Abstract
ATIC, SHMT2, and SLC46A1 have essential roles in one-carbon (1-C) transfer. The authors examined whether associations between ovarian carcinoma and 15 variants in these genes are modified by regular multivitamin use, a source of 1-C donors, among Caucasian participants from two US case–control studies. Using a phased study design, variant-by-multivitamin interactions were tested, and associations between variants and ovarian carcinoma were reported stratified by multivitamin supplement use. Per-allele risk associations were modified by multivitamin use at six variants among 655 cases and 920 controls (Phase 1). In a larger sample of 968 cases and 1,265 controls (Phases 1 and 2), interactions were significant (P ≤ 0.03) for two variants, particularly among regular multivitamin users: ATIC rs7586969 [odds ratio (OR) = 0.7, 95% confidence interval (CI) = 0.6–0.9] and ATIC rs16853834 (OR = 1.5, 95% CI = 1.1–2.0). The two ATIC single nucleotide polymorphisms (SNPs) did not share the same haplotype; however, the haplotypes they comprised mirrored their SNP risk associations among regular multivitamin supplement users. A multi-variant analysis was also performed by comparing the observed likelihood ratio test statistic from adjusted models with and without the two ATIC variant-by-multivitamin interaction terms with a null distribution of test statistics generated by permuting case status 10,000 times. The corresponding observed P value of 0.001 was more extreme than the permutation-derived P value of 0.009, suggesting rejection of the null hypothesis of no association. In summary, there is little statistical evidence that the 15 variants are independently associated with risk of ovarian carcinoma. However, the statistical interaction of ATIC variants with regular multivitamin intake, when evaluated at both the SNP and gene level, may support these findings as relevant to ovarian health and disease processes.
Keywords: autophagy, effect modification, haplotype, one-carbon transfer, permutation, purines
Introduction
The one-carbon (1-C) transfer pathway refers to biologic reactions that depend on 1-C transfers mediated by folate coenzymes: these transfers play essential roles in many major cellular processes including nucleic acid biosynthesis and methyl group generation (Ulrich et al., 2008). Consequently, perturbations in this pathway caused by folate or 1-C donor nutrient deficiency can compromise DNA synthesis and methylation and can lead to tumorigenesis (Choi and Mason, 2000). Polymorphisms in genes involved in the 1-C transfer pathway may mimic folate or 1-C donor deficiency and influence the risk of cancers. However, risk may be modified by dietary factors such as folate and other 1-C donor nutrients (Ma et al., 1997, 2009; Maruti et al., 2009; Levine et al., 2010).
We previously examined the association between single nucleotide polymorphisms (SNPs) in several genes in the 1-C transfer pathway and ovarian carcinoma risk, but found little evidence in secondary analyses for interactions with regular multivitamin supplement use as a source of folate and other B-vitamins (Kelemen et al., 2008). In that study, the smallest false positive report probabilities (FPRP) for interaction analyses were between 0.54 and 0.66 for associations between three SNPs in DNMT3A and risk of ovarian carcinoma among multivitamin supplement users (Kelemen et al., 2008).
In the present study, we elucidated the potential contribution of variants in three genes for which we hypothesized, a priori, would have interactive effects with multivitamin supplement use as a source of folate and other 1-C donor nutrients. Aminoimidazole carboxamide ribonucleotide transformylase/inosine monophosphate cyclohydrolase (ATIC) is a single bifunctional protein that catalyzes the penultimate and final steps of the de novo purine biosynthesis via a 1-C transfer from reduced folate (Wolan et al., 2004). The ATIC gene is highly conserved across several species from Escherichia coli to humans (Cheong et al., 2004). Serine hydroxymethyltransferase 2 (SHMT2) is the mitochondrial isoenzyme of cytoplasmic SHMT1. Both enzymes catalyze the reversible transfer of a single carbon from serine to tetrahydrofolate to form glycine and 5,10 methylene-tetrahydrofolate, which is required for de novo pyrimidine synthesis (Tibbetts and Appling, 2010). Polymorphisms in SHMT1 have been reported to alter the risk of ovarian carcinoma (Kelemen et al., 2008), squamous cell carcinoma of the head and neck (Zhang et al., 2005), and lung carcinoma (Wang et al., 2007). Solute carrier protein 46A1 (SLC46A1) plays a critical role in the absorption of folate from diet by functioning as a proton-coupled folate transporter and as a mediator of folate receptor endocytosis (Qiu et al., 2006; Zhao et al., 2009). Nucleotide substitutions in SLC46A1 that result in point mutations or a splice variant in the corresponding protein have recently been described in patients exhibiting hereditary folate malabsorption (Zhao et al., 2007).
We present here our findings for the associations between 15 common SNPs in ATIC, SHMT2, and SLC46A1 and ovarian carcinoma risk, stratified by regular multivitamin supplement use as an estimate of B-vitamin intake, using data from two case–control studies including 968 cases and 1,265 controls.
Materials and Methods
Study population
The study population consisted of a combination of two individual studies of epithelial ovarian cancer from Mayo Clinic and Duke University. Details of the study protocols have been published previously (Sellers et al., 2005). Briefly, participants included Caucasians and African-Americans enrolled from June 1999 onward. Mayo Clinic cases were 20 years of age or older and resident in a six-state region (Minnesota, Iowa, Wisconsin, Illinois, North Dakota, and South Dakota) that defined the primary catchment area. Controls were women seeking general medical care at Mayo Clinic. Duke University cases were between 20 and 74 years of age and identified from a 48-county region in North Carolina. Controls were identified from the same region using population-based list-assisted random digit dialing. In both studies, cases were newly diagnosed, histologically confirmed, of either borderline or invasive behavior, and enrolled within 1 year of diagnosis. Controls had at least one intact ovary, no history of ovarian cancer and were frequency-matched to cases on age (5-year age categories), race, and state of residence. The response among cases and controls, respectively, was high at Mayo Clinic (83 and 74%) and Duke University (75 and 64%). All participants provided written informed consent and procedures followed were in accordance with the Institutional Review Board of Mayo Clinic and the Institutional Review Board of Duke University Medical Center.
Assessment of regular multivitamin supplement use and other risk factors
Multivitamin supplement use and information on established risk factors such as reproductive history, family history of cancer, medical history, and lifestyle habits were obtained from all subjects through in-person interviews using study-specific questionnaires. Variables were harmonized across studies for this analysis. The Mayo Clinic study began collecting information on multivitamin intake 3 years after the start of the study, while the Duke University study elicited this information from the start of the study. At Duke University, participants indicated their regularity of multivitamin use (none, <1 pill per week, 1–2 per week, 3–4 per week, 5–6 per week, or daily) during the previous 5 years. We defined users as those who consumed at least three pills per week for at least 48 months during the past 5 years (e.g., allowing 80% adherence). At Mayo Clinic, participants responded yes or no to whether they consumed multivitamins at least four times per week during the previous year for controls and 1 year before diagnosis for cases; they also estimated their total number of years’ duration of use. To create a “multivitamin user” definition that compared to the Duke University subjects, we defined Mayo Clinic users as women consuming at least four pills per week for at least 48 months’ duration of lifetime use.
Genetic analysis
DNA was obtained from peripheral blood samples using salt extraction protocols (Sellers et al., 2005; Cunningham et al., 2008). In addition, whole genome amplification (WGA) using the REPLI-G WGA protocol (Qiagen Inc., Valencia, CA, USA) was used to enrich DNA samples from Duke University due to limited quantities. Excellent (>99%) concordance between genomic and WGA DNA genotype calls for these samples has been reported previously (Cunningham et al., 2008).
TagSNPs were chosen using SNP information from unrelated Caucasian samples within HapMap Consortium’s release 22 (The International HapMap Consortium, 2003) as previously described (Kelemen et al., 2008), and also for their predicted likelihood of successful genotyping using the Illumina Golden Gate Assay™. We selected nine tagSNPs in ATIC (rs3772078, rs2372536, rs1880586, rs1983462, rs16853826, rs7586969, rs16853834, rs1404772, and rs7604984) from among 45 individual SNPs, two tagSNPs in SHMT2 (rs7301155 and rs2229716) from among two individual SNPs and four tagSNPs in SLC46A1 (rs9894260, rs739439, rs2239908, and rs17719944) from among 10 individual SNPs using criteria of minor allele frequency (MAF) ≥ 0.05 and pairwise linkage disequilibrium (LD) of r2 < 0.8. The SNPs were located within, and 5 kb upstream and downstream of, each gene region.
The 15 tagSNPs were genotyped using a phased study design. In the first phase (Phase 1), we genotyped the SNPs as part of a larger investigation of 1,152 SNPs in a variety of pathways using the Illumina GoldenGate™ assay and Illumina BeadStudio software (Oliphant et al., 2002; Fan et al., 2005; Steemers and Gunderson, 2005). Genotyping was attempted on 897 genomic DNA samples from Mayo Clinic and 1,279 WGA samples from Duke University (total = 2,176 including 129 duplicate samples) for subjects recruited through March 2006 as well as 65 laboratory controls. Of these samples, we excluded 44 (one Mayo Clinic sample and 43 Duke University samples) with call rates < 90% and Illumina quality control (GenCall) scores < 0.25, and 22 samples where the cancer was not of epithelial origin or the samples were mislabeled, resulting in 1,981 unique samples that were successfully genotyped. The sample call rate was 99.74% and the concordance for duplicate samples was 99.99%. All 15 tagSNPs in ATIC, SHMT2, and SLC46A1 were genotyped successfully, except SHMT2 rs7301155 that failed in Duke University samples. In Phase 2, we genotyped or imputed the 15 tagSNPs as part of a genome-wide association study (GWAS) of ovarian carcinoma using the Illumina 610 K chip on 186 genomic DNA samples from Mayo Clinic and 477 WGA samples from Duke University for subjects recruited after March 2006. Genotyping call rates for these SNPs were ≥ 99%. Imputation was performed using the MACH software (Li et al., 2009) and the phase II HapMap (release 22) referent genotypes for Caucasian samples for eight SNPs [ATIC (rs2372536, rs1880586, rs1983462, rs16853826, and rs7586969), SHMT2 (rs7301155 and rs2229716), and SLC46A1 (rs9894260)]. Because SHMT2 rs7301155 failed in Duke University samples from Phase 1, we included the imputed genotypes for this SNP from both Phase 1 and Phase 2 Duke University samples represented on the GWAS. Squared correlations between imputed and true genotypes were between 0.93 and 0.99, except r2 = 0.43 for SHMT2 rs2229716, which was likely due to its rare frequency (Table A1 in Appendix).
