Abstract
Linking the molecular aberrations of cancer to drug responses could guide treatment choice and identify new therapeutic applications. However, there has been no systematic approach for analyzing gene-drug interactions in human cells. We establish a multiplexed assay to study the cellular fitness of a panel of engineered isogenic cancer cells in response to a collection of drugs, enabling the systematic analysis of thousands of gene-drug interactions. Applying this approach to breast cancer revealed various synthetic-lethal interactions and drug resistance mechanisms, some of which were known, thereby validating the method. NOTCH pathway activation, which occurs frequently in breast cancer, unexpectedly conferred resistance to PI3K inhibitors, which are currently undergoing clinical trials in breast cancer patients. NOTCH1 and downstream induction of c-MYC overrode the dependency of cells on the PI3K/mTOR pathway for proliferation. These data reveal a novel mechanism of resistance to PI3K inhibitors with direct clinical implications.
INTRODUCTION
Many factors contribute to patients’ responses to anti-cancer therapy, including pharmacogenetics, tumor microenvironment, vascularity and genetic aberrations 1-5. Identifying the molecular mechanisms that influence response to anti-cancer drugs can improve therapy by identifying those individuals who will benefit most while avoiding unnecessary treatment. However, due partly to the heterogeneity between tumors, identifying robust biomarkers and functionally linking cancer genes to drug sensitivity has been challenging. Nonetheless, catalogues describing the molecular changes in the major tumor types, currently emerging from sequencing efforts, will theoretically enable systematic studies into the molecular aberrations underpinning treatment response 4, 6, 7.
Another important objective of cancer research is to develop new anti-cancer treatments with increased specificity for cancer cells. For example, the monoclonal antibody Trastuzumab directly targets HER2/NEU-positive breast cancer and BRAF kinase inhibitors have recently shown promise in melanoma carrying BRAF mutations 8, 9. However, it is not often possible to directly translate known molecular aberrations of cancer cells into targeted therapies. For instance, the oncogenic transcription factor c-MYC is overexpressed in a variety of malignancies, but because it lacks critical hydrophobic pockets it is challenging to target by small-molecule compounds 10, 11. Alternative approaches for identifying drugs that specifically target cancer cells are urgently needed.
The molecular changes that occur in cancer cells can result in a dependency on gene products that are not essential in normal cells 12-14. Inhibition of these proteins would thus result in cell cycle arrest or death of the cancer cell but would not affect fitness of their normal counterparts. This notion, which is termed synthetic sickness or lethality, induced essentiality or non-oncogene addiction, provides a framework to identify drugs that do not target the cancer gene directly yet are specific for cells that contain the aberration. Indeed, the observation that cells containing BRCA mutations are hypersensitive to inhibition of the enzyme PARP has found its way into the clinic and represents the paradigm for synthetic lethality-based therapy 15, 16. However, there are currently only a few cancer-relevant synthetic-lethal interactions that have been identified 17. Thus, a systematic analysis of the effect of individual cancer genes on the cellular response to existing and experimental drugs may identify new targeted anti-cancer therapies directly relevant for the clinic. The challenge of such a systematic approach is the large number of combinations among drugs and genes that would have to be analyzed. The promise of insight into drug actions as exemplified by similar screens in model organisms, most notably yeast, warrants development of suitable methods in human cells 18, 19.
We developed a method to multiplex cellular fitness measurements of up to one hundred isogenic cell lines using molecular barcodes to facilitate the quantitative assessment of functional drug-gene interactions in human cells. This method assists the systematic assessment of the impact of cancer aberrations on proliferation in response to a collection of drugs. Here, we present the approach and use it to query a 70 × 87 drug-gene interaction matrix in breast cancer cells, which allowed the interrogation of over 6 thousand drug-gene pairs. In addition to several previously identified drug-gene interactions, we report a novel mechanism of resistance to PI3K inhibitors, which are currently in clinical trials 20. This is of particular importance given the large fraction of breast tumors with activating mutations in the PI3K pathway 21.
RESULTS
A platform for combinatorial fitness screens
The first step in building a platform to multiplex large numbers of combinations of genetic and chemical perturbations was to develop a sensitive and quantitative method using molecular barcodes to allow the identification of populations of cells carrying specific genetic modifications within a complex mixture. Molecular barcodes are short non-transcribed stretches of DNA, which when integrated into the genomic DNA of a cell line introduce a molecular beacon that can be selectively quantified by PCR. In a mixed population of cells, each containing a unique barcode, the relative number of cells containing a particular vector can therefore be determined by quantification of the barcodes. By pairing genetic modifications of cells (e.g. the expression of an oncogene or knockdown of a tumor suppressor) with these barcodes, the cellular fitness upon drug treatment can be followed in a multiplexed fashion. Thus, we first generated one hundred lentiviral vectors carrying unique molecular barcodes flanked by common primer sites for efficient delivery into human cells (Supplementary Results, Supplementary Fig. 1).
We used an isogenic cell line approach to identify the effect of individual genetic changes on cell growth (i.e. fitness) in response to a specific drug, and bypass the difficulty of comparing heterogeneous cell lines with their multitudes of genetic changes14. Individual genetic modifications were introduced into cells with the same genetic background using overexpression and RNA interference (RNAi). To systematically analyze the effects of a drug library on this heterogeneous population of cells, each unique barcode was then paired with one genetic modification, so that the cellular fitness upon drug treatment could be followed in a multiplexed fashion (Fig. 1a).
