Abstract
We previously demonstrated that black bean (BB) and soy flour (SF)-based diets inhibit azoxymethane (AOM)-induced colon cancer. The objective of this study was to identify genes altered by carcinogen treatment in normal-appearing colonic mucosa and those attenuated by bean feeding. Ninety-five male F344 rats were fed control (AIN) diets upon arrival. At 4 and 5 weeks, rats were injected with AOM (15 mg/kg) or saline and one week later administered an AIN, BB-, or SF-based diet. Rats were sacrificed after 31 weeks, and microarrays were conducted on RNA isolated from the distal colonic mucosa. AOM treatment induced a number of genes involved in immunity, including several MHC II-associated antigens and innate defense genes (RatNP-3, Lyz2, Pla2g2a). BB- and SF-fed rats exhibited a higher expression of genes involved in energy metabolism and water and sodium absorption and lower expression of innate (RatNP-3, Pla2g2a, Tlr4, Dmbt1) and cell cycle-associated (Cdc2, Ccnb1, Top2a) genes. Genes involved in the extracellular matrix (Col1a1, Fn1) and innate immunity (RatNP-3, Pla2g2a) were induced by AOM in all diets, but to a lower extent in bean-fed animals. This profile suggests beans inhibit colon carcinogenesis by modulating cellular kinetics and reducing inflammation, potentially by preserving mucosal barrier function.
1. Introduction
Colorectal cancer (CRC) is one of the most common neoplasms afflicting industrialized societies [1]. In 2008, there were 609,051 deaths due to colorectal cancer worldwide, with 50,640 cases in the United States alone [1]. Both genetic and environmental exposures have been implicated in the etiology of CRC, and it has been estimated that up to 75% of cases may be preventable by adequate diets and regular exercise [2–4]. Consumption of diets low in red meat and alcohol and high in vegetables and cereal grains is generally associated with a decreased risk of developing CRC [4–6]. Additionally, populations consuming higher intakes of legumes (peas, beans, lentils, peanuts) are reported to have a lower risk of [6–12] and mortality from CRC [13].
It has long been known that dietary patterns modulate the incidence and mortality of colorectal cancer [2, 3], however, identification of specific mechanisms has been limited. The azoxymethane- (AOM-) induced colon cancer model in rodents has been utilized extensively to examine dietary influences on colon cancer. Tumors develop almost exclusively in the colon, primarily in the distal region, similar to the distribution observed in humans from high-risk areas. Additionally, many of the common genetic and pathogenic changes contributing to human colon cancers are also observed during AOM-induced colon carcinogenesis [14–19]. Although Apc mutations are infrequently detected [20], mutations in GSK-3β phosphorylation consensus sites on β-catenin are present in up to 77% of AOM-induced colon cancers as well as in early preneoplastic lesions [17, 21–23]. These sites are important for downregulation of β-catenin by ubiquitination and result in stabilization and nuclear localization of the protein [23]. Activating mutations on codon 12 and 13 of the k-ras gene [14, 15], upregulation of cyclo-oxygenase 2 (COX-2), and inducible nitric oxide synthase (iNOS), as well as alterations in transforming growth factor β signaling, are also common features to both human and AOM-induced colon cancers [24–29]. Using this model, experiments conducted in our laboratory [30–33] and by others [34–36] have demonstrated the potential of bean-based diets to inhibit AOM-induced colon cancer. For example, Hughes et al. [35] and Hangen and Bennink [32] found that rats fed dry beans (pinto, navy, or black beans) had a 50–57% lower incidence of colon cancer than rats fed a casein-based diet. Similarly, in a series of experiments, Bennink et al. [30, 31, 33] reported a significant reduction in colon tumor incidence and tumor burden in rats fed defatted soy flour compared to casein-fed control animals.
The purpose of the current investigation was to elucidate cellular mechanisms underlying colon cancer inhibition by beans in vivo. Microarrays were performed on mRNA isolated from distal colonic epithelium of saline and AOM-injected F344 rats fed either a casein (AIN), black bean (BB), or defatted soy flour (SF) diet for 31 weeks. We chose to focus on the distal segment because most tumors develop in this area using standard protocol (15 mg/kg AOM) and there is evidence for site-specific effects of food constituents on tumorigenesis [37]. It was anticipated that genes most important to dietary suppression of colon cancer would be similarly affected by black beans and soy flour and have altered expression (increased or decreased) that corroborated tumor incidence. The profile of genes altered in this experiment suggests aberrant activation of innate and adaptive immune responses are permissive for colon carcinogenesis, whereas inhibition of tumor promotion by bean feeding is associated with modulation of genes involved in crypt cell homeostasis, innate defense, and extracellular matrix components.
2. Materials and Methods
2.1. Animal Care and Experimental Diets
This study was conducted in conformity with the regulatory guidelines of the Michigan State University Institutional Animal Care and Use Committee. Ninety-five male Fischer (F344) rats were obtained from Harlan Sprague-Dawley (Indianapolis, IL) at 3 weeks of age and housed in plastic cages (2-3 rats/cage) in temperature-(23°C ± 2°) and humidity- (40–60%) controlled rooms with a 12-hour light/dark cycle. Throughout the experiment, animals had free access to food and distilled water and were assessed daily for health status and monthly for weight gain.
One control and two experimental diets were formulated based on the AIN-93G rodent diet with modifications [38], to contain the majority (i.e., >85%) of protein from either (1) high nitrogen casein (AIN), (2) black beans (BB), or (3) defatted soy flour (SF) (Archer Daniels Midland; Decatur, IL) and matched to have similar nutrient : energy ratios (Table 1). Black beans were soaked overnight in distilled water, cooked in a steam jacket kettle for 30 minutes, dried at 58°C, and then finely ground to pass through a 1.6 mm diameter screen prior to mixing with other diet ingredients. All diets contained approximately 18.9% (wt/wt) total protein, 11.3% dietary fiber, and 16.7% fat (wt/wt). Casein and tryptophan were added to black bean diet and methionine was added to all diets to increase the amino acid score to >90%. Lard, corn, and soybean oil were added to all diets, adjusted so the total saturated (SFA) : monounsaturated (MUFA) : polyunsaturated (PUFA) fatty acid ratios were 1.0 : 1.2 : 1.1, respectively.
Table 1.
Nutrient composition of experimental diets1.
| Ingredient | g/100 g diet | ||
|---|---|---|---|
| AIN | Black bean | Soy flour | |
| Casein | 20 | 2.7 | — |
| Black bean flour | — | 74 | — |
| Defatted soy flour | — | — | 34 |
| Cornstarch | 45 | — | 36 |
| Sucrose | 1.8 | 1.8 | 1.8 |
| Total lipid2 | 17 | 17 | 17 |
| Total fiber in diet3 | 11 | 11 | 11 |
| Mineral mix | 3.9 | 3.9 | 3.9 |
| Vitamin mix | 1.1 | 1.1 | 1.1 |
| Methionine | 0.33 | 0.40 | 0.33 |
| Tryptophan | — | 0.004 | — |
| Calcium carbonate | 0.25 | 0.25 | 0.25 |
| Choline bitartrate | 0.28 | 0.28 | 0.28 |
| Tert-butylhydroquinone | 0.002 | 0.002 | 0.002 |
1Nutrient compositions were calculated from the USDA nutrient database and Reeves [38].
2Total lipid content in diets calculated based on natural occurring lipids and added fat. The SFA : MUFA : PUFA composition of all diets was 1 : 1.2 : 1.
3Total fiber content (11.25%) in all diets based on the amount present from individual dietary components as well as added fiber (cellulose).
2.2. Experimental Design
Upon arrival, animals were fed the control (AIN) diet and allowed one week to acclimatize to new conditions. At 4 and 5 weeks of age, rats received subcutaneous injections (100 μL) of either 15 mg/kg of azoxymethane (AOM) prepared in saline (n = 75; Ash Stevens, Detroit, MI) or saline (saline, n = 20). Animals were fed the control (AIN) diet until one week after the second injection, when they were randomized by weight to either continue on the control (AIN) diet or to be fed one of the experimental diets (BB or SF). At 36 weeks of age, animals were sacrificed by CO2 inhalation and exsanguination, and the colon was immediately excised, opened longitudinally, and rinsed briefly in tap water to remove debris. Macroscopic tumors, when present, were excised and stored at −80°C to be analyzed as a separate part of this study. The colon was then transected into proximal and distal segments and epithelial cells were collected by gently scraping normal-appearing mucosa from the distal half of the colon (excluding the lowermost 1 cm) with a glass slide. Samples were snap frozen and stored at –80°C until RNA extraction could be performed.
2.3. Microarray Target Preparation and Hybridization
Affymetrix RU34A rat genome chips (Santa Clara, CA) were used in this experiment. For total RNA isolation, the distal colonic mucosa was homogenized using a Tekmar homogenizer in TRIzol reagent containing RNase-free glycogen according to the manufacturer's instructions (Gibco, Carlsbad, CA). After RNA extraction, samples were cleaned with RNeasy minicolumns (Qiagen, Valencia, CA), quantified using a UV spectrophotometer (A 260/A 280), and the quality of RNA assessed by agarose-formaldehyde gel electrophoresis. Only high quality RNA was used in subsequent steps.
Biotinylated cRNA was prepared in accordance with instructions supplied in the GeneChip Expression Manual (Affymetrix, Santa Clara, CA). Double-stranded cDNA was synthesized from 10 μg of total RNA, pooled from 2–4 animals/treatment, using T7-(dT)24 primers containing a T7 RNA polymerase promoter site (Proligo, Boulder, CO) and the Superscript II system (Invitrogen, Carlsbad, CA). Biotinylated cRNA was prepared using the Enzo BioArray HighYield RNA Transcript Labeling Kit (Affymetrix, Santa Clara, CA) and then purified with RNeasy minicolumns. Approximately 15 μg cRNA was fragmented at 94°C for 35 minutes and hybridized to RGU34A rat genome chips for 16 hours at 45°C. Following hybridization, arrays were washed and stained with a streptavidin-phycoerythrin conjugate on an Affymetrix Fluidics station according to standard protocol. Processed arrays were scanned at 570 nm using a Hewlett Packard GeneArray Scanner.
