrate interactions. II. Computation of the binding energy for substrates in the active site of -chymotrypsin. Int J Pept Protein Res. 1972;4(3):201–219. doi: 10.1111/j.1399-3011.1972.tb03420.x. [DOI] [PubMed] [Google Scholar]
- Poon R., Beerman T. A., Goldberg I. H. Characterization of DNA strand breakage in vitro by the antitumor protein neocarzinostatin. Biochemistry. 1977 Feb 8;16(3):486–493. doi: 10.1021/bi00622a023. [DOI] [PubMed] [Google Scholar]
- Povirk L. F., Dattagupta N., Warf B. C., Goldberg I. H. Neocarzinostatin chromophore binds to deoxyribonucleic acid by intercalation. Biochemistry. 1981 Jul 7;20(14):4007–4014. doi: 10.1021/bi00517a009. [DOI] [PubMed] [Google Scholar]
- Povirk L. F., Goldberg I. H. Base substitution mutations induced in the cI gene of lambda phage by neocarzinostatin chromophore: correlation with depyrimidination hotspots at the sequence AGC. Nucleic Acids Res. 1986 Feb 11;14(3):1417–1426. doi: 10.1093/nar/14.3.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Povirk L. F., Goldberg I. H. Endonuclease-resistant apyrimidinic sites formed by neocarzinostatin at cytosine residues in DNA: evidence for a possible role in mutagenesis. Proc Natl Acad Sci U S A. 1985 May;82(10):3182–3186. doi: 10.1073/pnas.82.10.3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Povirk L. F., Houlgrave C. W. Effect of apurinic/apyrimidinic endonucleases and polyamines on DNA treated with bleomycin and neocarzinostatin: specific formation and cleavage of closely opposed lesions in complementary strands. Biochemistry. 1988 May 17;27(10):3850–3857. doi: 10.1021/bi00410a049. [DOI] [PubMed] [Google Scholar]
- Povirk L. F., Houlgrave C. W., Han Y. H. Neocarzinostatin-induced DNA base release accompanied by staggered oxidative cleavage of the complementary strand. J Biol Chem. 1988 Dec 25;263(36):19263–19266. [PubMed] [Google Scholar]
- Takeshita M., Kappen L. S., Grollman A. P., Eisenberg M., Goldberg I. H. Strand scission of deoxyribonucleic acid by neocarzinostatin, auromomycin, and bleomycin: studies on base release and nucleotide sequence specificity. Biochemistry. 1981 Dec 22;20(26):7599–7606. doi: 10.1021/bi00529a039. [DOI] [PubMed] [Google Scholar]
- Tidor B., Irikura K. K., Brooks B. R., Karplus M. Dynamics of DNA oligomers. J Biomol Struct Dyn. 1983 Oct;1(1):231–252. doi: 10.1080/07391102.1983.10507437. [DOI] [PubMed] [Google Scholar]
- Tsai C. C., Jain S. C., Sobell H. M. Visualization of drug-nucleic acid interactions at atomic resolution. I. Structure of an ethidium/dinucleoside monophosphate crystalline complex, ethidium:5-iodouridylyl (3'-5') adenosine. J Mol Biol. 1977 Aug 15;114(3):301–315. doi: 10.1016/0022-2836(77)90252-2. [DOI] [PubMed] [Google Scholar]
- Wu J. C., Kozarich J. W., Stubbe J. The mechanism of free base formation from DNA by bleomycin. A proposal based on site specific tritium release from Poly(dA.dU). J Biol Chem. 1983 Apr 25;258(8):4694–4697. [PubMed] [Google Scholar]
- Zein N., Poncin M., Nilakantan R., Ellestad G. A. Calicheamicin gamma 1I and DNA: molecular recognition process responsible for site-specificity. Science. 1989 May 12;244(4905):697–699. doi: 10.1126/science.2717946. [DOI] [PubMed] [Google Scholar]
- Zein N., Sinha A. M., McGahren W. J., Ellestad G. A. Calicheamicin gamma 1I: an antitumor antibiotic that cleaves double-stranded DNA site specifically. Science. 1988 May 27;240(4856):1198–1201. doi: 10.1126/science.3240341. [DOI] [PubMed] [Google Scholar]