Figure 1. Barcode screen set-up, detection and performance.
(a) Isogenic cell lines infected with a lentiviral vector carrying a unique 24 base pair barcode sequence and specific genetic modification (e.g. cDNA or RNAi) are pooled, seeded in multi-well plates and subsequently treated with drug or DMSO control (left). The relative abundance of the barcodes in the population of cells is a proxy for the cellular fitness. In the example the cells with the “orange” barcode display a synthetic sick/lethal interactions with Drug X. After drug treatment the pooled isogenic cell lines are harvested, genomic DNA (gDNA) is isolated and barcodes are amplified (right). Labeled product is then hybridized to Luminex microspheres and the mixture is measured on a Luminex machine to determine the relative abundance for each of the 100 barcode sequences. (b) Barcoded cells expressing the inactive FANCD2-K561R cDNA were mixed into a pool of barcoded cells expressing wild-type FANCD2 and treated with MMC (15 ng/ml) for 5 days. Shown are the median signals for all barcodes of 4 independent drug treatments compared to DMSO control.
To quantify the barcodes we used the hybridization-based Luminex xMAP technology, which uses a set of fluorescent microspheres coupled to antisense DNA barcodes that are analyzed by flow cytometry 22. Advantages of this methodology over massive parallel sequencing are that it is fast and the cost per sample is independent of the size of the experiment, making the method highly flexible and affordable (about 2 cents per data point). Briefly, barcodes were amplified from genomic DNA by PCR, fluorescently labeled and hybridized to microspheres that are coupled to the antisense barcode sequence. Subsequent analysis of the beads then reveals the relative abundance of each barcode (Fig. 1a).
We subjected the screening platform to specific tests to determine its reliability and power for identifying drug-gene interactions. The typical dynamic range and linearity of the barcode detection extended over two orders of magnitude and the relative signals were maintained upon reamplification, indicating limited PCR bias (Supplementary Figs. 2, 3) Furthermore, the method was highly robust as illustrated by the high correlation coefficients of both technical and biological replicates (Pearson correlation coefficient r2 >0.98; Supplementary Fig. 4).
Because the quantification method is hybridization-based, we needed to exclude any cross-hybridization of barcode sequences as this could obscure the detection of individual barcodes. For this purpose we assembled one hundred pools of barcoded vectors in which a single vector was omitted and performed barcode measurements on PCR amplified material. In all cases the absence of the correct barcode was confirmed, indicating limited cross hybridization under these conditions (Supplementary Fig. 5).
Next, we determined if the method was able to detect differences in cellular fitness in a complex mixture of barcoded cells. We used drug hypersensitivity as a benchmark as it is technically more challenging to detect the absence of a cell within a population than the increase in proliferation occurring in drug resistance. Cells were infected with one of 95 barcoded vectors carrying a puromycin resistance gene or a barcoded vector lacking this cassette (#96). As expected, treatment with puromycin only killed the cells without the resistance gene, leaving all others unaffected (Supplementary Fig. 6). In addition, when all cells were pooled and subsequently treated with puromycin, a strong and highly significant (2-tailed t-test, p< 0.0001) depletion of the barcode associated with the puromycin-less vector was detectable whereas all other barcodes remained unchanged (Supplementary Fig. 6). Thus, the approach was sensitive enough to detect the loss of one individual cell population within a complex mixture.
As an additional proof-of-principle experiment, we measured the known hypersensitivity of Fanconi Anemia complementation group D2 (FANCD2) patient cells for the DNA cross-linking agent Mitomycin C (MMC) in the multiplexed assay 23. A patient-derived cell line (PD20) stably transduced with a vector expressing wild-type FANCD2 or an inactive point mutant (K561R) were infected with barcoded lentiviruses, pooled and subsequently exposed to MMC. As predicted, the barcode derived from the cells expressing the inactive mutant protein was depleted from the population, which could be clearly detected with our screening approach, thus confirming the MMC hypersensitivity of FANCD2 mutant cells (Fig. 1b, Supplementary Fig. 7). Together, these experiments show that the screening platform provides a semi-quantitative method to determine cellular fitness in a multiplexed format.
A synthetic lethal and drug resistance screen
We applied our screening platform to interrogate drug-gene interactions in breast cancer cells. We first established an isogenic cell line model based on the non-tumorigenic human breast epithelial cell line MCF10A. The cell line was selected because it has a relatively normal karyotype and is thought to represent a multi-lineage progenitor as it has transcriptional characteristics of both basal and luminal cell types 24. Furthermore, the cells are responsive to most signaling pathways present in normal breast epithelial cells. A previously reported deletion of the INK4A locus and some other chromosomal aberrations could be confirmed by high-density SNP array (data not shown)25. We selected breast cancer-relevant genetic aberrations using an extensive literature and database search. This yielded a list of seventy genes that have been clearly linked to breast cancer, including HER2, BRCA1/2, c-MYC, NOTCH1 and PTEN, which were selected for the drug-gene interaction screen (Supplementary Fig. 8, Supplementary Table 1). To mimic the aberrations of these genes in cancer we manipulated their expression using cDNA overexpression or RNAi, and unique barcodes were introduced by lentiviral transduction, yielding a total of 89 isogenic cell lines (Supplementary Tables 1-5). All cDNAs and the majority of knockdowns were confirmed using immunoblot and qRT-PCR and for a number of stable cell lines a marked morphological change was observed indicative of oncogenic transformation (Supplementary Fig. 9, Supplementary Table 6 and data not shown).