2.4. Confirmation of Gene Changes by Quantitative Reverse Transcriptase PCR (qRT-PCR)
Select genes were confirmed using qRT-PCR. Gene-specific primers for cell division cycle 2 (Cdc2), cyclin B1 (Ccnb1), topoisomerase II alpha (Top2A), group IIA secretory phospholipase A2 (Pla2g2a), fibronectin 1 (Fn1), collagen, type I, alpha 1 (Col1a1), rat neutrophil (NP) defensin 3 (RatNP-3), aquaporin 8 (Aqp8), and 3-hydroxy-3-methylglutaryl-Coenzyme A synthase 2 (Hmgcs2) were designed with the Primer Express 2.0 program (Applied Biosystems, Foster City, CA). β-actin was used as an internal control. The sequences of the primer pairs used are available in Supplementary Table 1 available at doi:10.1155/2012/351796. Single-stranded (ss) cDNA was synthesized from 2.5 μg of total RNA using T7-(dT)24 primers (Proligo, Boulder, CO) and the Superscript II system (Invitrogen, Carlsbad, CA). Reverse transcription was performed in a thermocycler following the Superscript first strand synthesis protocol (Invitrogen, Carlsbad, California).
Quantitative determination of gene expression was performed with the ABI Prism7000 (Perkin Elmer Corp., Foster City, CA) using the SYBR Green Universal Master Mix (Applied Biosystems, Foster City, CA). The reaction mixture (25 μL total volume) contained 20 ng ss cDNA, 12.5 μL SYBR Green Universal Master mix, and 10.5 μL of diluted primers (300–600 nM). The real-time cycle conditions were as follows: PCR initial activation step at 95°C for 15 min and a total of 40 cycles for melting (95°C, 15 s) and annealing/extension (60°C, 1 min). All assays were performed in duplicate, using 3-4 samples per group (representing 9–12 animals/group) and relative fold-changes were quantified by using the comparative CT (ΔΔCT) method (User bulletin number 2, Applied Biosystems, Foster City, CA).
2.5. Statistical Analyses
Data for weight gain, microarrays, and qRT-PCR were analyzed using the General Linear Models procedure of SAS (SAS Institute, Cary, NC, Version 7.0). When statistical differences were detected with the F statistic, individual comparisons were made using the least significant difference (LSD) method. Tumor incidence data were analyzed with a χ 2 test using the Proc Freq procedure in SAS. Prior to statistical analyses, fluorescence intensity data from microarrays were globally scaled to a target intensity of 500 in Affymetrix Microarray Suite, Version 5.0 to control within-chip variations. Globally scaled data were then imported into GeneSpring (Silicon Genetics, Inc., Redwood City, CA, Version 6.0) for normalization and filtering. All chips were normalized to the median intensity of a set of invariant genes whose expression across all conditions (injection type, diet) after global scaling showed less than a 30% coefficient of variation (CV). A filtering step excluded genes not considered “Present” or “Marginal” in at least 42% of the individual samples. An additional filtering step limited the genes further to those exhibiting greater than 1.3 or less than 0.7 fold-change difference between diets or injection type. Normalized expression values were exported then analyzed using the GLM procedure of SAS. When present, duplicate transcripts were averaged prior to statistical analysis.
Differentially expressed transcripts (P < 0.05) were broadly grouped into categories based on known gene ontologies and biological functions reported in the literature. Gene ontologies were retrieved using the Affymetrix NetAffix Analysis Center (http://www.affymetrix.com/analysis/index.affx) and DAVID 6.7 functional analysis tool [39, 40]. Data are presented as mean fold-change differences standardized to the AIN (control) group for diet-dependent differences or to the saline-injected group for injection-dependent changes.
3. Results
3.1. Weight Gain and Tumor Incidence
There were no significant effects of diet (AIN, BB, SF) or injection regime (AOM versus saline) on body weight gain. The total weight gain (g, LSM ± SEM) of rats while on experimental diets was AIN = 304 ± 7.4, BB = 287 ± 8.3, SF = 299 ± 8.2. There was a significant effect of diet on tumor incidence (P = 0.03). As previously established, bean-based diets inhibited tumor incidence by ~60% compared to rats fed the control diet (Figure 1).
Figure 1.
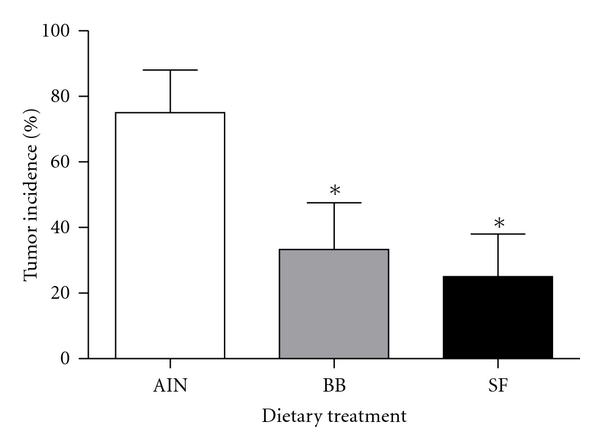
Tumor incidence in rats treated with the carcinogen azoxymethane and fed either a casein (AIN control), black bean- (BB-), or soy flour- (SF-) based diet. *Denotes significance compared to AIN controls (P < 0.05).
3.2. Biological Classification of Gene Changes in Distal Colonic Mucosa during AOM-Induced Carcinogenesis
Among the 8799 genes and ESTs present on the rat genome UG34A array, a total of 155 transcripts were significantly affected by injection regime (AOM versus saline), 257 by dietary treatment (AIN, BB, SF), and 5 were affected by both (P < 0.05). Transcripts differentially expressed by either carcinogen or diet were broadly classified into one of twelve functional categories and results are depicted in Figure 2.
Figure 2.
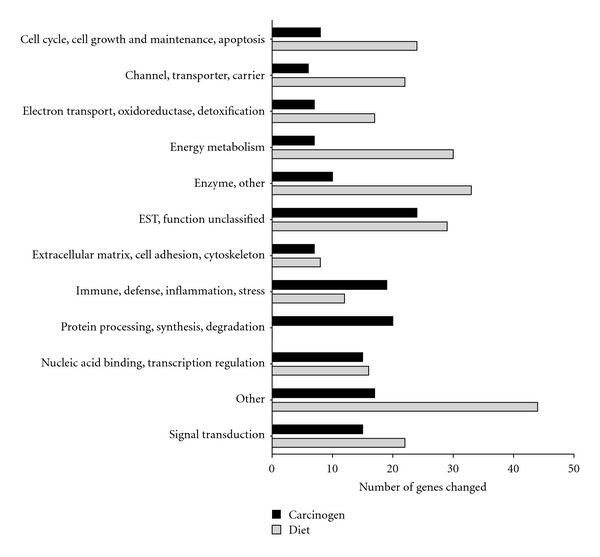
Functional classification of genes significantly altered by carcinogen (AOM) and by dietary treatment in the distal colon mucosa of rats detected by microarrays. A total of 155 genes were altered by carcinogen (AOM) and 257 by dietary treatment (AIN versus BB versus SF, P < 0.05).
3.3. Genes Differentially Expressed by Carcinogen (AOM) in Distal Colonic Mucosa
A total of 108 transcripts were higher and 47 lower in the colon of rats injected with carcinogen (AOM) compared to saline-injected controls (P < 0.05). As shown in Figure 2, a majority (55%) of transcripts affected were associated with immune, defense, inflammation (n = 19, 12%), signal transduction (n = 15, 10%), other (n = 17, 11%), or protein processing, synthesis, degradation (n = 20, 13%). Genes involved in antigen presentation (RT1-Ba, RT1-Da, RT1-Bb, RT1-DMb, RT1-M3-1, RT1-Db1, CD74), immune, defense, inflammation (RatNP-3, Lyz2, Pla2g2a), and components of the extracellular matrix (Col1a1, Fn1, Col3a1) were among those most highly induced by AOM-treatment (Table 2). Several of these genes have previously been shown to be overexpressed in carcinogen-induced colon cancer [41] and inflammatory conditions of the colon [42]. Several ribosomal proteins, including components of the 40S ribosomal protein subunit (Rps7, Rps15, Rps17, Rps9, Rps4x) and the 60S unit (Rpl37, Rpl4, Rpl36a) were also moderately induced by carcinogen compared to saline-injected controls.
Table 2.
Select genes significantly affected by carcinogen (AOM) treatment in the distal colonic epithelium of male F344 rats1.
| Gene symbol | Gene title | AOM (fold change) |
|---|---|---|
| (I) Extracellular matrix, cell adhesion, cytoskeleton | ||
|
| ||
| Fn1 | fibronectin 1 | 1.7 |
| Col1a1 | collagen, type I, alpha 1 | 1.6 |
| Vim | vimentin | 1.5 |
| Tmsb10 | thymosin, beta 10 | 1.3 |
| Col3a1 | collagen, type III, alpha 1 | 1.3 |
|
| ||
| (II) Immune, defense, inflammation, stress | ||
|
| ||
| Pla2g2a | phospholipase A2, group IIA (platelets, synovial fluid) | 2.4 |
| Cd74 | Cd74 molecule, major histocompatibility complex, class II invariant chain | 2.1 |
| Lyz2 | lysozyme 2 | 2.1 |
| RT1-Db1 | RT1 class II, locus Db1 | 2.0 |
| RT1-DMb | RT1 class II, locus DMb | 1.7 |
| RatNP-3 | defensin RatNP-3 precursor | 1.7 |
| RT1-Da | histocompatibility 2, class II antigen E alpha | 1.6 |
| RT1-Ba | RT1 class II, locus Ba | 1.5 |
| Cxcl13 | chemokine (C-X-C motif) ligand 13 | 1.5 |
| RT1-Bb | RT1 class II, locus Bb | 1.5 |
| Ccl2 | chemokine (C-C motif) ligand 2 | 1.4 |
| Mif | macrophage migration inhibitory factor | 1.3 |
| RT1-M3-1 | RT1 class Ib, locus M3, gene 1 | 1.3 |
| Irf7 | interferon regulatory factor 7 | 1.3 |
| Cxcr4 | chemokine (C-X-C motif) receptor 4 | 1.3 |
| Il15 | interleukin 15 | 0.6 |
|
| ||
| (III) Protein processing, synthesis, degradation | ||
|
| ||
| Rps7 | ribosomal protein S7 | 1.4 |
| Rps15 | ribosomal protein S15 | 1.4 |
| Pfdn2 | prefoldin subunit 2 | 1.4 |
| Hspe1 | heat shock protein 1 (chaperonin 10) | 1.3 |
| Rpl37 | ribosomal protein L37 | 1.3 |
| Rpl4 | ribosomal protein L4 | 1.3 |
| Psmb4 | proteasome (prosome, macropain) subunit, beta type 4 | 1.3 |
| Rpl3l | ribosomal protein L3-like | 1.3 |
| Rps4x | ribosomal protein S4, X-linked | 1.3 |
| Rps17 | ribosomal protein S17 | 1.3 |
| Rpl36al | ribosomal protein L36a-like | 1.3 |
| Rps9 | ribosomal protein S9 | 1.3 |
1Data expressed as mean-fold change normalized to saline-injected animals (n = 12/group). All genes presented were significantly altered compared to saline-injected animals (P < 0.05).