Statistical analysis
Analyses were restricted to subjects who were self-reported Caucasian controls or cases with invasive epithelial ovarian carcinoma, resulting in a final sample size of 1,575 participants in Phase 1 from Mayo Clinic (312 cases and 439 controls) and Duke University (343 cases and 481 controls) and 658 participants in Phase 2 from Mayo Clinic (89 cases and 96 controls) and Duke University (224 cases and 249 controls). Genotypes of participants were used to estimate allele frequencies and chi squared tests were used to examine deviations of genotype frequencies from those expected under Hardy–Weinberg equilibrium (HWE) in control subjects. Pairwise LD was examined across all SNPs with the r2 statistic using Haploview (Barrett et al., 2005). Data from Mayo Clinic and Duke University were combined following confirmation of no statistical heterogeneity in SNP associations between study sites.
Our primary hypothesis was that SNP associations would be modified by regular multivitamin supplement use as a source of folate or other nutrients involved 1-C transfer. We evaluated this first in Phase 1 samples. Interactions were evaluated with the Wald test in models that included a one degree-of-freedom product term for regular multivitamin supplement use (yes/no) and the ordinal coding for genotype, in addition to the covariates age (<40, 40–49, 50–59, 60–69, and 70+ years), region of residence (Minnesota, Iowa, Wisconsin, Illinois, North Dakota, South Dakota, and North Carolina), SNP, regular multivitamin supplement use, body mass index, age at menarche, oral contraceptive use, postmenopausal status, postmenopausal hormone use, parity/age at first birth, smoking, and education (see Table 2 for categories). To determine the degree of falsely positive findings due to the number of interaction tests that were performed, we calculated q-values to estimate false discovery rates (FDR; Hochberg and Benjamini, 1990). Associations between SNPs and ovarian carcinoma were stratified by regular multivitamin supplement use and estimated as odds ratios (OR) with 95% confidence intervals (CI) using multivariable-adjusted unconditional logistic regression under ordinal genetic models. For SNPs where the tests for interaction had higher FDR q-values (e.g., >0.23 in our data), suggesting a higher likelihood of falsely positive tests, we estimated associations between the genetic main effects and ovarian carcinoma under both co-dominant and ordinal genetic models. Models evaluating the main effects of SNPs were adjusted for age and region of residence. Adjustment for additional covariates did not change the ORs (data not shown).
Table 2.
Odds ratios (OR) and 95% CIs for the association between variants in ATIC, SHMT2, and SLC46A1and ovarian carcinoma stratified by multivitamin supplement use among 1,575 Caucasian subjects (Phase 1), Mayo Clinic, and Duke University.
| Gene/SNP rsID | Multivitamin non-users |
Multivitamin usersa |
Pintc | FDRd | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Homozygous major allele (referent) | Heterozygous | Homozygous minor allele | Ordinal (per minor allele) | Homozygous major allele (referent) | Heterozygous | Homozygous minor allele | Ordinal (per minor allele) | |||
| Cases/controls | Cases/controls | Cases/controls | OR (95%CI)b | Cases/controls | Cases/controls | Cases/controls | OR (95%CI)b | |||
| ATIC | ||||||||||
| rs3772078 | 212/319 | 95/160 | 6/14 | 0.9 (0.7–1.2) | 122/241 | 58/110 | 7/16 | 1.0 (0.7–1.4) | 0.54 | 0.63 |
| rs2372536 | 137/225 | 156/226 | 28/55 | 1.0 (0.8–1.2) | 98/167 | 64/162 | 25/44 | 0.9 (0.7–1.2) | 0.52 | 0.63 |
| rs1880586 | 76/131 | 176/268 | 71/108 | 1.1 (0.9–1.3) | 52/107 | 84/188 | 51/83 | 1.1 (0.9–1.4) | 0.79 | 0.79 |
| rs1983462 | 147/235 | 146/221 | 31/51 | 1.0 (0.8–1.3) | 97/166 | 73/180 | 18/32 | 0.8 (0.6–1.1) | 0.27 | 0.45 |
| rs16853826 | 258/378 | 65/122 | 1/7 | 0.7 (0.5–1.0) | 139/291 | 46/80 | 3/7 | 1.2 (0.8–1.7) | 0.09 | 0.23 |
| rs7586969 | 115/199 | 161/235 | 47/72 | 1.1 (0.9–1.4) | 81/135 | 83/185 | 24/58 | 0.8 (0.6–1.0) | 0.07 | 0.23 |
| rs16853834 | 227/356 | 85/140 | 12/11 | 1.0 (0.8–1.4) | 119/281 | 60/87 | 9/10 | 1.5 (1.1–2.1) | 0.07 | 0.23 |
| rs1404772 | 279/450 | 43/55 | 2/2 | 1.3 (0.9–1.9) | 167/325 | 20/51 | 1/2 | 0.8 (0.5–1.3) | 0.17 | 0.31 |
| rs7604984 | 106/189 | 172/241 | 46/77 | 1.1 (0.9–1.3) | 81/128 | 77/187 | 30/62 | 0.8 (0.6–1.1) | 0.12 | 0.26 |
| SHMT2 | ||||||||||
| rs7301155e | 46/76 | 48/101 | 16/25 | 1.0 (0.7–1.4) | 23/104 | 42/93 | 8/20 | 1.7 (1.1–2.6) | 0.08 | 0.23 |
| rs2229716 | 300/462 | 23/44 | 1/1 | 0.8 (0.5–1.4) | 174/348 | 14/29 | 0/0 | 0.9 (0.5–1.9) | 0.70 | 0.75 |
| SLC46A1 | ||||||||||
| rs9894260 | 265/419 | 57/86 | 1/2 | 1.0 (0.7–1.4) | 151/319 | 32/58 | 4/1 | 1.5 (1.0–2.3) | 0.08 | 0.23 |
| rs739439 | 216/350 | 93/148 | 14/9 | 1.2 (0.9–1.5) | 126/237 | 56/124 | 5/17 | 0.8 (0.6–1.1) | 0.09 | 0.23 |
| rs2239908 | 98/159 | 163/253 | 62/94 | 1.0 (0.8–1.3) | 57/102 | 96/203 | 34/72 | 0.9 (0.7–1.2) | 0.54 | 0.63 |
| rs17719944 | 270/426 | 50/75 | 4/6 | 1.0 (0.7–1.5) | 152/298 | 34/77 | 1/3 | 0.9 (0.6–1.3) | 0.46 | 0.63 |
aAt least 4 pills per week during the previous year for at least 48 months’ duration of lifetime use (Mayo Clinic subjects) or at least three pills per week for at least 48 months during the past 60 months (Duke University subjects).
bAdjusted for age (<40, 40–49, 50–59, 60–69, and 70+ years), region of residence (Minnesota, Iowa, Wisconsin, Illinois, North Dakota, South Dakota, and North Carolina), body mass index, age at menarche, oral contraceptive use, postmenopausal status, postmenopausal hormone use, parity/age at first birth, smoking, and education (see Table 1 for categories). SNP effect was determined using a log-additive logistic regression model.
cP for interaction.
dFalse discovery rate q-value.
eMayo Clinic subjects only.
We performed the same analyses using Phase 2 samples as an independent replication, and then combined the Phase 1 and Phase 2 samples to increase statistical power. To account for the potential correlations among SNPs within genes, we performed both a multi-variant analysis and haplotype analysis. The multi-variant analysis was performed by calculating a likelihood ratio test (LRT) statistic comparing a regression model consisting of the main effects of those SNPs exhibiting significant interactions with multivitamin supplement use, their corresponding interaction terms, and potential confounding variables to a model without the SNP-by-multivitamin interaction terms. SNP main effects and their interaction terms were regressed under the ordinal (log-additive) genetic model. Permutation-based tests were then used to compute P values from a null distribution of the LRT statistic generated by permuting case status 10,000 times. The corresponding P value threshold was calculated as the probability of observing a permuted LRT statistic as extreme or more extreme than the observed LRT statistic. We also estimated haplotype frequencies for each gene, stratified and unstratified by regular multivitamin supplement use, by computing the posterior probabilities of all possible haplotypes for an individual, conditioned on the observed genotypes, using an expectation–maximization algorithm (Schaid et al., 2002). We used these probabilities to define haplotype design variables that estimated the number of each of the haplotypes carried by an individual, and retained only those with estimated frequencies of >1%. We first tested whether differences among the haplotypes within each gene were associated statistically with ovarian carcinoma overall (P < 0.016, Bonferroni-corrected for three genes). Next, associations between individual haplotypes and risk were estimated under ordinal genetic models (e.g., carriage of 0, 1, or 2 copies of a particular haplotype; Schaid, 2011). Individual associations were not considered statistically significant in the absence of global significance.
Statistical tests were two-sided, and were implemented with SAS (SAS Institute, NC), R (R Development Core Team, 2011), and Haplo.stats (Schaid, 2011) software.
Results
There was no evidence of deviation from HWE in controls from both studies for any of the SNPs. The MAFs among controls ranged from 0.04 to 0.48 and were similar across study sites in Phase 1 (Table A1 in Appendix), Phase 2 (data not shown), and Phases 1 and 2 combined, with imputed SNPs in Phases 1 and 2 combined showing similar MAFs as typed SNPs in Phase 1 (Table A1 in Appendix). The distribution of some covariates varied among cases and controls at both Mayo Clinic and Duke University in Phase 1 (Table A2 in Appendix), Phase 2 (data not shown), and Phases 1 and 2 combined (Table 1); however, histological subtypes of tumors were distributed comparably for both studies.
Table 1.