After pooling all barcoded cells they were screened against a custom compound library, which was selected to maximize the chance of identifying a drug-gene interaction that could be useful in the clinic. The library mainly consisted of clinically relevant kinase inhibitors and several tool compounds, together comprising 87 small molecules (Supplementary Fig. 10, Supplementary Table 7). The library was screened at various concentrations in quadruplicate, which yielded over thirty thousand data points (Fig. 2a). Data analysis revealed several gene-drug interactions including synthetic lethal interactions between three components of the NOTCH signaling pathway (i.e. JAG1, NOTCH1 and c-MYC) and the Aurora kinase drugs AT9283 and SNS-314 (Fig. 2a, Supplementary Table 8). Validation experiments with cells expressing the intracellular active domain of NOTCH1 (ICN1) or c-MYC confirmed the exquisite sensitivity to these compounds and four additional Aurora kinase inhibitors (Fig. 2b, Supplementary Fig. 11). NOTCH1 and its putative direct target gene c-MYC have recently been shown to display a synthetic lethal interaction with Aurora B kinase in retinal epithelial cells, corroborating our findings and further validating the approach 26. Furthermore, the observation that multiple components of a single pathway cluster with two drugs targeting the same gene product illustrates how large-scale drug-gene screens in human cells could be used to elucidate drug action and gene function, and is reminiscent of the synthetic lethal screens in yeast 18, 19.
Figure 2. Combinatorial breast cancer gene small compound screen.
(a) Radial gene-drug interaction plot displaying the 7743 (89 isogenic cell lines × 87 drugs) pairwise drug-gene measurements. Distance from the center indicates significance and dot size is proportional to the magnitude of the drug versus control effect. P-values for selected hits are indicated. (b) Dose-response analysis of c-MYC, ICN1 and control MCF10A cells with the Aurora kinase inhibitor AT9283. Cells were treated with the indicated concentrations for 5 days and relative cell number was assessed. The experiment was repeated three times in triplicate and standard deviations are indicated. (c) Dose-response analysis of ICN1 and control MCF10A cells. Cells were treated with the indicated concentrations BEZ-235 for 5 days and relative cell numbers were measured. The graph is based on 4 independent experiments in triplicate and standard deviations are indicated. (d) Crystal violet stained culture dishes of colony formation experiment. Cells were seeded at low density and treated with 30 pg/μl BEZ-235 for 10 days.
NOTCH1 activation confers resistance to PI3K inhibition
Importantly, our screen revealed several novel drug-gene interactions. The highest scoring resistance hit in the screen was the intracellular active domain of NOTCH1 (ICN1), conferring resistance to the dual PI3K/mTOR inhibitor BEZ-235 (Fig. 2a) 27. Given the clinical relevance of both PI3K inhibitors and NOTCH1 in breast cancer, and no reported connection between the two, we decided to study this observation further 20, 21.
A marked resistance to BEZ-235 in ICN1 expressing cells was observed in short-term dose-response analysis and long-term growth assays, confirming the results from the screen (Figs. 2c,d; Supplementary Figs. 12, 13). Furthermore, in cells expressing a NOTCH1 mutant that lacks the extracellular domain (NOTCH-delta E) BEZ-235 sensitivity could be restored by inhibiting γ-secretase, indicating that naturally cleaved NOTCH1 also confers resistance to PI3K/mTOR inhibition (Supplementary Fig. 14) 28. Although our initial analysis revealed that ICN1 only showed a significant interaction with BEZ-235, we reasoned ICN1 cells might also be resistant to some of the other PI3K inhibitors used in the screen. Indeed, when all remaining PI3K inhibitors were analyzed as a group, the interaction with ICN1 was also significant (1-tailed t-test, p < 0.05), indicating that the resistance could be extended to other PI3K inhibitors (Supplementary Fig. 15). Consistent with this, we found that resistance to PIK90, a selective PI3K inhibitor, could be confirmed in dose-response experiments (Supplementary Fig. 16).
To begin to uncover the mechanism whereby activation of NOTCH1 in cells confers resistance to PI3K inhibitors we analyzed one of the main downstream effector pathways of PI3K: the serine-threonine kinase mTOR, which resides in the two distinct protein complexes mTORC1 and mTORC2 29. We found that ICN1 expressing cells were also less sensitive to PP242, an mTOR kinase inhibitor, and Everolimus or Rapamycin, non-ATP competitive mTOR inhibitors that may affect mTORC1 more potently than mTORC2 (Figs. 3a,b, Supplementary Fig. 17) 30. Similarly, ICN1 cells were much less affected by mTOR knockdown than control cells (Supplementary Fig. 18). Together, this indicates that activation of NOTCH1 can bypass the cellular requirement for this growth pathway and that consistent with previous reports, in these cells PI3K inhibitors mainly exert their effect by acting on the mTOR pathway 31.