3.4. Genes Differentially Affected by Dietary Treatment in Distal Colonic Mucosa
Compared to the AIN diet, feeding rats black beans (BB) significantly affected 188 genes (102 upregulated, 86 downregulated), and soy flour (SF) affected 140 genes (97 upregulated, 43 downregulated). Fifty three genes were significantly coinduced and 34 corepressed by BB and SF, representing 34% of gene changes, although an additional 24% showed the same direction of change. A majority of known transcripts affected by dietary treatment (68%) fell into one of five categories including other (n = 44, 17%), enzymes (n = 33, 13%), energy metabolism (n = 30, 12%), cell cycle, cell growth and maintenance, and apoptosis (n = 24, 9.3%), channel, transporter, carrier proteins (n = 22, 8.6%), and signal transduction (n = 22, 8.6%) see Figure 2.
A select listing of transcripts similarly affected the colon of bean-fed compared to casein-fed rats is presented in Table 3. As shown, bean-based diets coinduced a number of genes involved in fatty acid metabolism and gluconeogenesis (Hmgcs2, Aldob, Pck1, Ech1), electron transport, oxidoreductase, detoxification (Cyp4b1, Gstm5, Gstm1, Cyp27a1, Prdx6), and solute, ion transport (Aqp8, Scnn1g, Slc12a7, Slc5a1, Slc16a1). Among genes corepressed by beans included those involved in cell cycle (Top2a, Ccnb1, Cdc2), fatty acid desaturation (Scd1, Scd), extracellular matrix (Col1a1, Fn1), immune, defense, inflammation (Dmbt1, Pla2g2a, RatNP-3, Tlr4, Ccxl14), and nucleic acid binding, transcription regulation (Egr1, Egr2).
Table 3.
Genes similarly affected by bean-feeding (BB and SF) in the distal colonic epithelium of male F344 rats1.
| Gene symbol | Gene title | BB | SF |
|---|---|---|---|
| (I) Cell cycle, cell growth and maintenance, apoptosis | |||
|
| |||
| Ceacam1 | carcinoembryonic antigen-related cell adhesion molecule 1 | 2.3* | 1.9* |
| Rb1 | retinoblastoma 1 | 2.2* | 2.2* |
| Gadd45a | growth arrest and DNA-damage-inducible, alpha | 1.5* | 1.5* |
| Bax | Bcl2-associated X protein | 1.3 | 1.4* |
| Egln3 | EGL nine homolog 3 (C. elegans) | 0.67* | 0.79 |
| Wfdc1 | WAP four-disulfide core domain 1 | 0.67* | 0.50* |
| Ccnb1 | cyclin B1 | 0.66* | 0.85 |
| Rfc4 | replication factor C (activator 1) 4 | 0.59* | 1.1 |
| Cdc2 | cell division cycle 2, G1 to S and G2 to M | 0.58* | 0.83 |
| Top2a | topoisomerase (DNA) II alpha | 0.57* | 0.85 |
| Bub1b | budding uninhibited by benzimidazoles 1 homolog, beta (S. cerevisiae) | 0.49* | 0.60* |
|
| |||
| (II) Channel, transporters, & carriers | |||
|
| |||
| Aqp8 | aquaporin 8 | 2.7* | 2.9* |
| Scnn1g | sodium channel, nonvoltage-gated 1 gamma | 2.2* | 2.0* |
| Apoa1 | apolipoprotein A-I | 1.6 | 2.4* |
| Slc12a7 | solute carrier family 12 (potassium/chloride transporters), member 7 | 1.6* | 1.4* |
| Slc5a1 | solute carrier family 5 (sodium/glucose cotransporter), member 1 | 1.5* | 1.3 |
| Slc16a1 | solute carrier family 16, member 1 (monocarboxylic acid transporter 1) | 1.5* | 1.5* |
| Atp1a1 | ATPase, Na+/K+ transporting, alpha 1 polypeptide | 1.4* | 1.3* |
| Lgals9 | lectin, galactoside-binding, soluble, 9 | 1.3* | 1.3* |
| Kcnk1 | potassium channel, subfamily K, member 1 | 1.3* | 1.3* |
| Slco1a1 | solute carrier organic anion transporter family, member 1a1 | 0.68 | 0.35* |
| SLC16A6 | solute carrier family 16, member 6 (monocarboxylic acid transporter 7) | 0.61* | 0.82 |
| Fabp5 | fatty acid binding protein 5, epidermal | 0.48* | 0.64* |
| Ttpa | tocopherol (alpha) transfer protein | 0.47* | 0.66* |
|
| |||
| (III) Electron transport, oxidoreductase, detoxification | |||
|
| |||
| Cyp4b1 | cytochrome P450, family 4, subfamily b, polypeptide 1 | 3.6* | 2.2* |
| Gstm5 | glutathione S-transferase, mu 5 | 1.5* | 1.5* |
| Gstm1 | glutathione S-transferase mu 1 | 1.4* | 1.3 |
| Cyp27a1 | cytochrome P450, family 27, subfamily a, polypeptide 1 | 1.4* | 1.5* |
| Prdx6 | peroxiredoxin 6 | 1.4 | 1.5* |
| Cyp2d1 | cytochrome P450, family 2, subfamily d, polypeptide 1 | 1.4* | 1.4* |
| Por | P450 (cytochrome) oxidoreductase | 1.4* | 1.4* |
| Cyb5a | cytochrome b5 type A (microsomal) | 1.4* | 1.3* |
| Tst | thiosulfate sulfurtransferase, mitochondrial | 1.3* | 1.3* |
|
| |||
| (IV) Energy metabolism | |||
|
| |||
| Hmgcs2 | 3-hydroxy-3-methylglutaryl-Coenzyme A synthase 2 | 2.2* | 2.1* |
| Aldob | aldolase B, fructose-bisphosphate | 2.1* | 1.8* |
| Pck1 | phosphoenolpyruvate carboxykinase 1 (soluble) | 2.0* | 1.8* |
| Ech1 | enoyl coenzyme A hydratase 1, peroxisomal | 1.6* | 1.6* |
| Glul | glutamate-ammonia ligase | 1.4* | 1.2* |
| Hadhb | hydroxyacyl-Coenzyme A dehydrogenase/3-ketoacyl-Coenzyme A thiolase/enoyl-Coenzyme A hydratase (trifunctional protein), beta subunit | 1.3* | 1.3* |
| Cbr1 | carbonyl reductase 1 | 1.2 | 1.5* |
| Hadha | hydroxyacyl-Coenzyme A dehydrogenase/3-ketoacyl-Coenzyme A thiolase/enoyl-Coenzyme A hydratase (trifunctional protein), alpha subunit | 1.2* | 1.3* |
| Pfkp | phosphofructokinase, platelet | 0.67* | 0.76* |
| Gpd2 | glycerol-3-phosphate dehydrogenase 2, mitochondrial | 0.67* | 0.82 |
| Acsl4 | acyl-CoA synthetase long-chain family member 4 | 0.64* | 0.80 |
| Pyy | peptide YY | 0.64* | 0.72* |
| Gcg | glucagon | 0.57* | 0.81 |
| Scd | stearoyl-CoA desaturase (delta-9-desaturase) | 0.53* | 0.72 |
| Scd1 | stearoyl-Coenzyme A desaturase 1 | 0.47* | 0.65 |
|
| |||
| (V) Extracellular matrix, cell adhesion, cytoskeleton | |||
|
| |||
| Sdc1 | syndecan 1 | 1.3* | 1.3* |
| Sparc | secreted protein, acidic, cysteine-rich (osteonectin) | 1.0 | 0.48* |
| Fn1 | fibronectin 1 | 0.66* | 0.61* |
| Tubb5 | tubulin, beta 5 | 0.65* | 0.92 |
| Col1a1 | collagen, type I, alpha 1 | 0.53* | 0.52* |
|
| |||
| (VI) Immune, defense, inflammation, stress | |||
|
| |||
| Hspa1a | heat shock 70 kD protein 1A | 1.9* | 1.4 |
| RT1-EC2 | RT1 class Ib, locus Aw2 | 1.5* | 1.4* |
| Tlr4 | toll-like receptor 4 | 0.78* | 0.68* |
| Mif | macrophage migration inhibitory factor | 0.71* | 0.88 |
| Dmbt1 | deleted in malignant brain tumors 1 | 0.64* | 0.74* |
| Pla2g2a | phospholipase A2, group IIA (platelets, synovial fluid) | 0.50* | 0.39* |
| RatNP-3 | defensin RatNP-3 precursor | 0.48* | 0.69* |
| Cxcl14 | chemokine (C-X-C motif) ligand 14 | 0.41* | 0.48* |
|
| |||
| (VII) Nucleic acid binding, transcription regulation | |||
|
| |||
| Nr1d2 | nuclear receptor subfamily 1, group D, member 2 | 2.2* | 1.3 |
| Nfib | nuclear factor I/B | 1.5* | 1.4* |
| Vdr | vitamin D (1,25- dihydroxyvitamin D3) receptor | 1.4 | 1.6* |
| Syt4 | synaptotagmin IV | 0.82 | 0.65* |
| Egr2 | early growth response 2 | 0.77* | 0.60* |
| Nr4a2 | nuclear receptor subfamily 4, group A, member 2 | 0.70* | 0.75* |
| Egr1 | early growth response 1 | 0.50* | 0.66* |
|
| |||
| (VIII) Signal transduction | |||
|
| |||
| Gchfr | GTP cyclohydrolase I feedback regulator | 1.9* | 1.8* |
| Ppm1b | protein phosphatase 1B, magnesium dependent, beta isoform | 1.6* | 1.4 |
| Mapk14 | mitogen activated protein kinase 14 | 1.3* | 1.4* |
| Gucy2c | guanylate cyclase 2C | 1.3* | 1.4* |
| P2ry2 | purinergic receptor P2Y, G-protein coupled 2 | 0.81 | 0.61* |
| Ptpn3 | protein tyrosine phosphatase, non-receptor type 3 | 0.79* | 0.73* |
| P2ry6 | pyrimidinergic receptor P2Y, G-protein coupled, 6 | 0.69* | 0.68* |
| Pld1 | phospholipase D1 | 0.68* | 0.85 |
| Ptpn18 | protein tyrosine phosphatase, non-receptor type 18 | 0.63* | 0.83* |
| Ptpro | protein tyrosine phosphatase, receptor type, O | 0.62* | 0.72* |
| Fzd1 | frizzled homolog 1 (Drosophila) | 0.62* | 0.74* |
1Data expressed as mean-fold change normalized to the AIN group (n = 8/group). *denotes significant difference compared to the control (AIN) diet (P < 0.05).
