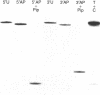
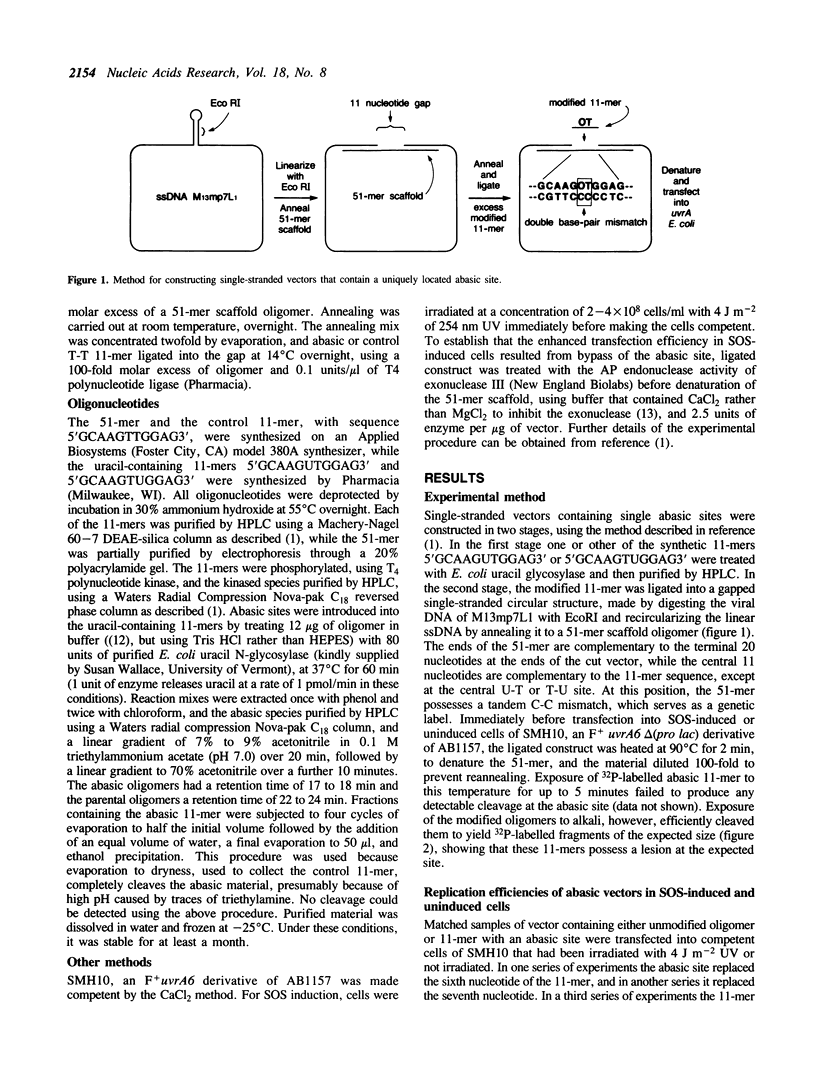
Abstract
We have investigated the mutagenic properties of an abasic site in DNA by transfecting SOS-induced and uninduced cells of E. coli with a single-stranded M13mp7-based vector that carries a single example of this lesion at one or other of two unique and adjacent sites. Random samples of progeny phage were sequenced to determine the nature of the replication events that occurred at and around these locations. 5% to 7% of the vectors could be replicated in SOS-induced cells, but only 0.1% to 0.7% of them gave plaques in the absence of SOS induction. In SOS-induced cells, 93% and 96% of the phage replicated resulted from the insertion of a nucleotide opposite the abasic site, while the remainder resulted from a targeted omission of a single nucleotide. At one of the sites, nucleotide insertions were 54% dAMP, 25% dTMP, 20% dGMP and 1% dCMP. At the other site they were 80% dAMP, 4% dTMP, 15% dGMP and 1% dCMP. The sequence variation in all but two of the 204 sequences analyzed was restricted to the abasic site itself. In the remaining two, a change at the abasic site was accompanied by a mutation at an immediately flanking nucleotide.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Banerjee S. K., Christensen R. B., Lawrence C. W., LeClerc J. E. Frequency and spectrum of mutations produced by a single cis-syn thymine-thymine cyclobutane dimer in a single-stranded vector. Proc Natl Acad Sci U S A. 1988 Nov;85(21):8141–8145. doi: 10.1073/pnas.85.21.8141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hevroni D., Livneh Z. Bypass and termination at apurinic sites during replication of single-stranded DNA in vitro: a model for apurinic site mutagenesis. Proc Natl Acad Sci U S A. 1988 Jul;85(14):5046–5050. doi: 10.1073/pnas.85.14.5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A. Mutational specificity of depurination. Proc Natl Acad Sci U S A. 1984 Mar;81(5):1494–1498. doi: 10.1073/pnas.81.5.1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A., Schaaper R. M., Loeb L. A. Depurination-induced infidelity of deoxyribonucleic acid synthesis with purified deoxyribonucleic acid replication proteins in vitro. Biochemistry. 1983 May 10;22(10):2378–2384. doi: 10.1021/bi00279a012. [DOI] [PubMed] [Google Scholar]