Figure 3. NOTCH activation renders breast cancer cells resistant to PI3K/mTORC1 inhibition.
(a) Bar graph showing relative viability of ICN1 or control MCF10A cells treated with PP242 (3.0 μM) for 5 days. Shown is the mean of a representative experiment performed in triplicate and standard deviations. (b) Dose-response analysis of MCF10A cells treated with the indicated concentrations of Everolimus (Rad001) for 5 days. Mean and standard deviations are indicated (c) Box plots of GFP positive cells transduced with an ICN1-ires-GFP virus and treated with BEZ-235 (10 pg/μl) or DMSO for 7 days. Data from three replicates each are shown. (d) Oncomine analysis (see Methods) of NUMB expression in 274 PI3K/mTOR inhibitor sensitive or resistant cell lines. The red boxed area indicates cell lines with lower than median expression of NUMB.
Next, we investigated if the NOTCH1-mediated resistance could also be observed in other human cancer cell lines. Importantly, the breast adenocarcinoma-like cell line MCF7 and the ductal carcinoma-like cell lines BT474, HCC70 and BT549 all showed resistance to BEZ-235 treatment upon expression of ICN1 (Fig. 3c, Supplementary Fig. 19) 24. To ask if NOTCH activation may also confer PI3K/mTOR inhibitor resistance in other tumor types we analyzed a publicly available dataset created by GlaxoSmithKline, comprising over 300 molecularly characterized and drug treated cell lines (see Methods). This revealed a significant (chi-square test, p < 0.01) correlation between low expression of NUMB, a negative regulator of NOTCH, and resistance to PI3K/mTOR inhibition in cell lines derived from various tumor types, including melanoma and hepatocellular carcinoma (Fig. 3d) 32. These results suggest that uncoupling proliferation from the PI3K/mTOR pathway via NOTCH1 activation may be a more general phenomenon across cancer cell lines.
ICN1 overrides mTORC1 signaling via c-MYC transcription
Ribosomal S6 Kinase (S6K) and the eukaryotic translation initiation factor 4E-binding protein 1 (4EBP1) are main effector molecules of mTORC1 and their phosphorylation stimulates protein translation 29. Interestingly, S6K and 4EBP1 phosphorylation was equally inhibited in ICN1 expressing cells as in control cells (Fig. 4a, Supplementary Fig. 21). This suggests that ICN1 uncouples mTORC1 signaling from proliferation by a downstream mechanism.
Figure 4. c-MYC induction confers resistance to PI3K/mTOR inhibition.
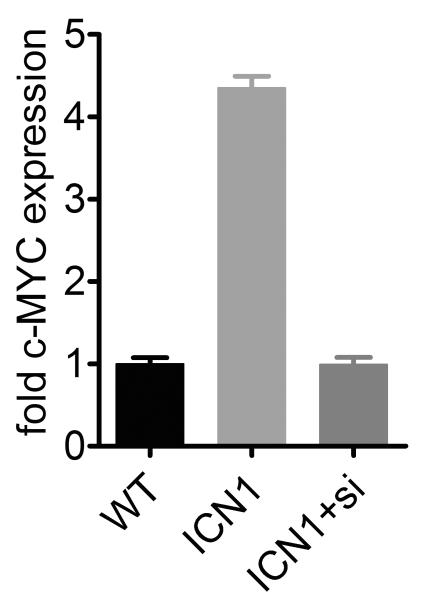
(a) Western blot analysis of ICN1 or control MCF10A cells treated with BEZ-235 (pg/μl) as indicated for 24 hours. Total lysates were probed with an antibody against phosphorylated ribosomal S6 kinase (Thr371) and total mTOR as a loading control (see Supplementary Fig. 20 for an uncropped version). (b) Data from the screen shows c-MYC as a significant hit for resistance to BEZ-235. (c) Relative c-MYC mRNA levels in ICN1 and c-MYC cells as determined by qRT-PCR. Shown is the fold change compared to wild-type MCF10A cells and standard deviations of 3 replicates. (d) Dose-response curve of c-MYC or control MCF10A cells treated with BEZ-235. Cells were treated for 5 days as indicated and relative cell number was measured. The data represent four independent experiments were performed in triplicate and error bars indicate standard deviations. (e) Quantitative RT-PCR of c-MYC expression in wild-type MCF10A or ICN1 cells transfected with Luciferase siRNA and ICN1 cells transfected with c-MYC siRNA pool (ICN1+si). Standard deviations of 3 replicates are indicated. (f) Dose-response curve of cells in (e) treated with BEZ-235 or vehicle for 5 days. Three replicates were performed; error bars indicate standard deviation. (g) Oncomine analysis (see Methods) of c-MYC gene copy number in PI3K/mTOR inhibitor sensitive or resistant cell lines. The red-boxed area indicates cell lines with c-MYC gene amplification (Chi square P value is indicated).