Characteristics of 2,233 Caucasian subjects (Phases 1 and 2), Mayo Clinic, and Duke University.
| Mayo Clinic |
Duke University |
|||
|---|---|---|---|---|
| Cases (n = 401) | Controls (n = 535) | Cases (n = 567) | Controls (n = 730) | |
| Age, year [mean (SD)] | 61.0 (12.5) | 60.7 (13.2) | 57.3 (10.3) | 55.3 (12.0) |
| BODY MASS INDEX, kg/m2 | ||||
| <23 [n (%)] | 80 (20.5) | 127 (24.8) | 152 (27.6) | 203 (28.4) |
| 23–26 [n (%)] | 100 (25.6) | 150 (29.3) | 131 (23.8) | 173 (24.2) |
| 26–29 [n (%)] | 75 (19.2) | 99 (19.3) | 113 (20.5) | 141 (19.7) |
| ≥29 [n (%)] | 136 (34.8) | 136 (26.6) | 155 (28.1) | 199 (27.8) |
| AGE AT MENARCHE, year | ||||
| <12 [n (%)] | 63 (19.0) | 77 (15.3) | 139 (24.7) | 132 (18.1) |
| 12 [n (%)] | 84 (25.3) | 111 (22.0) | 160 (28.4) | 210 (28.8) |
| 13 [n (%)] | 93 (28.0) | 149 (29.5) | 139 (24.7) | 206 (28.3) |
| ≥14 [n (%)] | 92 (27.7) | 168 (33.3) | 126 (22.3) | 181 (24.8) |
| ORAL CONTRACEPTIVE USE, months | ||||
| Never [n (%)] | 182 (47.5) | 200 (39.8) | 194 (40.3) | 207 (31.1) |
| 1–48 [n (%)] | 101 (26.4) | 107 (21.3) | 142 (29.5) | 181 (27.2) |
| ≥48 [n (%)] | 100 (26.1) | 196 (39.0) | 145 (30.2) | 278 (41.7) |
| POSTMENOPAUSAL STATUS | ||||
| Yes [n (%)] | 290 (74.7) | 393 (76.0) | 251 (46.9) | 502 (69.8) |
| POSTMENOPAUSAL HORMONE USE, months | ||||
| Never [n (%)] | 241 (61.6) | 287 (58.5) | 211 (39.2) | 407 (58.4) |
| 1–60 [n (%)] | 71 (18.2) | 91 (18.5) | 166 (30.9) | 147 (21.1) |
| ≥60 [n (%)] | 79 (20.2) | 113 (23.0) | 161 (29.9) | 143 (20.5) |
| PARITY, n/age at first birth, year | ||||
| Nulliparous [n (%)] | 69 (17.4) | 77 (15.0) | 113 (19.9) | 94 (12.9) |
| 1–2/≤20 [n (%)] | 29 (7.3) | 26 (5.1) | 80 (14.1) | 92 (12.6) |
| 1–2/≥20 [n (%)] | 115 (29.0) | 155 (30.1) | 208 (36.7) | 314 (43.0) |
| ≥3/≤20 [n (%)] | 77 (19.4) | 74 (14.4) | 80 (14.1) | 92 (12.6) |
| ≥3/≥20 [n (%)] | 106 (26.8) | 183 (35.5) | 86 (15.2) | 138 (18.9) |
| FAMILY HISTORY OF OVARIAN CANCERa | ||||
| Yes [n (%)] | 50 (12.6) | 42 (8.1) | 56 (9.9) | 46 (6.3) |
| SMOKING, pack-years | ||||
| None [n (%)] | 242 (63.7) | 333 (68.5) | 303 (55.7) | 376 (53.5) |
| ≤20 [n (%)] | 75 (19.7) | 100 (20.6) | 137 (25.2) | 187 (26.6) |
| >20 [n (%)] | 63 (16.6) | 53 (10.9) | 104 (19.1) | 140 (19.9) |
| EDUCATION | ||||
| No diploma [n (%)] | 22 (5.8) | 21 (4.1) | 49 (8.6) | 66 (9.0) |
| High School diploma [n (%)] | 136 (35.9) | 133 (25.7) | 179 (31.6) | 192 (26.3) |
| Post-high school education [n (%)] | 221 (58.3) | 364 (70.3) | 339 (59.8) | 472 (64.7) |
| MULTIVITAMIN USEb | ||||
| Yes [n (%)] | 119 (44.9) | 259 (52.4) | 210 (38.1) | 260 (36.4) |
| TUMOR HISTOLOGY, CASES | ||||
| Serous [n (%)] | 255 (63.9) | 329 (58.1) | ||
| Mucinous [n (%)] | 11 (2.8) | 26 (4.6) | ||
| Endometrioid [n (%)] | 77 (19.3) | 85 (15.0) | ||
| Clear cell [n (%)] | 25 (6.3) | 60 (10.6) | ||
| Other [n (%)] | 31 (7.8) | 66 (11.7) | ||
Counts do not total to 2,233 subjects due to missing data for some variables.
aIn first- or second-degree relative.
bAt least four pills per week during the previous year for at least 48 months’ duration of lifetime use (Mayo Clinic subjects) or at least three pills per week for at least 48 months during the past 60 months (Duke University subjects).
In Phase 1, the associations between six SNPs and ovarian carcinoma risk were modified by regular multivitamin supplement use at the smallest FDR q-value of 0.23: ATIC rs16853826, rs7586969 and rs16853834, SHMT2 rs7301155, and SLC46A1 rs9894260 and rs739439 (Table 2). In stratified analyses, among women who took multivitamins regularly, the strongest associations were observed between ovarian carcinoma and ATIC rs16853834 (OR = 1.5, 95% CI = 1.1–2.1, P = 0.01) and SHMT2 rs7301155 (OR = 1.7, 95% CI = 1.1–2.6, P = 0.01). Among non-regular supplement users, a trend was observed between ovarian carcinoma and ATIC rs16853826 (OR = 0.7, 95% CI = 0.5–1.0, P = 0.06). Comparable stratified analyses are shown in Table A3 in Appendix for Phase 2 samples, with none of the aforementioned SNPs showing statistically significant interactions in this smaller sample, although the direction of association among multivitamin supplement users was consistent for ATIC rs16853834 (OR = 1.6, 95% CI = 0.7–3.7). Combining Phase 1 and Phase 2 samples (968 cases and 1,265 controls; Table 3), statistically significant interactions were observed between regular multivitamin supplement use and ATIC rs7586969 (OR = 0.7, 95% CI = 0.6–0.9) and ATIC rs16853834 (OR = 1.5, 95% CI = 1.1–2.0).
Table 3.
Odds ratios (OR) and 95% CIs for the association between variants in ATIC, SHMT2, and SLC46A1and ovarian carcinoma stratified by multivitamin supplement use among 2,233 subjects (Phases 1 and 2), Mayo Clinic and Duke University.
| Gene/SNP rsID | Multivitamin non-users |
Multivitamin usersa |
Pintc | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Homozygous major allele (referent) | Heterozygous | Homozygous minor allele | Ordinal (per minor allele) | Homozygous major allele (referent) | Heterozygous | Homozygous minor allele | Ordinal (per minor allele) | ||
| Cases/controls | Cases/controls | Cases/controls | OR (95%CI)b | Cases/controls | Cases/controls | Cases/controls | OR (95%CI)b | ||
| ATIC | |||||||||
| rs3772078 | 323/438 | 141/217 | 12/20 | 0.9 (0.7–1.1) | 214/337 | 102/149 | 12/22 | 1.0 (0.7–1.3) | 0.51 |
| rs2372536d | 224/323 | 213/289 | 47/76 | 1.0 (0.8–1.2) | 159/232 | 122/222 | 47/60 | 1.0 (0.8–1.3) | 0.87 |
| rs1880586d | 125/191 | 253/342 | 108/156 | 1.0 (0.9–1.2) | 83/146 | 149/249 | 96/124 | 1.2 (0.9–1.4) | 0.47 |
| rs1983462d | 208/315 | 222/298 | 57/76 | 1.1 (0.9–1.3) | 174/230 | 125/237 | 30/52 | 0.8 (0.6–1.0) | 0.05 |
| rs16853826d | 384/516 | 98/164 | 5/9 | 0.8 (0.6–1.0) | 245/400 | 80/111 | 4/8 | 1.1 (0.8–1.6) | 0.11 |
| rs7586969d | 166/261 | 239/323 | 81/104 | 1.1 (0.9–1.3) | 142/185 | 149/252 | 38/82 | 0.7 (0.6–0.9) | 0.01 |
| rs16853834 | 349/483 | 121/186 | 16/18 | 1.0 (0.8–1.3) | 214/384 | 102/121 | 13/14 | 1.5 (1.1–2.0) | 0.03 |
| rs1404772 | 425/609 | 59/77 | 3/3 | 1.2 (0.8–1.7) | 290/447 | 38/70 | 1/2 | 0.7 (0.5–1.2) | 0.20 |
| rs7604984 | 179/275 | 239/306 | 69/108 | 1.0 (0.9–1.3) | 130/184 | 142/248 | 57/85 | 0.9 (0.7–1.2) | 0.35 |
| SHMT2 | |||||||||
| rs7301155d | 204/283 | 187/296 | 42/66 | 0.9 (0.7–1.1) | 133/239 | 147/211 | 28/42 | 1.3 (1.0–1.6) | 0.08 |
| rs2229716d | 458/638 | 28/49 | 1/2 | 0.8 (0.5–1.3) | 309/488 | 20/30 | 0/0 | 1.0 (0.5–2.0) | 0.51 |
| SLC46A1 | |||||||||
| rs9894260d | 405/575 | 78/111 | 3/3 | 1.0 (0.7–1.3) | 272/433 | 50/83 | 6/3 | 1.1 (0.7–1.6) | 0.62 |
| rs739439 | 323/475 | 143/201 | 20/13 | 1.1 (0.9–1.4) | 206/336 | 109/160 | 13/23 | 1.0 (0.7–1.3) | 0.46 |
| rs2239908 | 139/213 | 249/348 | 98/127 | 1.1 (0.9–1.3) | 82/144 | 178/268 | 68/106 | 1.0 (0.8–1.2) | 0.64 |
| rs17719944 | 404/576 | 78/104 | 5/9 | 1.0 (0.7–1.3) | 261/421 | 65/94 | 1/4 | 1.1 (0.7–1.6) | 0.66 |
aAt least four pills per week during the previous year for at least 48 months’ duration of lifetime use (Mayo Clinic subjects) or at least three pills per week for at least 48 months during the past 60 months (Duke University subjects).
bAdjusted for age (<40, 40–49, 50–59, 60–69, and 70+ years), region of residence (Minnesota, Iowa, Wisconsin, Illinois, North Dakota, South Dakota, and North Carolina), body mass index, age at menarche, oral contraceptive use, postmenopausal status, postmenopausal hormone use, parity/age at first birth, smoking, and education (see Table 1 for categories). SNP effect was determined using a log-additive logistic regression model.
cP for interaction.
dSNP imputed from GWAS for all Duke University subjects (Phase 1 and Phase 2).