The relative expression of select genes (Cdc2, Ccnb1, Top2a, Hmgcs2, and Aqp8) was further evaluated by qRT-PCR. As shown in Figure 3, the mRNA for Cdc2, Ccnb1, Top2a were all significantly lower, whereas Hmgcs2 and Aqp8 were higher in the colon of BB- and SF-fed rats compared to controls (AIN P < 0.05). The direction of change was generally consistent with those obtained using microarrays (Table 3).
Figure 3.
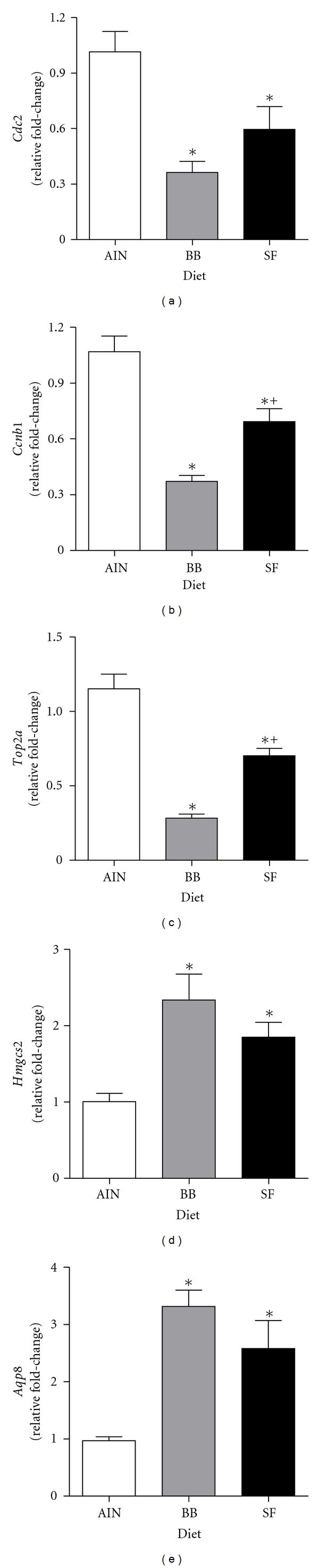
qRT-PCR analysis of genes for (a) Cdc2, (b) Ccnb1, (c) Top2a, (d) Hmgcs2, and (e) Aqp8 in the distal colonic mucosa of rats fed either an AIN (control), black bean- (BB-), or soy flour- (SF-) based diet. Results were normalized to the housekeeping gene β-Actin and are presented as relative fold-changes (LSM ± SEM) standardized to the AIN (control) diet. *Denotes significance compared to AIN controls; +denotes significance between BB and SF-fed animals (P < 0.05). Abbreviations: Cdc2, cell division cycle 2; Ccnb1, cyclin B1; Top2a, topoisomerase II alpha; Hmgcs2, 3-hydroxy-3-methylglutaryl-Coenzyme A synthase 2; Aqp8, Aquaporin 8.
3.5. Genes Affected by Both Dietary Treatment and Carcinogen (AOM) in Distal Colonic Mucosa
Five genes were found to be influenced both by diet and carcinogen and are presented in Table 4. Transcripts for phospholipase A2, group IIA (Pla2g2a), rat neutrophil defensin 3 (RatNP-3), collagen, type I, alpha 1 (Col1a1), and fibronectin 1 (Fn1) were all induced by carcinogen treatment, but to a lower extent in rats fed either BB or SF. Changes in expression of these genes were further evaluated by qRT-PCR and results presented in Figure 4. In accordance with microarray data, there were significant main effects for both diet (P < 0.05) and carcinogen (P < 0.05) treatment for each gene examined. Generally, expression was lowest in bean-fed rats but increased in all diets with carcinogen injections. Rats injected with AOM and fed the control (AIN) diet had the highest overall expression level coinciding with the higher tumor incidence observed in these animals. Somatostatin 2 receptor was also influenced by both diet and carcinogen treatment, being basally higher in bean-fed animals and decreasing in all groups following carcinogen administration (Table 4).
Table 4.
Genes significantly affected both by carcinogen (AOM) and dietary treatment in the distal colonic epithelium of male F344 rats1.
| Gene symbol | Gene title | Saline-treated | AOM-treated | ||||
|---|---|---|---|---|---|---|---|
| AIN | BB | SF | AIN | BB | SF | ||
| Pla2g2a | phospholipase A2, group IIA | 1.0 | 0.64 | 0.33 | 2.5 | 0.97 | 1.2 |
| RatNP-3 | NP defensin 3 | 1.0 | 0.44 | 0.70 | 1.7 | 0.78 | 1.0 |
| Col1a1 | collagen, type I, alpha 1 | 1.0 | 0.67 | 0.59 | 2.3 | 0.94 | 0.97 |
| Fn1 | fibronectin 1 | 1.0 | 0.72 | 0.77 | 2.0 | 1.2 | 1.0 |
| Sstr2 | somatostatin receptor 2 | 1.0 | 1.4 | 1.7 | 0.69 | 1.1 | 1.2 |
1Data expressed as mean fold-change differences standardized to the AIN (saline-injected) group (n = 4/group). There were significant main effects for injection type (saline versus AOM) and dietary treatment (AIN versus BB or SF) for each gene listed (P < 0.05).
Figure 4.
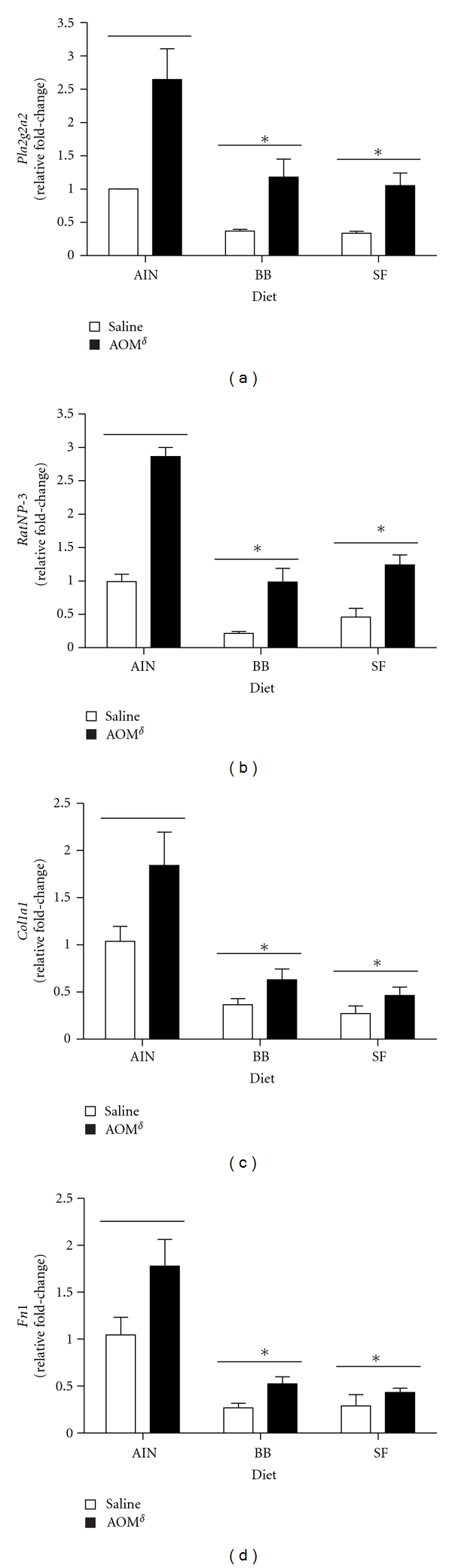
Relative fold-changes in (a) Pla2g2a, (b) RatNP-3, (c) Col1a1, and (d) Fn1 detected with qRT-PCR. Results were normalized to the housekeeping gene β-Actin and are presented as mean fold-changes (LSM ± SEM) relative to the AIN(saline-injected) group. There were significant main effects for both diet and carcinogen for each gene presented (P < 0.05). *Denotes significant effect of diet compared to AIN controls; δ denotes significant effect of carcinogen treatment compared to saline controls (P < 0.05). Abbreviations: BB, black bean; SF, soy flour; Pla2g2a, phospholipase A2, group IIA; RatNP-3, rat neutrophil defensin 3; Col1a1, collagen, type I, alpha 1; Fn1, fibronectin 1.