Upon closer inspection of the screening data we found that cells transduced with c-MYC also displayed remarkable resistance to BEZ-235 and other PI3K inhibitors (Fig. 4b, Supplementary Fig. 22). Notably, the c-MYC expression level and shift in the BEZ-235 dose-response curve was comparable to ICN1 expressing cells, indicating that c-MYC may be the main transcriptional target conferring the resistance (Figs. 4c,d) 33-35. In agreement with this, overexpression of the NOTCH canonical target genes HES1, HEY1 or HEY2 did not confer BEZ-235 resistance to MCF10A cells (Supplementary Fig. 23). Furthermore, c-MYC induction in NOTCH-deltaE expressing cells was γ-secretase sensitive and the NOTCH3 intracellular domain—that in these cells did not induce c-MYC expression—also did not confer resistance (Supplementary Fig. 24).
To investigate directly if c-MYC induction was required for resistance to BEZ-235 inhibition, we inhibited c-MYC expression by RNAi in ICN1 cells (Fig. 4e). As predicted, knockdown of c-MYC to levels comparable to control MCF10A cells completely reversed the resistance to BEZ-235 (Fig. 4f). This was not due to a general cytotoxic effect of c-MYC knockdown as the increased sensitivity to Aurora kinase inhibitors (i.e. synthetic lethality) was also reverted (Supplementary Fig. 25). These experiments show that c-MYC induction by ICN1 is necessary and sufficient for the PI3K/mTOR resistance.
Finally, the notion that c-MYC upregulation confers resistance to PI3K/mTOR inhibition prompted us to investigate if cell lines with c-MYC gene amplification also displayed this characteristic. Indeed, c-MYC amplification was observed significantly more often (chi-square test, p < 0.01) among PI3K/mTOR inhibitor resistant cell lines (Fig. 4g). This effect was specific as c-MYC amplified cells lines were not resistant for Aurora kinase inhibition but rather showed a trend towards synthetic lethality, which is in agreement with our previous findings (Supplementary Fig. 26, p=0.07).
Thus, we conclude that NOTCH pathway activation uncouples PI3K-mTOR signaling from proliferation by induction of c-MYC and this may have direct implications for patients treated with drugs targeting this pathway.
DISCUSSION
We identified a novel mechanism of resistance to PI3K inhibitors in breast cancer cell lines by activating NOTCH signaling and induction of c-MYC. NOTCH activation occurs in a subset of breast cancers and is associated with tumor progression and poor prognosis and MYC amplification is a relative frequent event 10, 36. PI3K and mTOR targeting drugs have received much attention as the pathway is frequently hijacked in a variety of malignancies, including breast cancer 21. As tumors invariably acquire resistance to single agent treatments, the ability to anticipate drug resistance has enormous clinical and economic value. However mechanisms of resistance in human tumors to PI3K inhibitors have not yet been reported.
We could show that resistance occurs by the transcriptional activation of c-MYC and that this seems to uncouple mTOR regulation of translation from proliferation. The stimulation of translation by c-MYC through the induction of eukaryotic initiation factor 4F (eIF4) family members is a known mechanism whereby c-MYC drives protein translation and is implicated in c-MYC-driven tumorigenesis 37, 38. This mechanism of how NOTCH1 activation could induce resistance to PI3K inhibitors is an attractive model but remains to be confirmed. Together, these observations position NOTCH and MYC activation as potential mechanisms of resistance to PI3K inhibitors with direct clinical implications.
We established a screening platform to systematically search for synthetic lethal interactions and mechanisms of drug resistance in cancer cells. The ability to pair tumor genotype with cancer treatment is receiving increasing attention as rising cost of cancer treatment is placing a burden on the health care system 39. The multiplexed assay allowed the interrogation of thousands of gene-drug combinations with the potential to identify clinically relevant interactions that could lead to new patient-stratified medicine. The method is cost effective, highly flexible, can be used with cDNA overexpression, RNAi or any cellular perturbation of interest and is applicable to all cells transducible with lentiviral vectors.
A potential drawback of engineered cells is that they do not necessarily fully capture the tumor evolution process of primary tumor cells and this may explain the absence of some expected “oncogene addiction” hits in our screen. Furthermore, false-negatives due to for instance insufficient knockdown or other technical limitations cannot be excluded and this may explain, for example, the absence of PTEN as a hit for resistance to PI3K inhibitors in our screen 40. Nonetheless, the identification of mechanisms of resistance and synthetic lethal interactions that are conserved across many cell lines justifies the approach and illustrates the power of isogenic models. Furthermore, the NOTCH pathway interaction with Aurora kinase inhibitors provides an example of how “guilt by association” can shed light on the mechanism of action of drugs or function of cancer genes 18. In summary, the ability to efficiently measure large numbers of drug-gene interactions in human cells has the potential to provide insight into various aspects of chemical biology.