A multi-variant analysis was performed by calculating a LRT statistic comparing adjusted models that included ATIC rs7586969 and ATIC rs16853834, multivitamin use and the SNP-by-multivitamin interaction terms to a model without ATIC SNP-by-multivitamin interaction terms, and comparing this to a null distribution of the LRT statistic generated by permuting case status 10,000 times. The observed P value for ATIC (χ2 = 12.98, 2 df, P = 0.001) was more extreme than the P value derived from permutation testing (P = 0.009), suggesting the evidence is inconsistent with the null hypothesis of no association between ATIC SNP-by-multivitamin interactions and risk of ovarian carcinoma.
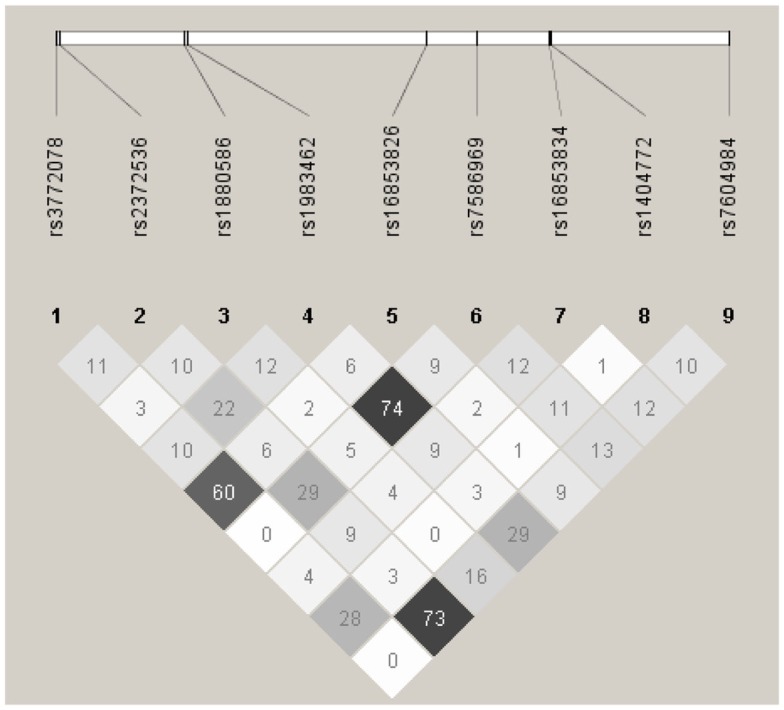
In haplotype analyses, only the ATIC gene was statistically significantly associated with ovarian carcinoma among regular multivitamin supplement users (Table 4: global haplotype test P = 0.01; data not shown for SHMT2 and SLC46A1). The two ATIC SNPs, rs7586969, and rs16853834, whose minor alleles showed opposing risks, did not share the same haplotype; however, the haplotypes they comprised mirrored their SNP risk associations among regular multivitamin supplement users. Haplotype #1 accounted for 24% of all estimated haplotypes, comprised the minor allele of ATIC rs7586969 and was associated with a 24% decreased risk. Haplotype #9 accounted for 13% of all estimated haplotypes, comprised the minor allele of ATIC rs16853834 and was associated with a 39% increased risk. A third haplotype, haplotype #10, with 8% frequency, comprised common alleles for ATIC rs7586969 and rs16853834 and was associated with a 42% increased risk. Among non-users, haplotype #5 was associated with a 27% decreased risk, but the global haplotype test was not statistically significant. None of the associations were statistically significant in the unstratified data. Squared correlations among the SNPs are shown in Figure A1 in Appendix.
Table 4.
Associationa between haplotypes derived from ATIC variants and ovarian carcinoma with and without stratification by multivitamin supplement use among 2,233 subjects (Phases 1 and 2), Mayo Clinic, and Duke University.
| Haplotypeb | Multivitamin non-users |
Multivitamin users |
All subjects |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Global haplotype test P value | Estimated haplotype frequency | OR (95% CI) | Global haplotype test P value | Estimated haplotype frequency | OR (95% CI) | Global haplotype test P value | Estimated haplotype frequency | OR (95% CI) | |
| 0.56 | 0.01 | 0.70 | |||||||
| 001101000 | 0.26 | 1.04 (0.86–1.26) | 0.24 | 0.76 (0.59–0.97) | 0.25 | 0.93 (0.81–1.07) | |||
| 010000001 | 0.26 | 0.98 (0.81–1.19) | 0.25 | 0.82 (0.65–1.04) | 0.26 | 0.94 (0.82–1.08) | |||
| 100001011 | 0.05 | 1.23 (0.84–1.79) | 0.05 | 0.74 (0.46–1.19) | 0.05 | 1.05 (0.80–1.38) | |||
| 000101000 | 0.07 | 1.16 (0.89–1.51) | 0.07 | 0.96 (0.68–1.35) | 0.06 | 1.01 (0.82–1.23) | |||
| 100010000 | 0.10 | 0.73 (0.55–0.98) | 0.10 | 1.10 (0.78–1.55) | 0.10 | 0.95 (0.78–1.16) | |||
| 101001011 | 0.01 | 0.69 (0.29–1.66) | 0.02 | 1.28 (0.53–3.05) | 0.01 | 0.77 (0.42–1.40) | |||
| 101010000 | 0.02 | 1.13 (0.63–2.03) | 0.03 | 1.50 (0.79–2.87) | 0.02 | 1.17 (0.78–1.75) | |||
| 000000100 | 0.03 | 0.87 (0.56–1.38) | 0.03 | 1.52 (0.90–2.56) | 0.03 | 1.02 (0.73–1.43) | |||
| 001000100 | 0.13 | 1.00 (0.78–1.29) | 0.13 | 1.39 (1.04–1.87) | 0.13 | 1.16 (0.97–1.38) | |||
| 011000001 | 0.06 | 1.07 (0.79–1.45) | 0.08 | 1.42 (1.04–1.95) | 0.06 | 1.12 (0.91–1.39) | |||
aAdjusted for age (<40, 40–49, 50–59, 60–69, and 70+ yrs), residence (Minnesota, Iowa, Wisconsin, Illinois, North Dakota, South Dakota, and North Carolina), body mass index, age at menarche, oral contraceptive use, postmenopausal status, postmenopausal hormone use, parity/age at first birth, smoking, and education (see Table 1 for categories). Haplotype associations were estimated using log-additive logistic regression models.
bEstimated using both cases and controls; SNPs that formed ATIC haplotypes were, in positional order, rs3772078, rs2372536, rs1880586, rs1983462, rs16853826, rs7586969, rs16853834, rs1404772, rs7604984, where 0 = common allele and 1 = rare allele. Bold indicates the two significant SNPs with opposing effects from Table 3.
For remaining SNPs with FDR q-values > 0.23 in Phase 1, we estimated main effect associations with ovarian carcinoma (Table A4 in Appendix). An increased risk was observed among 12 carriers homozygous for the rare variant SLC46A1 rs9894260 compared to carriers homozygous for the wildtype allele (OR = 5.4, 95% CI = 1.4–20.5). Similar results were seen using Phase 2 samples (data not shown), as well as Phase 1 and Phase 2 samples combined (Table A5 in Appendix). The main effect association between regular multivitamin supplement use and ovarian carcinoma in Phase 1 and Phase 2 samples combined was not statistically significant at P < 0.05 (multivariable OR = 0.86, 95% CI = 0.70–1.06, P = 0.15; data not shown).
Discussion
We evaluated the association between 15 SNPs in ATIC, SHMT2, and SLC46A1 in the 1-C transfer pathway and ovarian carcinoma risk, and the modification of risk by regular multivitamin supplement use among 968 cases and 1,265 controls. There was little statistical evidence that SNPs or haplotypes in these genes were independently associated with ovarian carcinoma risk. However, the associations of ATIC rs7586969 and ATIC rs16853834 with ovarian carcinoma, when stratified by regular multivitamin supplement use and evaluated further in multi-variant and haplotype analyses, suggest the potentially important role that lifestyle factors may have in modifying genetic susceptibility.
We calculated that six interactions between SNPs and regular multivitamin use had an FDR q-value of 0.23 indicating that, out of 100 SNPs with this FDR q-value, 23 will be falsely positive, or approximately one in four. Further analyses in a larger sample showed consistent associations for two of these SNPs at P < 0.05. ATIC rs7586969 and ATIC rs16853834 have MAF in our data of 0.39 and 0.16, respectively, but are weakly correlated with each other (pairwise LD r2 = 0.12). The power to detect these associations was relatively high: with an average exposure in our control group of 43% for regular multivitamin supplement use, we achieved close to 80% power needed with 1,054 cases using a ratio of 1:1.3 case–control pairs (actual: 968 cases and 1,265 controls) to detect an OR of 0.7 (ATIC rs7586969) at alpha = 0.05, and we would require 1,161 case–control pairs to detect an OR of 1.5 (ATIC rs16853834) under the log-additive genetic model (Quanto version 1.2, CA, USA, http://hydra.usc.edu/gxe). The power to detect the haplotype associations, where the greatest frequency was 24% (haplotype #1), was lower. The presence of two SNPs in the same gene with relatively independent (and opposing) effects was resolved with haplotype analysis: the two SNPs do not share the same haplotype. Both the haplotype analysis and the multi-variant, permutation-based test suggest the data are inconsistent with the null hypothesis of no association between ATIC SNP-by-multivitamin interactions and risk of ovarian carcinoma. The aforementioned ATIC SNPs are intronic and, unlike exons, little is known of their role in the regulation of the final functional protein. However, because we genotyped tagSNPs, which merely identify a region in the gene at which an association is found, additional genotyping of the region (finemapping) and association testing is required to determine the precise causal variant(s). In vitro cell culture studies, where the nutrient medium can be manipulated, could then begin to unravel the functional impact of SNP-by-nutrient interactions.