4. Discussion
The focus of the current research was to identify potential cellular and molecular events underlying suppression of tumorigenesis by beans using a highly relevant animal model of colon cancer. We profiled global changes in gene expression affected by AOM treatment in normal-appearing colon mucosa to determine early events permissive for tumor formation and whether these changes could be attenuated by dietary treatment. Although not a primary focus of this study, tumor incidence was also assessed. As previously demonstrated, both BB- [32] and SF-fed [30, 31, 43] rats developed significantly fewer tumors overall, confirming that these diets inhibit experimental colon carcinogenesis. In the study by Hangen and Bennink [32], however, black and navy bean- fed-rats ate less and as a result had significantly lower body weights at termination of the study. Because of the inverse association between energy restriction and tumorigenesis, the tryptophan and methionine content in the black bean diets was adjusted to raise the amino acid score comparable to that of the AIN and SF diets. As a result, no significant differences in final weight gain were detected, indicating that black beans inhibit tumorigenesis by a mechanism other than energy restriction.
We found that AOM treatment most notably affected genes involved in innate defense and immunity. For example, the antimicrobial genes lysozyme, group IIA phospholipase A2 (Pla2g2a; sPLA2), and neutrophil (NP) defensin 3 (RatNP-3) were approximately 2-fold higher than in saline-injected controls. Several major histocompatibility class (MHC) II-associated antigens as well as CD74, the class II MHC-associated invariant chain, were also induced in the colon of AOM-injected rats. Epithelial cells, activated dendritic cells, and/or macrophages underlying the intestinal cell layer can function as antigen-presenting cells [44–48], and specific upregulation of these genes implies that carcinogen treatment alters immune responsiveness to luminal and/or bacterial antigens. Additionally, CD74, aside from its classical antigen transporting role, has also been reported to bind macrophage inhibitory factor (MIF), leading to nuclear factor kappa B (NFκB) activation, inhibition of p53 phosphorylation, and cell proliferation [49, 50]. Another cluster of genes influenced by AOM treatment involved moderate induction of several ribosomal proteins. RNA and protein synthesis decrease as cells terminally differentiate [51], and enhanced presence of ribosomal protein transcripts is consistent with findings from other studies in the colon of rats susceptible to PhIP-induced colon cancer [52] and in animals during aging [53].
Chronic inflammation creates an environment permissive to carcinogenesis through enhanced production of lipid mediators, cytokines, and chemokines that influence cell proliferation and apoptosis [54–58]. Additionally generation of reactive oxygen and nitrogen species either directly or through activation of phagocytic cells can lead to oxidative damage of DNA [55, 59, 60]. The mucosal barrier, comprised of goblet cell-derived mucin and reinforced by tight junctions normally protects the epithelium and limits activation of immune cells within the lamina propria [61–64]. Dysfunctions in one or more components of the mucosal barrier have been implicated in the pathogenesis of inflammatory bowel diseases (IBD) [65–67] as well as in CRC [68–72]. For example, increased intestinal permeability and altered structure of tight junction proteins precedes relapse in individuals with IBD [73]. Additionally, Soler et al. [70] noted defects in tight-junction permeability in normal mucosa and in colon tumors from carcinogen-treated animals, and the surface epithelial cells of aberrant crypt foci are reported to be deficient in mature goblet cells, have altered mucin composition, and contain irregular microvilli [74]. Alterations in one or more components of the mucosal barrier would support our findings of higher immune responsive genes observed in carcinogen treated animals and is likely an early and permissive event in promotion of colon carcinogenesis.
Because both black beans and soy flour reduced tumorigenesis to a similar extent in this study, we next evaluated genes that were either co-induced or suppressed compared to AIN-fed animals. We identified several transcripts associated with proliferation and apoptosis to be regulated in a diet-dependent manner. An interesting finding was a 2-fold higher expression of Ceacam1 in rats fed either BB or SF. Ceacam1 encodes a cell-surface glycoprotein expressed in the differentiated cell compartment of colonic crypts and expression correlates positively with normal rates of apoptosis [75, 76]. A tumor suppressive function for Ceacam1 has been suggested due to loss of expression in hyperplastic polyps, adenomas, as well as in human colon cancers that precedes defects in the APC pathway [75]. Somatostatin receptor 2 (Sst2), which mediates anti-proliferative responses to the hormone somatostatin [77, 78], was also more abundant in bean-fed animals and levels tended to decrease following AOM treatment. These results are consistent with decreased expression of sst2 in human colon tumors [79] and with findings from Xiao et al. [80] who demonstrated enhanced colonic mRNA and serum protein levels of somatostatin in rats fed either whey or soy protein isolate. Bean-fed rats also exhibited a lower expression of the mitotic genes Ccnb1, Cdc2, and Top2a. Top2a is involved in a variety of processes including DNA replication, chromosome segregation, and maintenance of chromosome structure [81]. Protein expression has been detected primarily in the actively proliferating cells at the base of the crypt and levels increase during tumorigenesis [82]. Binding of cyclin B1 to Cdc2 is required for cells to enter mitosis at the G2 checkpoint [83, 84]. Although these genes can be regulated at the transcriptional level in a p53-dependent manner [84], the fold change differences between diets would more likely suggest an increase in the proportion of cells undergoing terminal differentiation. This is further supported by the anatomical distribution of Topoisomerase II alpha and CEACAM1 along the crypt-lumen axis and implies a general effect of bean-feeding on maintaining normal crypt cell homeostasis.
Other clusters of genes similarly affected by bean-feeding highlight differences in fiber sources on colon cell physiology. For example, the most highly induced class of genes affected by bean diets included those involved in water channel and ion transport (Aqp8, Scnn1g, Slc12a7, Tfrc, Slc5a1, Slc16a1) and energy metabolism (Hmgcs2, Aldob, Pck1, Hadhb, Ech1). These changes are consistent with the physiological effects of fermentable fibers on intestinal function. Bacterial fermentation of dietary fibers and resistant starch produces the short-chain fatty acids (SCFA), acetate, propionate, and butyrate [85, 86]. SCFAs are trophic to the normal colonic epithelium, enhance water and sodium absorption, increase mucosal blood flow, and modulate enterohormone release [87, 88]. Butyrate in particular is an important energy source for colonocytes and can induce growth arrest, differentiation, and/or apoptosis of colon epithelial cells in vitro [89, 90]. Augenlicht et al. [90, 91] demonstrated that butyrate metabolism, through mitochondrial β-oxidation, is important for induction of apoptosis both in vitro and in vivo. Dsyregulation of genes involved in energy metabolism, such as Hmgcs2, have been observed in inflammatory conditions of the colon [92] and during tumorigenesis [42, 93], suggesting a potential link between enhanced colonic expression and reduced cancer risk.
Despite the large number of genes influenced by carcinogen or diet alone, only 5 transcripts were significantly affected by both treatments. It was originally hypothesized that gene changes within this group would be the most important to understanding dietary modulation of tumorigenesis, with particular interest in those that paralleled tumor incidence data. We identified that transcripts for antimicrobial genes (Pla2g2a, RatNP-3) and extracellular matrix components (Col1a1, Fn1) were induced by AOM treatment in all diets, but to a much lesser extent in bean-fed animals, whereas somatostatin receptor 2 (Sstr2) showed the opposite trend. sPLA2 (Pla2g2a) and NP defensin 3 (RatNP-3) exhibit antimicrobial activity and together with other proteins play an important role in mucosal epithelial defense [94–100]. sPLA2 is a multifunctional protein induced in a variety of inflammatory and neoplastic conditions [101–107]. Enhanced expression has been reported in colon tumors of patients with familial adenomatous polyposis [108], in areas adjacent to sporadic colon tumors [109], in inflammatory bowel disease [101, 104, 105, 110], as well as in carcinogen-induced tumors in rodents [41, 111], suggesting a promoting role in colonic neoplasia. Aside from antimicrobial activity [95], sPLA2 may coordinate immune defenses by enhancing neutrophil function [112] and contributing to eicosanoid synthesis [103, 113, 114]. Similar to sPLA2, NP defensin 3 exhibits antimicrobial activity as well as other immune modulatory roles [99, 115, 116]. Normal colonic expression of alpha defensins is low [96, 97], but expression is induced during inflammation [96, 97, 117, 118], potentially by the proinflammatory cytokines IL-1β, TNF-α, and IL-6 [119]. Recently, increased levels of human neutrophil (HNP) defensins 1–3 were identified in individuals with colorectal tumors [120–122] as well as in the serum and colon from patients with active IBD [117], indicating that alpha defensins may be a plausible and early biomarker for gastrointestinal disease. Dietary modulation of these genes in addition to a lower expression of other innate immune genes, including toll-like receptor 4 (Tlr4) and Dmbt1, further suggest bean feeding inhibits tumor promotion by limiting microbially induced inflammation.
Alterations in extracellular matrix (ECM) components are commonly observed during inflammation and carcinogenesis [123–126] and contribute to number of processes including cell adhesion and migration, wound healing, angiogenesis, and immune cell migration and activation [123, 127]. Additionally, changes in the distribution and expression of different ECM proteins have also been reported along the crypt axis [128–130] suggesting a role in normal epithelial migration and differentiation [130–132]. Col1a1 expression is upregulated in colon cancer and in other hyperproliferative disorders [133], but normal colonic expression has not been previously reported. Fibronectin has been identified as a downstream target of the Wnt/β-catenin pathway, which is frequently altered in colorectal carcinogenesis [134, 135]. Additionally, Kolachala et al. [136] reported expression is localized to surface epithelial cells and protein levels increase during the active and restitution phase of dextran sodium-induced colitis in mice. This was associated with induction of the α5-integrin receptor, increased cell attachment, and activation of the NFκB signaling pathway. Although important in restitution to injury, several bacteria also contain binding sites for fibronectin, and increased apical secretion may influence adherence to mucosal surfaces, thereby potentiating inflammation [137–140]. The lower expression of these genes in bean-fed animals treated with carcinogen may represent fewer preneoplastic lesions and/or a more general effect of diet on maintaining mucosal integrity.