METHODS
Cell culture, antibodies, compounds and RNAi
MCF10A cells (ATCC) were cultured in DMEM/F12 supplemented with 5% horse serum (Gibco), penicillin/streptomycin, insulin (10 ug/ml), cholera toxin (100 ng/ml), EGF (20 ng/ml) and hydrocortisone (500 ng/ml) (Sigma). All other cells were grown in DMEM supplemented with 10% FBS (Gibco) and penicillin/streptomycin. PDK1 antibody (E-3), anti-GFP and anti-p53 (DO-1) were purchased from Santa Cruz Biotechnology. Anti-beta-actin and anti-c-Myc antibody were obtained from Sigma-Aldrich. All other antibodies were acquired from Cell Signaling. Compounds were obtained from SynThesis Medchem (China) except for Rapamycin, Everolimus, Mitomycin C and PP242 (Sigma). Compound purity was >/= 95% according to the manufacturer except for PP242 (>/=98%). The γ-secretase inhibitor dibenzazepine was kindly provided by James Bradner. Purity and identity of this compound was verified by mass spectrometry and matched published standards.
siRNA experiments were performed by transfecting MCF10A cells with siLentfect (Bio-Rad) and 10 nM siRNA. c-MYC siRNA SMARTPool sequences (Dharmacon): 5′-CGAUGUUGUUUCUGUGGAA, 5′-AACGUUAGCUUCACCAACA, 5′-GAACACACAACGUCUUGGA, 5′-ACGGAACUCUUGUGCGUAA; Luciferase: 5′-UCGAAGUAUUCCGCGUACG. The previously validated shRNA targeting mTOR was obtained by cloning oligos into pLKO.1 and verified by sequencing 41.
Barcoded vectors and generation of isogenic cell lines
The stuffer fragment in the lentiviral vector pLKO.1 42 was replaced with a short linker sequence and barcodes (Flexmap barcode tags; Supplementary Table 5) flanked by primer sites and inserted 5′ of the U6 promoter. This vector (pLKO.2, see Supplementary Fig. 1) was then used to introduce stable DNA barcodes into cells by lentiviral transduction. Cloning oligos into pLKO.2 using the AgeI and EcoRI restriction sites generated short hairpin RNA expressing vectors. An overview of all vectors used in the screen is provided in Supplementary Table 1.
MCF10A isogenic cell lines overexpressing cDNAs or shRNAs were produced by lenti- or retroviral transduction and selection. Stable lines were cultured for approximately 4 weeks prior to the screen and barcoded by a second infection, when applicable. Prior to siRNA SMARTPool transfections MCF10A were infected with barcoded lentivirus.
Screen set-up and Luminex assay
For each compound a 4-point dose-response curve was determined in MCF10A cells using the Celltiter Glo assay (Promega). From these data, concentrations were selected for the screen. All barcoded cell lines were pooled, counted and seeded in multiwell plates in quadruplicate. Compound or DMSO was added 16 h after seeding using a liquid handling robot (Cybio). Medium was refreshed every second day and cells were cultured for a total of 9 days (split once) after which genomic DNA was isolated and barcodes were amplified. Genomic DNA extraction was performed with a liquid handler (Cybio) using the Genfind v2.0 kit (Agencourt). In brief, medium was removed and cells were washed twice with PBS. After lysis (1% SDS, 10 mM EDTA, 10 mM NaCl 10 mM Tris-HCl pH 8.0,), 100 μl raw lysate was transferred into 96-deepwell plates and 60 μl Agencourt binding buffer was added. Beads were washed six times with 70% ethanol and purified genomic DNA was eluted in dH2O. Barcodes were amplified in a 2-step protocol by PCR (Fwd 5′-CGATTAGTGAACGGATCTC, Rev 5′-GAAGGTGAGAACAGGAGC) and linear amplification was performed with a 5′ biotinylated primer (5′ Biotin-TGAGGATAGCAGAGAAGG). The single stranded product was hybridized to pre-coupled Luminex xMAP beads (as described 43) for 1.5 h at 40°C in 384 well plates and streptavidin coupled phycoerythrin (SAPE, Invitrogen) was added for 30 min. at 40°C. Finally, beads were washed once and samples were measured in a Flexmap 3D plate reader (Luminex) at 40°C.
Quantitative real-time PCR
RNA was isolated from sub-confluent cells using Trizol (Invitrogen). After purification and DNase treatment (Turbo-DNA free, Ambion) reverse transcription was performed using random hexamer primers and RevertAid reverse transcriptase (Fermentas). Quantitative real-time PCR was carried out using the iTaq SYBR Green Supermix (Bio-Rad) according to the manufacturer’s instructions. Measurements were performed in triplicate and related to GAPDH as a reference gene. All primer sequences are listed in Supplementary Table 6.
GFP competition assay
Cells were infected with vectors carrying the cDNAs for ICN1 and GFP (EF-hICN1-CMV-GFP) or an empty control vector. After infection, cells were pooled and distributed among multiple 6-well plates for BEZ-235 or DMSO treatment. GFP positive cells were measured by FACS or microscopy (Leica DMI6000B). For the microscopy analysis, 10 randomly chosen fields were imaged for each cell line-drug combination and cells were quantified using CellProfiler (The Broad Institute). Uninfected cells were used to determine background fluorescence levels.
Oncomine analysis
The Wooster cell-line dataset consists of over 300 cell lines (GSK300) that have been profiled for gene expression, copy number (CHG) and sensitivity to 19 compounds, including the PI3K/mTOR inhibitors BEZ-235, GSK1059615, Temsirolimus and the Aurora kinase inhibitor GSK1070916 (also see https://cabig.nci.nih.gov/caArray_GSKdata/). The analysis was done by grouping the drugs based on target pathway (i.e. PI3K/mTOR or Aurora kinase). A c-MYC copy number >4 was considered evidence for c-MYC gene amplification and the resistant/sensitive classification and median NUMB expression was used as defined by Oncomine.