ATIC catalyzes the final two steps in de novo purine biosynthesis, which is the main source of purine nucleotides (Kondo et al., 2000; Wolan et al., 2004). The final atom of the purine ring is supplied by folate, producing 5-aminoimidazole-4-carboxamide ribonucleotide (AICAR). Not only can AICAR induce apoptosis under certain conditions (Hallstrom et al., 2008), but as the substrate for ATIC, it can be catabolized to the purine “parent” nucleotide, inosine monophosphate (IMP), that ultimately generates two of the four bases, adenine and guanine. The bases are incorporated into DNA and RNA and also function as important sources of energy that drive most biologic reactions (e.g., ATP, GTP, UTP, CTP; Rodwell, 2000). Conceivably, ATIC may link nutrient metabolism vis-à-vis the 1-C transfer pathway to enable autophagy, which is the cellular catabolic degradation response to starvation or stress whereby cellular proteins, organelles, and cytoplasm are engulfed, digested, and recycled to sustain cellular metabolism (Mathew et al., 2007). For example, amino acid starvation is a potent inducer of autophagy (Mizushima, 2007), and defects in autophagy have been linked to tumorigenesis (Mathew et al., 2007).
Most individuals do not meet their Dietary Reference Intakes (DRI) for certain nutrients from foods alone. For example, among non-supplement users, folate intake (on average, <300 μg/day; Zhang et al., 1999; Feigelson et al., 2003; Kelemen et al., 2004; Ericson et al., 2007) is less than the DRI (400 μg/day). A typical multivitamin contains a minimum of 400 μg of folic acid, thereby increasing some individuals’ intakes to >600 μg/day (Zhang et al., 1999; Feigelson et al., 2003). Pharmacologic doses of vitamin intake are hypothesized to be both chemo-preventive and chemo-progressive, and the mechanism(s) for these alterations may be related to the presence of existing pre-cancerous lesions, where vitamins like folate are required for growth by proliferating cells including tumors (Kim, 2003), and/or genetic susceptibility. Perhaps the best known example where nutrients influence gene-product function is that of high folate levels physically stabilizing, and thereby rescuing, MTHFR enzyme function in the presence of the MTHFR C677T SNP (Frosst et al., 1995; Anonymous, 1999; Guenther et al., 1999). Individuals with this SNP who also have higher folate intakes have lower risks of CVD (Klerk et al., 2002) and cancers of the colon (Giovannucci, 2002; Sharp and Little, 2004) than those with the SNP who have lower folate intakes.
Candidate SNP studies and GWAS examining main effect associations between SNPs and cancer risk have revealed relatively few causal variants. Given that some environmental exposures are known to play a role in the development of cancer (World Cancer Research Fund/American Institute for Cancer Research, 2007), it is entirely possible that the genetic effects assumed present but not yet found may be hidden within specific subsets of individuals defined by their environmental exposures, as shown in the current study. The SNPs in this investigation would not have been identified had we only focused on main effects, supporting the need for more studies such as this one to fully discern the interplay between genes and environment on risk of cancer. Large international consortia with well-annotated exposure and phenotype information, such as the international Ovarian Cancer Association Consortium, are well-positioned to provide the large sample sizes needed to detect effect modification, especially for SNPs with low MAF.
The strengths of this analysis include the use of stringent quality control for genotyping that resulted in few excluded or failed samples, with remaining call rates over 99%, and the large sample to investigate our findings for the SNP-by-multivitamin interactions, especially for ATIC SNPs. Population structure is unlikely, since analyses were restricted to Caucasians and there was no statistical heterogeneity in SNP associations between study sites. There are also potential limitations. For most other SNPs, the power to detect gene–environment interactions was low. Although we applied the FDR method to estimate the number of falsely positive interactions, the application of FDR has been traditionally to high dimensionality data and may not be applicable to smaller datasets. Nevertheless, the consistent associations in the combined Phases 1 and 2 sample of two of the six SNPs with lower FDR values from Phase 1, in addition to the multi-variant and haplotype analyses of ATIC SNP-by-multivitamin interactions, suggests the associations may not be due to chance. Another potential limitation is that we did not have information on dietary intake to verify which nutrient(s) was related to the modifying effects of multivitamins. Although self-reported multivitamin supplement use adequately represents the higher nutrient status among consumers compared to non-consumers (Zhang et al., 1999, 2003; Kelemen et al., 2004), the potential for misclassification of this exposure may still exist in our study. The reference period for multivitamin use differed by site resulting in a slightly different definition in the Mayo Clinic and the Duke University studies. Our results for the interaction P values were somewhat attenuated compared to analyses that disregarded the information on duration of use (data not shown). Ostensibly, misclassification of exposure, although present, did not appear to influence our findings substantially.
In summary, this report provides little statistical evidence that SNPs in ATIC, SHMT2, and SLC46A1 are independently associated with ovarian carcinoma risk. The statistical interaction of ATIC SNPs with regular multivitamin intake and ATIC’s role in cellular metabolism and autophagy may support these findings as relevant to ovarian health and disease processes.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by Alberta Cancer Research Institute Projects 23905 and 24258, and NIH/NCI R01 CA88868 and R01 CA122443. Linda E. Kelemen is supported by a Canadian Institutes of Health Research New Investigator award MSH-87734.
Appendix
Figure A1.
Linkage disequilibrium plot for tagSNPs in AITC using 2,233 Caucasian subjects, Mayo Clinic, and Duke University. Numbers in squares indicate the correlation (r2) between SNPs.
Table A1.
Descriptive information and test statistics for SNPs in ATIC, SHMT2, and SLC46A1 among Caucasian controls, Mayo Clinic, and Duke University.
| Gene | Chr | Chr Position (bp)a | SNP rsID | Location | Phase 1 |
Phases 1 and 2 |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P HWEb |
MAFc |
P HWEb |
MAFc |
|||||||||||||
| Mayo | Duke | All | Mayo | Duke | All | Mayo | Duke | All | Mayo | Duke | All | |||||
| ATIC | 2 | 215898090 | rs3772078 | Intron | 0.88 | 0.88 | 0.74 | 0.19 | 0.19 | 0.19 | 1.00 | 0.80 | 0.85 | 0.20 | 0.18 | 0.19 |
| 2 | 215898265 | rs2372536d | Exone | 0.23 | 0.54 | 0.71 | 0.32 | 0.34 | 0.33 | 0.32 | 0.40 | 0.18 | 0.32 | 0.33 | 0.32 | |
| 2 | 215903416 | rs1880586d | Intron | 0.77 | 0.36 | 0.35 | 0.48 | 0.48 | 0.48 | 0.94 | 0.46 | 0.51 | 0.49 | 0.48 | 0.48 | |
| 2 | 215903526 | rs1983462d | Intron | 0.58 | 0.30 | 0.23 | 0.32 | 0.32 | 0.32 | 0.77 | 0.61 | 0.90 | 0.32 | 0.33 | 0.33 | |
| 2 | 215913412 | rs16853826d | Intron | 1.00 | 0.69 | 0.77 | 0.13 | 0.13 | 0.13 | 1.00 | 0.30 | 0.38 | 0.13 | 0.12 | 0.13 | |
| 2 | 215915470 | rs7586969d | Intron | 0.42 | 0.38 | 0.94 | 0.39 | 0.37 | 0.38 | 0.47 | 0.53 | 1.00 | 0.39 | 0.39 | 0.39 | |
| 2 | 215918481 | rs16853834 | Intron | 0.36 | 0.49 | 1.00 | 0.15 | 0.15 | 0.15 | 0.61 | 0.89 | 0.60 | 0.15 | 0.16 | 0.16 | |
| 2 | 215918544 | rs1404772 | Intron | 1.00 | 0.65 | 0.78 | 0.07 | 0.06 | 0.06 | 1.00 | 0.73 | 1.00 | 0.07 | 0.06 | 0.06 | |
| 2 | 215925884 | rs7604984 | 3′ Flanking | 0.84 | 0.70 | 0.94 | 0.40 | 0.40 | 0.40 | 0.53 | 0.24 | 0.19 | 0.39 | 0.39 | 0.39 | |
| SHMT2 | 12 | 55909088 | rs7301155d | 5′ Flanking | 0.67 | – | 0.67 | 0.34 | – | 0.34 | 0.70 | 0.39 | 0.39 | 0.34 | 0.30 | 0.32 |
| 12 | 55912849 | rs2229716d | Exonf | 1.00 | 1.00 | 1.00 | 0.04 | 0.04 | 0.04 | 1.00 | 0.22 | 0.67 | 0.04 | 0.04 | 0.04 | |
| SLC46A1 | 17 | 23743752 | rs9894260d | Intron | 0.34 | 0.56 | 0.19 | 0.08 | 0.09 | 0.08 | 1.00 | 0.36 | 0.36 | 0.08 | 0.09 | 0.08 |
| 17 | 23747949 | rs739439 | 3′ UTR | 0.72 | 0.16 | 0.44 | 0.17 | 0.20 | 0.18 | 0.66 | 0.38 | 0.71 | 0.18 | 0.19 | 0.18 | |
| 17 | 23749392 | rs2239908 | 3′ UTR | 0.24 | 0.65 | 0.23 | 0.44 | 0.46 | 0.45 | 0.67 | 0.50 | 0.40 | 0.45 | 0.45 | 0.45 | |
| 17 | 23753580 | rs17719944 | Intron | 0.55 | 1.00 | 0.70 | 0.09 | 0.10 | 0.10 | 0.80 | 0.51 | 0.51 | 0.09 | 0.09 | 0.09 | |
aBase pairs on genome build 36; UTR, untranslated region.
bP values from the chi-square test assessing deviation of genotype frequencies among controls from those expected under Hardy–Weinberg Equilibrium.
cMinor allele frequency.
dImputed SNPs in Phase 2 samples.
eNon-synonymous polymorphism.
fSynonymous polymorphism.
Table A2.