Our findings provide a strong basis for future studies on legumes and colon cancer prevention, however, there are a few limitations to the current study. First, although microarrays are a powerful tool for biomedical research, one limitation to this technology is decreased sensitivity for the detection of low-abundance genes [141]. Additionally, differences in gene expression may not directly reflect changes in protein levels nor account for other posttranslational modifications such as protein phosphorylation that may also be involved in dietary modulation of colon cancer [141]. Additional studies should be undertaken to address the functional involvement of gene changes and corresponding proteins from this study to further confirm postulated roles in chemoprevention. Another potential limitation was the use of colonic mucosal scrapings rather than whole colon tissue. Mucosal scrapings yield a heterogeneous cell population consisting primarily of colonocytes, with lesser amounts of intraepithelial lymphocytes, macrophages, and endothelial cells. Although cancer evolves from genetic and epigenetic alterations arising in epithelial cells, there is increasing recognition for the importance of the microenvironment in the carcinogenic process [142]. For example, infiltrating immune cells in the lamina propria may contribute to tumor development through generation of reactive oxygen species [143, 144] as well as local production of cytokines, chemokines, and other lipid mediators which can influence carcinogenesis by promoting angiogenesis and disrupting cell cycle regulation [142]. Although our primary interests were in epithelial gene changes, gene expression patterns in other cell types were likely underrepresented and should be considered in future studies to more fully understand the impact of legumes on colon cancer development.
5. Conclusions
In summary, dietary habits are strongly associated with colon cancer risk, and this research lends further support to epidemiological and experimental data that consumption of bean-based diets inhibits colon cancer development. The finding that beans reduce markers of colonic inflammation is consistent with the inverse association of long-term non-steroidal anti-inflammatory drug (NSAID) use on CRC risk [145, 146]. Further, it is well recognized that individuals previously treated for colon cancer are at a higher risk of recurrence of the disease [147]. This observation is proposed to be related to molecular abnormalities in areas surrounding cancer tissue [148, 149]. Results from this study suggest that some abnormalities may be related to changes in cytokinetics, the innate immune system, and extracellular matrix components. We speculate dietary modulation of these genes is associated with reduced inflammation, possibly through enhancing mucosal barrier function. Confirming some of these gene changes in humans, further identifying what causes these changes to occur and determining if bean consumption can reverse these changes would strengthen the relationship of bean consumption on colon cancer inhibition and may provide useful adjunct therapy for those at risk.
Supplementary Material
The sequences of the primer pairs used to confirm gene changes with qRT-PCR. Gene specific primers for cell division cycle 2 (Cdc2), cyclin B1 (Ccnb1), topoisomerase II alpha (Top2A), group IIA secretory phospholipase A2 (Pla2g2a), fibronectin 1 (Fn1), collagen, type I, alpha 1 (Col1a1), rat neutrophil (NP) defensin 3 (RatNP-3), aquaporin 8 (Aqp8), and 3-hydroxy-3-methylglutaryl-Coenzyme A synthase 2 (Hmgcs2) were designed with the Primer Express 2.0 program (Applied Biosystems, Foster City, CA). β-actin was used as an internal control.
Conflict of Interests
The authors declare no conflict of interests.
Acknowledgments
Financial support was provided by the United States Agency for International Development through the Dry Grain Pulse Collaborative Research Support Program and the Michigan State University AgBioResearch Unit. The authors would like to sincerely thank Annette Thelen and the Genomics Technology Support Facility at Michigan State University for technical assistance and support with microarray hybridization and processing.
References
- 1.Ferlay J, Shin H, Bray F, et al. GLOBOCAN 2008, Cancer Incidence and Mortality Worldwide. IARC CancerBase No. 10 [Internet] Lyon, France, 2010.
- 2.Armstrong B, Doll R. Environmental factors and cancer incidence and mortality in different countries, with special reference to dietary practices. International Journal of Cancer. 1975;15(4):617–631. doi: 10.1002/ijc.2910150411. [DOI] [PubMed] [Google Scholar]
- 3.Doll R, Peto R. The causes of cancer: quantitative estimates of avoidable risks of cancer in the United States today. Journal of the National Cancer Institute. 1981;66(6):1191–1308. [PubMed] [Google Scholar]
- 4.WCRF. Food, Nutrition, Physical Activity, and the Prevention of Cancer: A Global Perspective. Washington, DC, USA: World Cancer Research Fund & American Institute for Cancer Research; 2007. [Google Scholar]
- 5.Potter JD. Colorectal cancer: molecules and populations. Journal of the National Cancer Institute. 1999;91(11):916–932. doi: 10.1093/jnci/91.11.916. [DOI] [PubMed] [Google Scholar]
- 6.Steinmetz KA, Potter JD. Food-group consumption and colon cancer in the Adelaide case-control study. I. Vegetables and fruit. International Journal of Cancer. 1993;53(5):711–719. doi: 10.1002/ijc.2910530502. [DOI] [PubMed] [Google Scholar]
- 7.Fraser GE. Associations between diet and cancer, ischemic heart disease, and all- cause mortality in non-Hispanic white California Seventh-day Adventists. American Journal of Clinical Nutrition. 1999;70(3) doi: 10.1093/ajcn/70.3.532s. [DOI] [PubMed] [Google Scholar]
- 8.Singh PN, Fraser GE. Dietary risk factors for colon cancer in a low-risk population. American Journal of Epidemiology. 1998;148(8):761–774. doi: 10.1093/oxfordjournals.aje.a009697. [DOI] [PubMed] [Google Scholar]
- 9.Marchand LL, Hankin JH, Wilkens LR, Kolonel LN, Englyst HN, Lyu LC. Dietary fiber and colorectal cancer risk. Epidemiology. 1997;8(6):658–665. doi: 10.1097/00001648-199710000-00008. [DOI] [PubMed] [Google Scholar]
- 10.Deneo-Pellegrini H, Boffetta P, De Stefani E, Ronco A, Brennan P, Mendilaharsu M. Plant foods and differences between colon and rectal cancers. European Journal of Cancer Prevention. 2002;11(4):369–375. doi: 10.1097/00008469-200208000-00009. [DOI] [PubMed] [Google Scholar]
- 11.Lanza E, Hartman TJ, Albert PS, et al. High dry bean intake and reduced risk of advanced colorectal adenoma recurrence among participants in the polyp prevention trial. Journal of Nutrition. 2006;136(7):1896–1903. doi: 10.1093/jn/136.7.1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Witte JS, Longnecker MP, Bird CL, Lee ER, Frankl HD, Haile RW. Relation of vegetable, fruit, and grain consumption to colorectal adenomatous polyps. American Journal of Epidemiology. 1996;144(11):1015–1025. doi: 10.1093/oxfordjournals.aje.a008872. [DOI] [PubMed] [Google Scholar]
- 13.Correa P. Epidemiological correlations between diet and cancer frequency. Cancer Research. 1981;41(9):3685–3690. [PubMed] [Google Scholar]
- 14.Jacoby RF, Llor X, Teng BB, Davidson NO, Brasitus TA. Mutations in the K-ras oncogene induced by 1,2-dimethylhydrazine in preneoplastic and neoplastic rat colonic mucosa. Journal of Clinical Investigation. 1991;87(2):624–630. doi: 10.1172/JCI115039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vivona AA, Shpitz B, Medline A, et al. K-ras mutations in aberrant crypt foci, adenomas and adenocarcinomas during azoxymethane-induced colon carcinogenesis. Carcinogenesis. 1993;14(9):1777–1781. doi: 10.1093/carcin/14.9.1777. [DOI] [PubMed] [Google Scholar]
- 16.Pories SE, Ramchurren N, Summerhayes I, Steele G. Animal models for colon carcinogenesis. Archives of Surgery. 1993;128(6):647–653. doi: 10.1001/archsurg.1993.01420180045009. [DOI] [PubMed] [Google Scholar]
- 17.Takahashi M, Fukuda K, Sugimura T, Wakabayashi K. β-catenin is frequently mutated and demonstrates altered cellular location in azoxymethane-induced rat colon tumors. Cancer Research. 1998;58(1):42–46. [PubMed] [Google Scholar]
- 18.Perše M, Cerar A. Morphological and molecular alterations in 1,2 dimethylhydrazine and azoxymethane induced colon carcinogenesis in rats. Journal of Biomedicine and Biotechnology. 2011;2011 doi: 10.1155/2011/473964. Article ID 473964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen J, Huang XF. The signal pathways in azoxymethane-induced colon cancer and preventive implications. Cancer Biology & Therapy. 2009;8(14):1313–1317. doi: 10.4161/cbt.8.14.8983. [DOI] [PubMed] [Google Scholar]
- 20.De Filippo C, Caderni G, Bazzicalupo M, et al. Mutations of the Apc gene in experimental colorectal carcinogenesis induced by azoxymethane in F344 rats. British Journal of Cancer. 1998;77(12):2148–2151. doi: 10.1038/bjc.1998.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takahashi M, Mutoh M, Kawamori T, Sugimura T, Wakabayashi K. Altered expression of β-catenin, inducible nitric oxide synthase and cyclooxygenase-2 in azoxymethane-induced rat colon carcinogenesis. Carcinogenesis. 2000;21(7):1319–1327. [PubMed] [Google Scholar]