Statistical analysis
Tests for statistical significance (as indicated in the text) and distribution of the data (Kolmogorov-Smirnov test) were calculated in GraphPad Prism 5.0 (GraphPad software). Experiments were performed in triplicate unless otherwise noted. P<0.05 (alpha=5%) was accepted as statistically significant. Further information on screen data analysis can be found in the Supplementary Methods.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the Tiina Berg, Ashot Harutyunyan, Uwe Rix and Guenther Hofbauer for technical assistance, Florian Ganglberger, Benjamin Eizinger and Patrick Markt for assisting in data analysis and Helen Pickersgill, Menno Creyghton and Giulio Superti-Furga for critical reading and suggestions. We are grateful to Todd Golub and Channing Yu for support and discussions in the early phases of the project, Alan D’andrea, Helmut Dolznig, Cliff Tabin, Thijn Brummelkamp, Rene Bernards, Pieter Eichhorn, Daniel Peeper, William Hahn, Ronald and Joan Conaway, Robert Weinberg, Isabella Screpanti, Jean Zhao, Richard Hynes, Yue Xiong, Ross Basch, Bert Vogelstein, Dmitry Bulavin, Paul Yaswen, Raphael Kopan, Malcolm Parker, James Bradner and David Root for providing reagents. We thank Luminex for providing reagents and support. This work was made possible by research grants from the Austrian Science Fund (FWF, P21768-B13) and the Vienna Science and Technology Fund (WWTF, LS09-009).
Footnotes
COMPETING FINANCIAL INTERESTS The authors declare no competing financial interests.
REFERENCES
- 1.Gottesman MM. Mechanisms of cancer drug resistance. Annu Rev Med. 2002;53:615–627. doi: 10.1146/annurev.med.53.082901.103929. [DOI] [PubMed] [Google Scholar]
- 2.Tredan O, Galmarini CM, Patel K, Tannock IF. Drug resistance and the solid tumor microenvironment. J Natl Cancer Inst. 2007;99:1441–1454. doi: 10.1093/jnci/djm135. [DOI] [PubMed] [Google Scholar]
- 3.Sharma SV, et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010;141:69–80. doi: 10.1016/j.cell.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McDermott U, Downing JR, Stratton MR. Genomics and the continuum of cancer care. N Engl J Med. 2011;364:340–350. doi: 10.1056/NEJMra0907178. [DOI] [PubMed] [Google Scholar]
- 5.Bernards R. It’s diagnostics, stupid. Cell. 2010;141:13–17. doi: 10.1016/j.cell.2010.03.018. [DOI] [PubMed] [Google Scholar]
- 6.Chin L, Gray JW. Translating insights from the cancer genome into clinical practice. Nature. 2008;452:553–563. doi: 10.1038/nature06914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lander ES. Initial impact of the sequencing of the human genome. Nature. 2011;470:187–197. doi: 10.1038/nature09792. [DOI] [PubMed] [Google Scholar]
- 8.Vogel CL, et al. Efficacy and safety of trastuzumab as a single agent in first-line treatment of HER2-overexpressing metastatic breast cancer. J Clin Oncol. 2002;20:719–726. doi: 10.1200/JCO.2002.20.3.719. [DOI] [PubMed] [Google Scholar]
- 9.Flaherty KT, et al. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med. 2010;363:809–819. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deming SL, Nass SJ, Dickson RB, Trock BJ. C-myc amplification in breast cancer: a meta-analysis of its occurrence and prognostic relevance. Br J Cancer. 2000;83:1688–1695. doi: 10.1054/bjoc.2000.1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Verdine GL, Walensky LD. The challenge of drugging undruggable targets in cancer: lessons learned from targeting BCL-2 family members. Clin Cancer Res. 2007;13:7264–7270. doi: 10.1158/1078-0432.CCR-07-2184. [DOI] [PubMed] [Google Scholar]
- 12.Luo J, Solimini NL, Elledge SJ. Principles of cancer therapy: oncogene and non-oncogene addiction. Cell. 2009;136:823–837. doi: 10.1016/j.cell.2009.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaelin WG., Jr. The concept of synthetic lethality in the context of anticancer therapy. Nat Rev Cancer. 2005;5:689–698. doi: 10.1038/nrc1691. [DOI] [PubMed] [Google Scholar]
- 14.Nijman SM. Synthetic lethality: general principles, utility and detection using genetic screens in human cells. FEBS letters. 2011;585:1–6. doi: 10.1016/j.febslet.2010.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fong PC, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med. 2009;361:123–134. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- 16.Fong PC, et al. Poly(ADP)-ribose polymerase inhibition: frequent durable responses in BRCA carrier ovarian cancer correlating with platinum-free interval. J Clin Oncol. 28:2512–2519. doi: 10.1200/JCO.2009.26.9589. [DOI] [PubMed] [Google Scholar]
- 17.Reinhardt HC, Jiang H, Hemann MT, Yaffe MB. Exploiting synthetic lethal interactions for targeted cancer therapy. Cell Cycle. 2009;8:3112–3119. doi: 10.4161/cc.8.19.9626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hillenmeyer ME, et al. The chemical genomic portrait of yeast: uncovering a phenotype for all genes. Science. 2008;320:362–365. doi: 10.1126/science.1150021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lum PY, et al. Discovering modes of action for therapeutic compounds using a genome-wide screen of yeast heterozygotes. Cell. 2004;116:121–137. doi: 10.1016/s0092-8674(03)01035-3. [DOI] [PubMed] [Google Scholar]