- Kypr J. Possible reason for the preferential insertion of adenine opposite abasic lesions in DNA. J Theor Biol. 1988 Nov 8;135(1):125–126. doi: 10.1016/s0022-5193(88)80180-2. [DOI] [PubMed] [Google Scholar]
- Lindahl T. Uracil-DNA glycosylase from Escherichia coli. Methods Enzymol. 1980;65(1):284–290. doi: 10.1016/s0076-6879(80)65038-1. [DOI] [PubMed] [Google Scholar]
- Loeb L. A., Preston B. D. Mutagenesis by apurinic/apyrimidinic sites. Annu Rev Genet. 1986;20:201–230. doi: 10.1146/annurev.ge.20.120186.001221. [DOI] [PubMed] [Google Scholar]
- Randall S. K., Eritja R., Kaplan B. E., Petruska J., Goodman M. F. Nucleotide insertion kinetics opposite abasic lesions in DNA. J Biol Chem. 1987 May 15;262(14):6864–6870. [PubMed] [Google Scholar]
- Rogers S. G., Weiss B. Exonuclease III of Escherichia coli K-12, an AP endonuclease. Methods Enzymol. 1980;65(1):201–211. doi: 10.1016/s0076-6879(80)65028-9. [DOI] [PubMed] [Google Scholar]
- Sagher D., Strauss B. Insertion of nucleotides opposite apurinic/apyrimidinic sites in deoxyribonucleic acid during in vitro synthesis: uniqueness of adenine nucleotides. Biochemistry. 1983 Sep 13;22(19):4518–4526. doi: 10.1021/bi00288a026. [DOI] [PubMed] [Google Scholar]
- Schaaper R. M., Kunkel T. A., Loeb L. A. Infidelity of DNA synthesis associated with bypass of apurinic sites. Proc Natl Acad Sci U S A. 1983 Jan;80(2):487–491. doi: 10.1073/pnas.80.2.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss B., Rabkin S., Sagher D., Moore P. The role of DNA polymerase in base substitution mutagenesis on non-instructional templates. Biochimie. 1982 Aug-Sep;64(8-9):829–838. doi: 10.1016/s0300-9084(82)80138-7. [DOI] [PubMed] [Google Scholar]
- Takeshita M., Chang C. N., Johnson F., Will S., Grollman A. P. Oligodeoxynucleotides containing synthetic abasic sites. Model substrates for DNA polymerases and apurinic/apyrimidinic endonucleases. J Biol Chem. 1987 Jul 25;262(21):10171–10179. [PubMed] [Google Scholar]