- 22.Yamada Y, Yoshimi N, Hirose Y, et al. Frequent β-catenin gene mutations and accumulations of the protein in the putative preneoplastic lesions lacking macroscopic aberrant crypt foci appearance, in rat colon carcinogenesis. Cancer Research. 2000;60(13):3323–3327. [PubMed] [Google Scholar]
- 23.Takahashi M, Nakatsugi S, Sugimura T, Wakabayashi K. Frequent mutations of the β-catenin gene in mouse colon tumors induced by azoxymethane. Carcinogenesis. 2000;21(6):1117–1120. [PubMed] [Google Scholar]
- 24.DuBois RN, Radhika A, Reddy BS, Entingh AJ. Increased cyclooxygenase-2 levels in carcinogen-induced rat colonic tumors. Gastroenterology. 1996;110(4):1259–1262. doi: 10.1053/gast.1996.v110.pm8613017. [DOI] [PubMed] [Google Scholar]
- 25.Ohta T, Takahashi M, Ochiai A. Increased protein expression of both inducible nitric oxide synthase and cyclooxygenase-2 in human colon cancers. Cancer Letters. 2006;239(2):246–253. doi: 10.1016/j.canlet.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 26.Watanabe K, Kawamori T, Nakatsugi S, Wakabayashi K. Cox-2 and inos, good targets for chemoprevention of colon cancer. BioFactors. 2000;12(1–4):129–133. doi: 10.1002/biof.5520120120. [DOI] [PubMed] [Google Scholar]
- 27.Guda K, Giardina C, Nambiar P, Cui H, Rosenberg DW. Aberrant transforming growth factor-β signaling in azoxymethane-induced mouse colon tumors. Molecular Carcinogenesis. 2001;31(4):204–213. doi: 10.1002/mc.1055. [DOI] [PubMed] [Google Scholar]
- 28.Shao J, Sheng H, Aramandla R, et al. Coordinate regulation of cyclooxygenase-2 and TGF-β1 in replication error-positive colon cancer and azoxymethane-induced rat colonic tumors. Carcinogenesis. 1999;20(2):185–191. doi: 10.1093/carcin/20.2.185. [DOI] [PubMed] [Google Scholar]
- 29.Bellam N, Pasche B. Tgf-beta signaling alterations and colon cancer. Cancer Treatment and Research. 2010;155:85–103. doi: 10.1007/978-1-4419-6033-7_5. [DOI] [PubMed] [Google Scholar]
- 30.Bennink MR, Om AS. Inhibition of Colon Cancer (CC) by soy phytochemicals but not by soy protein. The FASEB Journal. 1998;12(5):p. A655. [Google Scholar]
- 31.Bennink MR, Om AS, Miyagi Y. Inhibition of colon cancer (CC) by soy flour but not by genistin or a mixture of isoflavones. The FASEB Journal. 1999;13:p. A50. [Google Scholar]
- 32.Hangen L, Bennink MR. Consumption of black beans and navy beans (Phaseolus vulgaris) reduced azoxymethane-induced colon cancer in rats. Nutrition and Cancer. 2002;44(1):60–65. doi: 10.1207/S15327914NC441_8. [DOI] [PubMed] [Google Scholar]
- 33.Rondini EA, Bennink MR. Defatted soy flour, but not soy concentrate, reduces azoxymethane-induced colon carcinogenesis. Journal of Nutrition. 2002;132:p. 589S. [Google Scholar]
- 34.Hakkak R, Korourian S, Ronis MJJ, Johnston JM, Badger TM. Soy protein isolate consumption protects against azoxymethane-induced colon tumors in male rats. Cancer Letters. 2001;166(1):27–32. doi: 10.1016/s0304-3835(01)00441-4. [DOI] [PubMed] [Google Scholar]
- 35.Hughes JS, Ganthavorn C, Wilson-Sanders S. Dry beans inhibit azoxymethane-induced colon carcinogenesis in F344 rats. Journal of Nutrition. 1997;127(12):2328–2333. doi: 10.1093/jn/127.12.2328. [DOI] [PubMed] [Google Scholar]
- 36.Bobe G, Barrett KG, Mentor-Marcel RA, et al. Dietary cooked navy beans and their fractions attenuate colon carcinogenesis in azoxymethane-induced Ob/Ob mice. Nutrition and Cancer. 2008;60(3):373–381. doi: 10.1080/01635580701775142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu T, Mokuolu AO, Rao CV, Reddy BS, Holt PR. Regional chemoprevention of carcinogen-induced tumors in rat colon. Gastroenterology. 1995;109(4):1167–1172. doi: 10.1016/0016-5085(95)90575-8. [DOI] [PubMed] [Google Scholar]
- 38.Reeves PG, Nielsen FH, Fahey GC. AIN-93 purified diets for laboratory rodents: final report of the American Institute of Nutrition ad hoc writing committee on the reformulation of the AIN-76A rodent diet. Journal of Nutrition. 1993;123(11):1939–1951. doi: 10.1093/jn/123.11.1939. [DOI] [PubMed] [Google Scholar]
- 39.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 40.Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Research. 2009;37(1):1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fujiwara K, Ochiai M, Ohta T, et al. Global gene expression analysis of rat colon cancers induced by a food-borne carcinogen, 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine. Carcinogenesis. 2004;25(8):1495–1505. doi: 10.1093/carcin/bgh155. [DOI] [PubMed] [Google Scholar]
- 42.Lawrance IC, Fiocchi C, Chakravarti S. Ulcerative colitis and Crohn’s disease: distinctive gene expression profiles and novel susceptibility candidate genes. Human Molecular Genetics. 2001;10(5):445–456. doi: 10.1093/hmg/10.5.445. [DOI] [PubMed] [Google Scholar]
- 43.Bennink MR. Dietary soy reduces colon carcinogenesis in human and rats: soy and colon cancer. Advances in Experimental Medicine and Biology. 2001;492:11–17. doi: 10.1007/978-1-4615-1283-7_2. [DOI] [PubMed] [Google Scholar]
- 44.Nagashima R, Maeda K, Imai Y, Takahashi T. Lamina propria macrophages in the human gastrointestinal mucosa: their distribution, immunohistological phenotype, and function. Journal of Histochemistry and Cytochemistry. 1996;44(7):721–731. doi: 10.1177/44.7.8675993. [DOI] [PubMed] [Google Scholar]
- 45.Maric I, Holt PG, Perdue MH, Bienenstock J. Class II MHC antigen (Ia)-bearing dendritic cells in the epithelium of the rat intestine. Journal of Immunology. 1996;156(4):1408–1414. [PubMed] [Google Scholar]
- 46.Baumgart DC, Carding SR. Inflammatory bowel disease: cause and immunobiology. The Lancet. 2007;369(9573):1627–1640. doi: 10.1016/S0140-6736(07)60750-8. [DOI] [PubMed] [Google Scholar]
- 47.Mowat AM, Viney JL. The anatomical basis of intestinal immunity. Immunological Reviews. 1997;156:145–166. doi: 10.1111/j.1600-065x.1997.tb00966.x. [DOI] [PubMed] [Google Scholar]
- 48.Tlaskalová-Hogenová H, Farré-Castany MA, Štěpánková R, et al. The gut as a lymphoepithelial organ: the role of intestinal epithelial cells in mucosal immunity. Folia Microbiologica. 1995;40(4):385–391. doi: 10.1007/BF02814746. [DOI] [PubMed] [Google Scholar]
- 49.Lue H, Thiele M, Franz J, et al. Macrophage migration inhibitory factor (MIF) promotes cell survival by activation of the Akt pathway and role for CSN5/JAB1 in the control of autocrine MIF activity. Oncogene. 2007;26(35):5046–5059. doi: 10.1038/sj.onc.1210318. [DOI] [PubMed] [Google Scholar]
- 50.Leng L, Metz CN, Fang Y, et al. MIF signal transduction initiated by binding to CD74. Journal of Experimental Medicine. 2003;197(11):1467–1476. doi: 10.1084/jem.20030286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Deschner EE, Lipkin M. Study of human rectal epithelial cells in vitro. III. RNA, protein, and DNA synthesis in polyps and adjacent mucosa. Journal of the National Cancer Institute. 1970;44(1):175–185. [PubMed] [Google Scholar]
- 52.Fujiwara K, Ochiai M, Ubagai T, et al. Differential gene expression profiles in colon epithelium of two rat strains with distinct susceptibility to colon carcinogenesis after exposure to PhIP in combination with dietary high fat. Cancer Science. 2003;94(8):672–678. doi: 10.1111/j.1349-7006.2003.tb01501.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee HM, Greeley GH, Englander EW. Age-associated changes in gene expression patterns in the duodenum and colon of rats. Mechanisms of Ageing and Development. 2001;122(4):355–371. doi: 10.1016/s0047-6374(00)00254-2. [DOI] [PubMed] [Google Scholar]
- 54.Khansari N, Shakiba Y, Mahmoudi M. Chronic inflammation and oxidative stress as a major cause of age-related diseases and cancer. Recent Patents on Inflammation and Allergy Drug Discovery. 2009;3(1):73–80. doi: 10.2174/187221309787158371. [DOI] [PubMed] [Google Scholar]
- 55.Federico A, Morgillo F, Tuccillo C, Ciardiello F, Loguercio C. Chronic inflammation and oxidative stress in human carcinogenesis. International Journal of Cancer. 2007;121(11):2381–2386. doi: 10.1002/ijc.23192. [DOI] [PubMed] [Google Scholar]
- 56.Macarthur M, Hold GL, El-Omar EM. Inflammation and Cancer II. Role of chronic inflammation and cytokine gene polymorphisms in the pathogenesis of gastrointestinal malignancy. American Journal of Physiology. 2004;286(4):G515–G520. doi: 10.1152/ajpgi.00475.2003. [DOI] [PubMed] [Google Scholar]
- 57.Van Der Woude CJ, Kleibeuker JH, Jansen PLM, Moshage H. Chronic inflammation, apoptosis and (pre-)malignant lesions in the gastro-intestinal tract. Apoptosis. 2004;9(2):123–130. doi: 10.1023/B:APPT.0000018794.26438.22. [DOI] [PubMed] [Google Scholar]
- 58.Shacter E, Weitzman SA. Chronic inflammation and cancer. Oncology. 2002;16(2):217–230. [PubMed] [Google Scholar]
- 59.Bartsch H, Nair J. Chronic inflammation and oxidative stress in the genesis and perpetuation of cancer: role of lipid peroxidation, DNA damage, and repair. Langenbeck’s Archives of Surgery. 2006;391(5):499–510. doi: 10.1007/s00423-006-0073-1. [DOI] [PubMed] [Google Scholar]
- 60.Grisham MB, Jourd'heuil D, Wink DA. Chronic inflammation and reactive oxygen and nitrogen metabolism—implications in DNA damage and mutagenesis. Alimentary Pharmacology and Therapeutics. 2000;14(supplement 1):3–9. doi: 10.1046/j.1365-2036.2000.014s1003.x. [DOI] [PubMed] [Google Scholar]