- 20.Workman P, Clarke PA, Raynaud FI, van Montfort RL. Drugging the PI3 kinome: from chemical tools to drugs in the clinic. Cancer Res. 2010;70:2146–2157. doi: 10.1158/0008-5472.CAN-09-4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu P, Cheng H, Roberts TM, Zhao JJ. Targeting the phosphoinositide 3-kinase pathway in cancer. Nat Rev Drug Discov. 2009;8:627–644. doi: 10.1038/nrd2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu J, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 23.Garcia-Higuera I, et al. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol Cell. 2001;7:249–262. doi: 10.1016/s1097-2765(01)00173-3. [DOI] [PubMed] [Google Scholar]
- 24.Neve RM, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iavarone A, Massague J. Repression of the CDK activator Cdc25A and cell-cycle arrest by cytokine TGF-beta in cells lacking the CDK inhibitor p15. Nature. 1997;387:417–422. doi: 10.1038/387417a0. [DOI] [PubMed] [Google Scholar]
- 26.Yang D, et al. Therapeutic potential of a synthetic lethal interaction between the MYC proto-oncogene and inhibition of aurora-B kinase. Proc Natl Acad Sci U S A. 107:13836–13841. doi: 10.1073/pnas.1008366107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maira SM, et al. Identification and characterization of NVP-BEZ235, a new orally available dual phosphatidylinositol 3-kinase/mammalian target of rapamycin inhibitor with potent in vivo antitumor activity. Mol Cancer Ther. 2008;7:1851–1863. doi: 10.1158/1535-7163.MCT-08-0017. [DOI] [PubMed] [Google Scholar]
- 28.Jarriault S, et al. Signalling downstream of activated mammalian Notch. Nature. 1995;377:355–358. doi: 10.1038/377355a0. [DOI] [PubMed] [Google Scholar]
- 29.Laplante M, Sabatini DM. mTOR signaling at a glance. J Cell Sci. 2009;122:3589–3594. doi: 10.1242/jcs.051011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feldman ME, et al. Active-site inhibitors of mTOR target rapamycin-resistant outputs of mTORC1 and mTORC2. PLoS Biol. 2009;7:e38. doi: 10.1371/journal.pbio.1000038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Serra V, et al. NVP-BEZ235, a dual PI3K/mTOR inhibitor, prevents PI3K signaling and inhibits the growth of cancer cells with activating PI3K mutations. Cancer Res. 2008;68:8022–8030. doi: 10.1158/0008-5472.CAN-08-1385. [DOI] [PubMed] [Google Scholar]
- 32.Pece S, et al. Loss of negative regulation by Numb over Notch is relevant to human breast carcinogenesis. J Cell Biol. 2004;167:215–221. doi: 10.1083/jcb.200406140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klinakis A, et al. Myc is a Notch1 transcriptional target and a requisite for Notch1-induced mammary tumorigenesis in mice. Proc Natl Acad of Sci USA. 2006;103:9262–9267. doi: 10.1073/pnas.0603371103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Palomero T, et al. NOTCH1 directly regulates c-MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth. Proc Natl Acad Sci USA. 2006;103:18261–18266. doi: 10.1073/pnas.0606108103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weng AP, et al. c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes Dev. 2006;20:2096–2109. doi: 10.1101/gad.1450406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reedijk M, et al. High-level coexpression of JAG1 and NOTCH1 is observed in human breast cancer and is associated with poor overall survival. Cancer Res. 2005;65:8530–8537. doi: 10.1158/0008-5472.CAN-05-1069. [DOI] [PubMed] [Google Scholar]
- 37.Lin CJ, Malina A, Pelletier J. c-Myc and eIF4F constitute a feedforward loop that regulates cell growth: implications for anticancer therapy. Cancer Res. 2009;69:7491–7494. doi: 10.1158/0008-5472.CAN-09-0813. [DOI] [PubMed] [Google Scholar]
- 38.Ruggero D, Pandolfi PP. Does the ribosome translate cancer? Nature Rev. Cancer. 2003;3:179–192. doi: 10.1038/nrc1015. [DOI] [PubMed] [Google Scholar]
- 39.Fojo T, Grady C. How much is life worth: cetuximab, non-small cell lung cancer, and the $440 billion question. J Natl Cancer Inst. 2009;101:1044–1048. doi: 10.1093/jnci/djp177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brachmann SM, et al. Specific apoptosis induction by the dual PI3K/mTor inhibitor NVP-BEZ235 in HER2 amplified and PIK3CA mutant breast cancer cells. Proc Natl Acad Sci USA. 2009;106:22299–22304. doi: 10.1073/pnas.0905152106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 42.Moffat J, et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124:1283–1298. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 43.Stegmaier K, et al. Signature-based small molecule screening identifies cytosine arabinoside as an EWS/FLI modulator in Ewing sarcoma. PLoS Med. 2007;4:e122. doi: 10.1371/journal.pmed.0040122. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.