Characteristics of 1,575 Caucasian subjects (Phase 1), Mayo Clinic, and Duke University.
| Mayo Clinic |
Duke University |
|||
|---|---|---|---|---|
| Cases (n = 312) | Controls (n = 439) | Cases (n = 343) | Controls (n = 481) | |
| Age, years [mean (SD)] | 60.8 (12.8) | 60.0 (12.9) | 56.8 (10.4) | 54.6 (12.0) |
| BODY MASS INDEX, kg/m2 | ||||
| <23 [n (%)] | 57 (18.5) | 103 (23.7) | 96 (29.0) | 132 (28.1) |
| 23–26 [n (%)] | 77 (24.9) | 125 (28.8) | 85 (25.7) | 117 (25.0) |
| 26–29 [n (%)] | 61 (19.7) | 88 (20.3) | 75 (22.7) | 110 (23.5) |
| ≥29 [n (%)] | 114 (36.9) | 118 (27.2) | 75 (22.7) | 110 (23.5) |
| AGE AT MENARCHE, year | ||||
| <12 [n (%)] | 46 (18.5) | 67 (15.7) | 88 (25.7) | 85 (17.7) |
| 12 [n (%)] | 60 (24.1) | 97 (22.8) | 99 (29.0) | 142 (29.5) |
| 13 [n (%)] | 66 (26.5) | 126 (29.6) | 82 (24.0) | 140 (29.1) |
| ≥14 [n (%)] | 77 (30.9) | 136 (31.9) | 73 (21.4) | 114 (23.7) |
| ORAL CONTRACEPTIVE USE, months | ||||
| Never [n (%)] | 150 (49.7) | 166 (38.9) | 116 (34.5) | 148 (31.0) |
| 1–48 [n (%)] | 75 (24.8) | 90 (21.1) | 105 (31.3) | 133 (27.8) |
| ≥48 [n (%)] | 77 (25.5) | 171 (40.1) | 115 (34.2) | 197 (41.2) |
| POSTMENOPAUSAL STATUS | ||||
| Yes [n (%)] | 222 (73.0) | 328 (75.1) | 251 (80.2) | 312 (66.1) |
| POSTMENOPAUSAL HORMONE USE, months | ||||
| Never [n (%)] | 193 (62.7) | 244 (58.5) | 108 (33.4) | 270 (59.3) |
| 1–60 [n (%)] | 54 (17.5) | 79 (18.9) | 114 (35.3) | 96 (21.1) |
| ≥60 [n (%)] | 61 (19.8) | 94 (22.5) | 101 (31.3) | 89 (19.6) |
| PARITY, n/age at first birth, year | ||||
| Nulliparous [n (%)] | 56 (18.0) | 64 (14.7) | 71 (20.7) | 64 (13.3) |
| 1–2/≤20 [n (%)] | 20 (6.4) | 24 (5.5) | 46 (13.4) | 57 (11.9) |
| 1–2/≥20 [n (%)] | 91 (29.2) | 130 (29.9) | 125 (36.4) | 211 (43.9) |
| ≥3/≤20 [n (%)] | 59 (18.9) | 63 (14.5) | 49 (14.3) | 61 (12.7) |
| ≥3/≥20 [n (%)] | 86 (27.6) | 154 (35.4) | 52 (15.2) | 88 (18.3) |
| FAMILY HISTORY OF OVARIAN CANCERa | ||||
| Yes [n (%)] | 41 (13.1) | 33 (7.5) | 33 (9.6) | 26 (5.4) |
| SMOKING, pack-years | ||||
| None [n (%)] | 194 (65.3) | 279 (68.2) | 190 (57.8) | 250 (54.1) |
| ≤20 [n (%)] | 56 (18.9) | 82 (20.1) | 80 (24.3) | 121 (26.2) |
| >20 [n (%)] | 47 (15.8) | 48 (11.7) | 59 (17.9) | 91 (19.7) |
| EDUCATION | ||||
| No diploma [n (%)] | 18 (6.1) | 19 (4.3) | 26 (7.6) | 42 (8.7) |
| High School diploma [n (%)] | 107 (36.2) | 116 (26.5) | 105 (30.6) | 131 (27.2) |
| Post-high school education [n (%)] | 171 (57.8) | 303 (69.2) | 212 (61.8) | 308 (64.0) |
| MULTIVITAMIN USEb | ||||
| Yes [n (%)] | 73 (39.9) | 217 (51.7) | 115 (35.0) | 161 (34.6) |
| TUMOR HISTOLOGY, CASES | ||||
| Serous [n (%)] | 191 (61.2) | 206 (60.2) | ||
| Mucinous [n (%)] | 8 (2.6) | 19 (5.7) | ||
| Endometrioid [n (%)] | 63 (20.2) | 53 (15.5) | ||
| Clear cell [n (%)] | 20 (6.4) | 28 (8.2) | ||
| Other [n (%)] | 30 (9.6) | 36 (10.5) | ||
Counts do not total to 1,575 subjects due to missing data for some variables.
aIn first- or second-degree relative.
bAt least four pills per week during the previous year for at least 48 months’ duration of lifetime use (Mayo Clinic subjects) or at least three pills per week for at least 48 months during the past 60 months (Duke University subjects).
Table A3.
Odds ratios (OR) and 95% CIs for the association between variants in ATIC, SHMT2, and SLC46A1and ovarian carcinoma stratified by multivitamin supplement use among 658 subjects included in the GWAS (Phase 2), Mayo Clinic, and Duke University.
| Gene/SNP rsID | Regular multivitamin usersa |
Regular multivitamin non-users |
Pintc | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Homozygous major allele (referent) | Heterozygous | Homozygous minor allele | Ordinal (per minor allele) | Homozygous major allele (referent) | Heterozygous | Homozygous minor allele | Ordinal (per minor allele) | ||
| Cases/controls | Cases/controls | Cases/controls | OR (95%CI)b | Cases/controls | Cases/controls | Cases/controls | OR (95%CI)b | ||
| ATIC | |||||||||
| rs3772078 | 92/96 | 44/39 | 5/6 | 0.9 (0.4–2.1) | 111/119 | 46/57 | 6/6 | 0.9 (0.4–1.7) | 0.92 |
| rs2372536 | 61/65 | 58/60 | 22/16 | 1.1 (0.6–2.1) | 87/98 | 57/63 | 19/21 | 1.2 (0.7–2.1) | 0.85 |
| rs1880586 | 31/39 | 65/61 | 45/41 | 0.9 (0.5–1.7) | 49/60 | 77/74 | 37/48 | 1.1 (0.6–1.9) | 0.90 |
| rs1983462 | 77/64 | 52/57 | 12/20 | 0.7 (0.4–1.4) | 61/80 | 76/77 | 26/25 | 1.0 (0.6–1.8) | 0.85 |
| rs16853826 | 106/109 | 34/31 | 1/1 | 0.9 (0.3–2.4) | 126/138 | 33/42 | 4/2 | 0.8 (0.3–1.8) | 0.77 |
| rs7586969 | 61/50 | 66/67 | 14/24 | 0.7 (0.4–1.4) | 51/62 | 78/88 | 34/32 | 1.0 (0.5–1.7) | 0.82 |
| rs16853834 | 95/103 | 42/34 | 4/4 | 1.6 (0.7–3.7) | 122/127 | 36/46 | 4/7 | 1.0 (0.4–2.1) | 0.73 |
| rs1404772 | 123/122 | 18/19 | 0/0 | 0.6 (0.1–2.8) | 146/159 | 16/22 | 1/1 | 0.8 (0.2–2.6) | 0.99 |
| rs7604984 | 49/56 | 65/61 | 27/23 | 1.1 (0.6–2.2) | 73/86 | 67/65 | 23/31 | 1.2 (0.7–2.1) | 0.60 |
| SHMT2 | |||||||||
| rs7301155 | 65/69 | 63/60 | 13/12 | 0.6 (0.3–1.3) | 85/80 | 64/82 | 14/20 | 0.6 (0.3–1.1) | 0.43 |
| rs2229716 | 135/140 | 6/1 | 0/0 | 5.8 (0.3–111.9) | 185/176 | 5/5 | 0/1 | 1.6 (0.2–13.2) | 0.68 |
| SLC46A1 | |||||||||
| rs9894260 | 121/114 | 18/25 | 2/2 | 0.7 (0.2–1.9) | 140/156 | 21/25 | 2/1 | 0.6 (0.2–1.8) | 0.56 |
| rs739439 | 80/99 | 53/36 | 8/6 | 1.0 (0.4–2.2) | 107/125 | 50/53 | 6/4 | 1.0 (0.5–2.1) | 0.39 |
| rs2239908 | 25/42 | 82/65 | 34/34 | 0.9 (0.5–1.8) | 41/54 | 86/95 | 36/33 | 1.6 (0.9–2.8) | 0.03 |
| rs17719944 | 109/123 | 31/17 | 0/1 | 4.0 (1.1–14.8) | 1,340/150 | 28/29 | 1/3 | 0.5 (0.2–1.4) | 0.02 |
aAt least four pills per week during the previous year for at least 48 months’ duration of lifetime use (Mayo Clinic subjects) or at least three pills per week for at least 48 months during the past 60 months (Duke University subjects).
bAdjusted for age (<40, 40–49, 50–59, 60–69, and 70+ years), region of residence (Minnesota, Iowa, Wisconsin, Illinois, North Dakota, South Dakota, and North Carolina), body mass index, age at menarche, oral contraceptive use, postmenopausal status, postmenopausal hormone use, parity/age at first birth, smoking, and education. SNP effect was determined using a log-additive logistic regression model.
cP for interaction.
Table A4.
Odds ratios (OR) and 95% CIs for the main effect association between variants in ATIC, SHMT2, and SLC46A1 and ovarian carcinoma among 1,575 Caucasian subjects (Phase 1), Mayo Clinic, and Duke University.
| Gene/SNP rsID | Homozygous major allele (referent) |
Heterozygous |
Homozygous minor allele |
P value (2 df) | Ordinal (per minor allele) |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cases | Controls | Cases | Controls | OR (95%CI)a | Cases | Controls | OR (95%CI)a | OR (95%CI)a | P-trend | ||
| ATIC | |||||||||||
| rs3772078 | 415 | 583 | 201 | 279 | 1.0 (0.8–1.3) | 20 | 30 | 0.9 (0.5–1.7) | 0.98 | 1.0 (0.8–1.2) | 0.93 |
| rs2372536 | 304 | 409 | 279 | 399 | 1.0 (0.8–1.2) | 67 | 103 | 0.9 (0.6–1.3) | 0.76 | 0.9 (0.8–1.1) | 0.46 |
| rs1880586 | 167 | 245 | 336 | 473 | 1.0 (0.8–1.3) | 149 | 201 | 1.1 (0.8–1.4) | 0.88 | 1.0 (0.9–1.2) | 0.61 |
| rs1983462 | 319 | 417 | 275 | 417 | 0.9 (0.7–1.1) | 60 | 86 | 0.9 (0.6–1.3) | 0.42 | 0.9 (0.8–1.1) | 0.30 |
| rs16853826 | 497 | 696 | 150 | 210 | 1.0 (0.8–1.3) | 8 | 14 | 0.8 (0.3–2.0) | 0.93 | 1.0 (0.8–1.2) | 0.84 |
| rs7586969 | 259 | 348 | 301 | 436 | 0.9 (0.7–1.1) | 93 | 134 | 0.9 (0.7–1.3) | 0.76 | 1.0 (0.8–1.1) | 0.55 |
| rs16853834 | 442 | 657 | 187 | 241 | 1.1 (0.9–1.4) | 26 | 22 | 1.7 (0.9–3.1) | 0.13 | 1.2 (1.0–1.4) | 0.06 |
| rs1404772 | 568 | 807 | 84 | 108 | 1.1 (0.8–1.5) | 3 | 4 | 1.1 (0.2–5.0) | 0.86 | 1.1 (0.8–1.4) | 0.59 |
| rs7604984 | 242 | 332 | 317 | 442 | 1.0 (0.8–1.2) | 96 | 144 | 0.9 (0.7–1.2) | 0.84 | 1.0 (0.8–1.1) | 0.64 |
| SHMT2 | |||||||||||
| rs7301155 | 121 | 187 | 153 | 202 | 1.2 (0.9–1.6) | 37 | 49 | 1.2 (0.7–1.9) | 0.56 | 1.1 (0.9–1.4) | 0.33 |
| rs2229716 | 601 | 838 | 51 | 79 | 0.9 (0.6–1.3) | 2 | 1 | 2.4 (0.2–27.0) | 0.63 | 0.9 (0.7–1.3) | 0.70 |
| SLC46A1 | |||||||||||
| rs9894260 | 531 | 768 | 111 | 148 | 1.1 (0.8–1.4) | 9 | 3 | 5.4 (1.4–20.5) | 0.04 | 1.2 (0.9–1.5) | 0.14 |
| rs739439 | 435 | 610 | 191 | 283 | 0.9 (0.8–1.2) | 26 | 27 | 1.3 (0.8–2.3) | 0.48 | 1.0 (0.8–1.2) | 0.87 |
| rs2239908 | 197 | 270 | 324 | 473 | 0.9 (0.7–1.2) | 131 | 175 | 1.0 (0.8–1.4) | 0.76 | 1.0 (0.9–1.2) | 0.99 |
| rs17719944 | 534 | 753 | 113 | 157 | 1.0 (0.8–1.3) | 7 | 9 | 1.1 (0.4–3.1) | 0.96 | 1.0 (0.8–1.3) | 0.81 |
aAdjusted for age (<40, 40–49, 50–59, 60–69, and 70+ years) and region of residence (Minnesota, Iowa, Wisconsin, Illinois, North Dakota, South Dakota, and North Carolina).
Table A5.
Odds ratios (OR) and 95% CIs for the main effect association between variants in ATIC, SHMT2, and SLC46A1 and ovarian carcinoma among 2,233 Caucasian subjects (Phases 1 and 2), Mayo Clinic, and Duke University.
| Gene/SNP rsID | Homozygous major allele (referent) |
Heterozygous |
Homozygous minor allele |
P value (2 df) | Ordinal (per minor allele) |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cases | Controls | Cases | Controls | OR (95%CI)a | Cases | Controls | OR (95%CI)a | OR (95%CI)a | P-trend | ||
| ATIC | |||||||||||
| rs3772078 | 623 | 810 | 294 | 384 | 1.0 (0.8–1.2) | 32 | 43 | 1.0 (0.6–1.6) | 0.98 | 1.0 (0.8–1.2) | 0.85 |
| rs2372536 | 455 | 583 | 399 | 530 | 1.0 (0.8–1.2) | 109 | 143 | 1.0 (0.7–1.3) | 0.95 | 1.0 (0.9–1.1) | 0.84 |
| rs1880586 | 248 | 348 | 482 | 619 | 1.1 (0.9–1.3) | 235 | 297 | 1.1 (0.9–1.4) | 0.69 | 1.0 (0.9–1.2) | 0.48 |
| rs1983462 | 463 | 573 | 406 | 559 | 0.9 (0.8–1.1) | 98 | 133 | 0.9 (0.7–1.2) | 0.54 | 0.9 (0.8–1.1) | 0.32 |
| rs16853826 | 734 | 957 | 220 | 291 | 1.0 (0.8–1.2) | 14 | 17 | 1.1 (0.5–2.3) | 0.92 | 1.0 (0.8–1.2) | 0.97 |
| rs7586969 | 377 | 469 | 448 | 602 | 0.9 (0.8–1.1) | 141 | 192 | 0.9 (0.7–1.2) | 0.66 | 0.9 (0.8–1.1) | 0.39 |
| rs16853834 | 665 | 902 | 268 | 328 | 1.1 (0.9–1.3) | 34 | 33 | 1.4 (0.8–2.2) | 0.32 | 1.1 (1.0–1.3) | 0.15 |
| rs1404772 | 846 | 1,107 | 118 | 152 | 1.0 (0.8–1.3) | 4 | 5 | 1.1 (0.3–4.1) | 0.99 | 1.0 (0.8–1.3) | 0.98 |
| rs7604984 | 367 | 483 | 455 | 577 | 1.0 (0.9–1.3) | 146 | 202 | 1.0 (0.7–1.2) | 0.75 | 1.0 (0.9–1.1) | 0.87 |
| SHMT2 | |||||||||||
| rs7301155 | 398 | 545 | 404 | 530 | 1.0 (0.9–1.3) | 86 | 114 | 1.0 (0.8–1.4) | 0.87 | 1.0 (0.9–1.2) | 0.64 |
| rs2229716 | 902 | 1,175 | 63 | 86 | 0.9 (0.7–1.3) | 2 | 2 | 1.1 (0.1–7.7) | 0.92 | 0.9 (0.7–3.3) | 0.73 |
| SLC46A1 | |||||||||||
| rs9894260 | 799 | 1,056 | 152 | 202 | 1.0 (0.8–1.2) | 13 | 6 | 3.1 (1.1–8.3) | 0.08 | 1.1 (0.9–1.3) | 0.44 |
| rs739439 | 627 | 845 | 298 | 281 | 1.0 (0.9–1.3) | 40 | 39 | 1.4 (0.9–2.2) | 0.36 | 1.1 (0.9–1.3) | 0.25 |
| rs2239908 | 267 | 370 | 495 | 642 | 1.1 (0.9–1.3) | 203 | 251 | 1.1 (0.9–1.4) | 0.73 | 1.1 (0.9–1.2) | 0.43 |
| rs17719944 | 784 | 1,040 | 174 | 211 | 1.1 (0.9–1.4) | 8 | 13 | 0.9 (0.4–2.1) | 0.66 | 1.1 (0.9–1.3) | 0.53 |
aAdjusted for age (<40, 40–49, 50–59, 60–69, and 70+ years) and region of residence (Minnesota, Iowa, Wisconsin, Illinois, North Dakota, South Dakota, and North Carolina).
References
- Anonymous. (1999). How folate fights disease. Nat. Struct. Biol. 6, 293–294 10.1038/7517 [DOI] [PubMed] [Google Scholar]
- Barrett J. C., Fry B., Maller J., Daly M. J. (2005). Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21, 263–265 10.1093/bioinformatics/bth457 [DOI] [PubMed] [Google Scholar]
- Cheong C. G., Wolan D. W., Greasley S. E., Horton P. A., Beardsley G. P., Wilson I. A. (2004). Crystal structures of human bifunctional enzyme aminoimidazole-4-carboxamide ribonucleotide transformylase/IMP cyclohydrolase in complex with potent sulfonyl-containing antifolates. J. Biol. Chem. 279, 18034–18045 10.1074/jbc.M313691200 [DOI] [PubMed] [Google Scholar]
- Choi S. W., Mason J. B. (2000). Folate and carcinogenesis: an integrated scheme. J. Nutr. 130, 129–132 [DOI] [PubMed] [Google Scholar]
- Cunningham J. M., Sellers T. A., Schildkraut J. M., Fredericksen Z. S., Vierkant R. A., Kelemen L. E., Gadre M., Phelan C. M., Huang Y., Meyer J. G., Pankratz V. S., Goode E. L. (2008). Performance of amplified DNA in an Illumina GoldenGate BeadArray assay. Cancer Epidemiol. Biomarkers Prev. 17, 1781–1789 10.1158/1055-9965.EPI-07-2849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ericson U., Sonestedt E., Gullberg B., Olsson H., Wirfalt E. (2007). High folate intake is associated with lower breast cancer incidence in postmenopausal women in the Malmo Diet and Cancer cohort. Am. J. Clin. Nutr. 86, 434–443 [DOI] [PubMed] [Google Scholar]
- Fan J. B., Hu S. X., Craumer W. C., Barker D. L. (2005). BeadArray-based solutions for enabling the promise of pharmacogenomics. BioTechniques 39, 583–588 10.2144/000112047 [DOI] [PubMed] [Google Scholar]
- Feigelson H. S., Jonas C. R., Robertson A. S., Mccullough M. L., Thun M. J., Calle E. E. (2003). Alcohol, folate, methionine, and risk of incident breast cancer in the American Cancer Society Cancer Prevention Study II Nutrition Cohort. Cancer Epidemiol. Biomarkers Prev. 12, 161–164 [PubMed] [Google Scholar]
- Frosst P., Blom H. J., Milos R., Goyette P., Sheppard C. A., Matthews R. G., Boers G. J., Den Heijer M., Kluijtmans L. A., Van Den Heuvel L. P., Rozen R. (1995). A candidate genetic risk factor for vascular disease: a common mutation in methylenetetrahydrofolate reductase. Nat. Genet. 10, 111–113 10.1038/ng0595-111 [DOI] [PubMed] [Google Scholar]
- Giovannucci E. (2002). Epidemiologic studies of folate and colorectal neoplasia: a review. J. Nutr. 132, 2350S–2355S [DOI] [PubMed] [Google Scholar]
- Guenther B. D., Sheppard C. A., Tran P., Rozen R., Matthews R. G., Ludwig M. L. (1999). The structure and properties of methylenetetrahydrofolate reductase from Escherichia coli suggest how folate ameliorates human hyperhomocysteinemia. Nat. Struct. Biol. 6, 359–365 10.1038/7594 [DOI] [PubMed] [Google Scholar]
- Hallstrom T. C., Mori S., Nevins J. R. (2008). An E2F1-dependent gene expression program that determines the balance between proliferation and cell death. Cancer Cell 13, 11–22 10.1016/j.ccr.2007.11.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochberg Y., Benjamini Y. (1990). More powerful procedures for multiple significance testing. Stat. Med. 9, 811–818 10.1002/sim.4780090710 [DOI] [PubMed] [Google Scholar]
- Kelemen L. E., Sellers T. A., Schildkraut J. M., Cunningham J. M., Vierkant R. A., Pankratz V. S., Fredericksen Z. S., Gadre M. K., Rider D. N., Liebow M., Goode E. L. (2008). Genetic variation in the one-carbon transfer pathway and ovarian cancer risk. Cancer Res. 68, 2498–2506 10.1158/0008-5472.CAN-07-5165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelemen L. E., Sellers T. A., Vierkant R. A., Harnack L., Cerhan J. R. (2004). Association of folate and alcohol with risk of ovarian cancer in a prospective study of postmenopausal women. Cancer Causes Control 15, 1085–1093 10.1007/s10552-004-1546-6 [DOI] [PubMed] [Google Scholar]
- Kim Y. I. (2003). Role of folate in colon cancer development and progression. J. Nutr. 133, 3731S–3739S [DOI] [PubMed] [Google Scholar]
- Klerk M., Verhoef P., Clarke R., Blom H. J., Kok F. J., Schouten E. G. (2002). MTHFR 677C→T polymorphism and risk of coronary heart disease: a meta-analysis. JAMA 288, 2023–2031 10.1001/jama.288.16.2023 [DOI] [PubMed] [Google Scholar]
- Kondo M., Yamaoka T., Honda S., Miwa Y., Katashima R., Moritani M., Yoshimoto K., Hayashi Y., Itakura M. (2000). The rate of cell growth is regulated by purine biosynthesis via ATP production and G(1) to S phase transition. J. Biochem. 128, 57–64 [DOI] [PubMed] [Google Scholar]
- Levine A. J., Figueiredo J. C., Lee W., Conti D. V., Kennedy K., Duggan D. J., Poynter J. N., Campbell P. T., Newcomb P., Martinez M. E., Hopper J. L., Le Marchand L., Baron J. A., Limburg P. J., Ulrich C. M., Haile R. W. (2010). A candidate gene study of folate-associated one carbon metabolism genes and colorectal cancer risk. Cancer Epidemiol. Biomarkers Prev. 19, 1812–1821 10.1158/1055-9965.EPI-09-0727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Willer C., Sanna S., Abecasis G. (2009). Genotype imputation. Annu. Rev. Genomics Hum. Genet. 10, 387–406 10.1146/annurev.genom.9.081307.164242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma E., Iwasaki M., Junko I., Hamada G. S., Nishimoto I. N., Carvalho S. M., Motola J., Jr., Laginha F. M., Tsugane S. (2009). Dietary intake of folate, vitamin B6, and vitamin B12, genetic polymorphism of related enzymes, and risk of breast cancer: a case-control study in Brazilian women. BMC Cancer 9, 122–131 10.1186/1471-2407-9-122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., Stampfer M. J., Giovannucci E., Artigas C., Hunter D. J., Fuchs C., Willett W. C., Selhub J., Hennekens C. H., Rozen R. (1997). Methylenetetrahydrofolate reductase polymorphism, dietary interactions, and risk of colorectal cancer. Cancer Res. 57, 1098–1102 [PubMed] [Google Scholar]
- Maruti S. S., Ulrich C. M., Jupe E. R., White E. (2009). MTHFR C677T and postmenopausal breast cancer risk by intakes of one-carbon metabolism nutrients: a nested case-control study. Breast Cancer Res. 11, R91. 10.1186/bcr2462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew R., Karantza-Wadsworth V., White E. (2007). Role of autophagy in cancer. Nat. Rev. Cancer 7, 961–967 10.1038/nrc2254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N. (2007). Autophagy: process and function. Genes Dev. 21, 2861–2873 10.1101/gad.1599207 [DOI] [PubMed] [Google Scholar]
- Oliphant A., Barker D. L., Stuelpnagel J. R., Chee M. S. (2002). BeadArray technology: enabling an accurate, cost-effective approach to high-throughput genotyping. Biotechniques June (Suppl. 56–58), 60–61 [PubMed] [Google Scholar]
- Qiu A., Jansen M., Sakaris A., Min S. H., Chattopadhyay S., Tsai E., Sandoval C., Zhao R., Akabas M. H., Goldman I. D. (2006). Identification of an intestinal folate transporter and the molecular basis for hereditary folate malabsorption. Cell 127, 917–928 10.1016/j.cell.2006.09.041 [DOI] [PubMed] [Google Scholar]
- R Development Core Team. (2011). A Language and Environment for Statistical Computing. Available at: http://www.R-project.org [Accessed 2012].
- Rodwell V. W. (2000). “Metabolism of purine and pyrimidine nucleotides,” in Harper’s Biochemistry, eds Murray R. K., Granner D. K., Mayes P. A., Rodwell V. W. (New York: McGraw-Hill; ), 386–401 [Google Scholar]
- Schaid D. J. (2011). Personal Web Page, Software List: Haplo.stats. Available at: http://mayoresearch.mayo.edu/schaid_lab/software.cfm [Accessed January 30, 2012].
- Schaid D. J., Rowland C. M., Tines D. E., Jacobson R. M., Poland G. A. (2002). Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am. J. Hum. Genet. 70, 425–434 10.1086/338688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellers T. A., Schildkraut J. M., Pankratz V. S., Vierkant R. A., Fredericksen Z. S., Olson J. E., Cunningham J., Taylor W., Liebow M., Mcpherson C., Hartmann L. C., Pal T., Adjei A. A. (2005). Estrogen bioactivation, genetic polymorphisms, and ovarian cancer. Cancer Epidemiol. Biomarkers Prev. 14, 2536–2543 10.1158/1055-9965.EPI-05-0142 [DOI] [PubMed] [Google Scholar]
- Sharp L., Little J. (2004). Polymorphisms in genes involved in folate metabolism and colorectal neoplasia: a HuGE review. Am. J. Epidemiol. 159, 423–443 10.1093/aje/kwh066 [DOI] [PubMed] [Google Scholar]
- Steemers F. J., Gunderson K. L. (2005). Illumina, Inc. Pharmacogenomics 6, 777–782 10.2217/14622416.6.7.777 [DOI] [PubMed] [Google Scholar]
- The International HapMap Consortium. (2003). The international HapMap project. Nature 426, 789–796 10.1038/nature02168 [DOI] [PubMed] [Google Scholar]
- Tibbetts A. S., Appling D. R. (2010). Compartmentalization of Mammalian folate-mediated one-carbon metabolism. Annu. Rev. Nutr. 30, 57–81 10.1146/annurev.nutr.012809.104810 [DOI] [PubMed] [Google Scholar]
- Ulrich C. M., Reed M. C., Nijhout H. F. (2008). Modeling folate, one-carbon metabolism, and DNA methylation. Nutr. Rev. 66(Suppl. 1), S27–S30 10.1111/j.1753-4887.2008.00062.x [DOI] [PubMed] [Google Scholar]
- Wang L., Lu J., An J., Shi Q., Spitz M. R., Wei Q. (2007). Polymorphisms of cytosolic serine hydroxymethyltransferase and risk of lung cancer: a case-control analysis. Lung Cancer 57, 143–151 10.1016/j.lungcan.2007.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolan D. W., Cheong C. G., Greasley S. E., Wilson I. A. (2004). Structural insights into the human and avian IMP cyclohydrolase mechanism via crystal structures with the bound XMP inhibitor. Biochemistry Mosc. 43, 1171–1183 [DOI] [PubMed] [Google Scholar]
- World Cancer Research Fund/American Institute for Cancer Research. (2007). Food, Nutrition, Physical Activity and the Prevention of Cancer: A Global Perspective. Washington, DC: AICR [Google Scholar]
- Zhang S., Hunter D. J., Hankinson S. E., Giovannucci E. L., Rosner B. A., Colditz G. A., Speizer F. E., Willett W. C. (1999). A prospective study of folate intake and the risk of breast cancer. JAMA 281, 1632–1637 10.1001/jama.281.17.1632 [DOI] [PubMed] [Google Scholar]
- Zhang S. M., Willett W. C., Selhub J., Hunter D. J., Giovannucci E. L., Holmes M. D., Colditz G. A., Hankinson S. E. (2003). Plasma folate, vitamin B6, vitamin B12, homocysteine, and risk of breast cancer. J. Natl. Cancer Inst. 95, 373–380 10.1093/jnci/95.5.373 [DOI] [PubMed] [Google Scholar]
- Zhang Z., Shi Q., Sturgis E. M., Spitz M. R., Wei Q. (2005). Polymorphisms and haplotypes of serine hydroxymethyltransferase and risk of squamous cell carcinoma of the head and neck: a case-control analysis. Pharmacogenet. Genomics 15, 557–564 10.1097/01.fpc.0000170915.19522.b2 [DOI] [PubMed] [Google Scholar]
- Zhao R., Min S. H., Qiu A., Sakaris A., Goldberg G. L., Sandoval C., Malatack J. J., Rosenblatt D. S., Goldman I. D. (2007). The spectrum of mutations in the PCFT gene, coding for an intestinal folate transporter, that are the basis for hereditary folate malabsorption. Blood 110, 1147–1152 10.1182/blood-2006-12-062927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R., Min S. H., Wang Y., Campanella E., Low P. S., Goldman I. D. (2009). A role for the proton-coupled folate transporter (PCFT-SLC46A1) in folate receptor-mediated endocytosis. J. Biol. Chem. 284, 4267–4274 10.1074/jbc.M109.011015 [DOI] [PMC free article] [PubMed] [Google Scholar]