- 61.Podolsky DK. Mucosal immunity and inflammation V. Innate mechanisms of mucosal defense and repair: the best offense is a good defense. American Journal of Physiology. 1999;277(3):G495–G499. doi: 10.1152/ajpgi.1999.277.3.G495. [DOI] [PubMed] [Google Scholar]
- 62.McGuckin MA, Lindén SK, Sutton P, Florin TH. Mucin dynamics and enteric pathogens. Nature Reviews Microbiology. 2011;9(4):265–278. doi: 10.1038/nrmicro2538. [DOI] [PubMed] [Google Scholar]
- 63.Turner JR. Molecular basis of epithelial barrier regulation: from basic mechanisms to clinical application. American Journal of Pathology. 2006;169(6):1901–1909. doi: 10.2353/ajpath.2006.060681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Turner JR. Intestinal mucosal barrier function in health and disease. Nature Reviews Immunology. 2009;9(11):799–809. doi: 10.1038/nri2653. [DOI] [PubMed] [Google Scholar]
- 65.Tang Y, Clayburgh DR, Mittal N, et al. Epithelial NF-κB enhances transmucosal fluid movement by altering tight junction protein composition after T cell activation. American Journal of Pathology. 2010;176(1):158–167. doi: 10.2353/ajpath.2010.090548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Matricon J, Barnich N, Ardid D. Immunopathogenesis of inflammatory bowel disease. Self/Nonself Immune Recognition and Signaling. 2010;1(4):299–309. doi: 10.4161/self.1.4.13560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Salim SY, Söderholm JD. Importance of disrupted intestinal barrier in inflammatory bowel diseases. Inflammatory Bowel Diseases. 2011;17(1):362–381. doi: 10.1002/ibd.21403. [DOI] [PubMed] [Google Scholar]
- 68.Schepens MAA, Schonewille AJ, Vink C, et al. Supplemental calcium attenuates the colitis-related increase in diarrhea, intestinal permeability, and extracellular matrix breakdown in HLA-B27 transgenic rats. Journal of Nutrition. 2009;139(8):1525–1533. doi: 10.3945/jn.109.105205. [DOI] [PubMed] [Google Scholar]
- 69.An G, Wei B, Xia B, et al. Increased susceptibility to colitis and colorectal tumors in mice lacking core 3-derived O-glycans. Journal of Experimental Medicine. 2007;204(6):1417–1429. doi: 10.1084/jem.20061929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Soler AP, Miller RD, Laughlin KV, Carp NZ, Klurfeld DM, Mullin JM. Increased tight junctional permeability is associated with the development of colon cancer. Carcinogenesis. 1999;20(8):1425–1431. doi: 10.1093/carcin/20.8.1425. [DOI] [PubMed] [Google Scholar]
- 71.Niv Y. Mucin and colorectal cancer. Israel Medical Association Journal. 2000;2(10):775–777. [PubMed] [Google Scholar]
- 72.Kim YS. Mucin glycoproteins in colonic neoplasia. Keio Journal of Medicine. 1998;47(1):10–18. doi: 10.2302/kjm.47.10. [DOI] [PubMed] [Google Scholar]
- 73.Bruewer M, Samarin S, Nusrat A. Inflammatory bowel disease and the apical junctional complex. Annals of the New York Academy of Sciences. 2006;1072:242–252. doi: 10.1196/annals.1326.017. [DOI] [PubMed] [Google Scholar]
- 74.Vaccina F, Scorcioni F, Pedroni M, et al. Scanning electron microscopy of aberrant crypt foci in human colorectal mucosa. Anticancer Research. 1998;18(5 A):3451–3456. [PubMed] [Google Scholar]
- 75.Nittka S, Günther J, Ebisch C, Erbersdobler A, Neumaier M. The human tumor suppressor CEACAM1 modulates apoptosis and is implicated in early colorectal tumorigenesis. Oncogene. 2004;23(58):9306–9313. doi: 10.1038/sj.onc.1208259. [DOI] [PubMed] [Google Scholar]
- 76.Nittka S, Böhm C, Zentgraf H, Neumaier M. The CEACAM1-mediated apoptosis pathway is activated by CEA and triggers dual cleavage of CEACAM1. Oncogene. 2008;27(26):3721–3728. doi: 10.1038/sj.onc.1211033. [DOI] [PubMed] [Google Scholar]
- 77.Buscail L, Estève JP, Saint-Laurent N, et al. Inhibition of cell proliferation by the somatostatin analogue RC-160 is mediated by somatostatin receptor subtypes SSTR2 and SSTR5 through different mechanisms. Proceedings of the National Academy of Sciences of the United States of America. 1995;92(5):1580–1584. doi: 10.1073/pnas.92.5.1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Buscail L, Saint-Laurent N, Chastre E, et al. Loss of sst2 somatostatin receptor gene expression in human pancreatic and colorectal cancer. Cancer Research. 1996;56(8):1823–1827. [PubMed] [Google Scholar]
- 79.Casini Raggi C, Calabrò A, Renzi D, et al. Quantitative evaluation of somatostatin receptor subtype 2 expression in sporadic colorectal tumor and in the corresponding normal mucosa. Clinical Cancer Research. 2002;8(2):419–427. [PubMed] [Google Scholar]
- 80.Xiao R, Badger TM, Simmen FA. Dietary exposure to soy or whey proteins alters colonic global gene expression profiles during rat colon tumorigenesis. Molecular Cancer. 2005;4, article 1 doi: 10.1186/1476-4598-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bakshi RP, Galande S, Muniyappa K. Functional and regulatory characteristics of eukaryotic type II DNA topoisomerase. Critical Reviews in Biochemistry and Molecular Biology. 2001;36(1):1–37. doi: 10.1080/20014091074165. [DOI] [PubMed] [Google Scholar]
- 82.Boman BM, Walters R, Fields JZ, et al. Colonic crypt changes during adenoma development in familial adenomatous polyposis: immunohistochemical evidence for expansion of the crypt base cell population. American Journal of Pathology. 2004;165(5):1489–1498. doi: 10.1016/S0002-9440(10)63407-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Castedo M, Perfettini JL, Roumier T, Kroemer G. Cyclin-dependent kinase-1: linking apoptosis to cell cycle and mitotic catastrophe. Cell Death and Differentiation. 2002;9(12):1287–1293. doi: 10.1038/sj.cdd.4401130. [DOI] [PubMed] [Google Scholar]
- 84.Taylor WR, Stark GR. Regulation of the G2/M transition by p53. Oncogene. 2001;20(15):1803–1815. doi: 10.1038/sj.onc.1204252. [DOI] [PubMed] [Google Scholar]
- 85.Cummings JH. Short chain fatty acids in the human colon. Gut. 1981;22(9):763–779. doi: 10.1136/gut.22.9.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cummings JH, Englyst HN. Fermentation in the human large intestine and the available substrates. American Journal of Clinical Nutrition. 1987;45(5):1243–1255. doi: 10.1093/ajcn/45.5.1243. [DOI] [PubMed] [Google Scholar]
- 87.Cook SI, Sellin JH. Review article: short chain fatty acids in health and disease. Alimentary Pharmacology and Therapeutics. 1998;12(6):499–507. doi: 10.1046/j.1365-2036.1998.00337.x. [DOI] [PubMed] [Google Scholar]
- 88.Miller SJ. Cellular and physiological effects of short-chain fatty acids. Mini-Reviews in Medicinal Chemistry. 2004;4(8):839–845. doi: 10.2174/1389557043403288. [DOI] [PubMed] [Google Scholar]
- 89.Hague A, Manning AM, Hanlon KA, Huschtscha LI, Hart D, Paraskeva C. Sodium butyrate induces apoptosis in human colonic tumour cell lines in a p53-independent pathway: implications for the possible role of dietary fibre in the prevention of large-bowel cancer. International Journal of Cancer. 1993;55(3):498–505. doi: 10.1002/ijc.2910550329. [DOI] [PubMed] [Google Scholar]
- 90.Heerdt BG, Houston MA, Augenlicht LH. Short-chain fatty acid-initiated cell cycle arrest and apoptosis of colonic epithelial cells is linked to mitochondrial function. Cell Growth and Differentiation. 1997;8(5):523–532. [PubMed] [Google Scholar]
- 91.Augenlicht LH, Anthony GM, Church TL, et al. Short-chain fatty acid metabolism, apoptosis, and Apc-initiated tumorigenesis in the mouse gastrointestinal mucosa. Cancer Research. 1999;59(23):6005–6009. [PubMed] [Google Scholar]
- 92.Thibault R, Blachier F, Darcy-Vrillon B, De Coppet P, Bourreille A, Segain JP. Butyrate utilization by the colonic mucosa in inflammatory bowel diseases: a transport deficiency. Inflammatory Bowel Diseases. 2010;16(4):684–695. doi: 10.1002/ibd.21108. [DOI] [PubMed] [Google Scholar]
- 93.Birkenkamp-Demtroder K, Christensen LL, Olesen SH, et al. Gene expression in colorectal cancer. Cancer Research. 2002;62(15):4352–4363. [PubMed] [Google Scholar]
- 94.Buckland AG, Heeley EL, Wilton DC. Bacterial cell membrane hydrolysis by secreted phospholipases A2: a major physiological role of human group IIa sPLA2 involving both bacterial cell wall penetration and interfacial catalysis. Biochimica et Biophysica Acta. 2000;1484(2-3):195–206. doi: 10.1016/s1388-1981(00)00018-4. [DOI] [PubMed] [Google Scholar]
- 95.Buckland AG, Wilton DC. The antibacterial properties of secreted phospholipases A2. Biochimica et Biophysica Acta. 2000;1488(1-2):71–82. doi: 10.1016/s1388-1981(00)00111-6. [DOI] [PubMed] [Google Scholar]
- 96.Cunliffe RN. α-Defensins in the gastrointestinal tract. Molecular Immunology. 2003;40(7):463–467. doi: 10.1016/s0161-5890(03)00157-3. [DOI] [PubMed] [Google Scholar]
- 97.Cunliffe RN, Mahida YR. Expression and regulation of antimicrobial peptides in the gastrointestinal tract. Journal of Leukocyte Biology. 2004;75(1):49–58. doi: 10.1189/jlb.0503249. [DOI] [PubMed] [Google Scholar]
- 98.Mahida YR, Cunliffe RN. Defensins and mucosal protection. Novartis Foundation Symposium. 2004;263:71–77. doi: 10.1002/0470090480.ch6. [DOI] [PubMed] [Google Scholar]
- 99.Hazlett L, Wu M. Defensins in innate immunity. Cell and Tissue Research. 2011;343:175–188. doi: 10.1007/s00441-010-1022-4. [DOI] [PubMed] [Google Scholar]
- 100.Yuan Q, Walker WA. Innate immunity of the gut: mucosal defense in health and disease. Journal of Pediatric Gastroenterology and Nutrition. 2004;38(5):463–473. doi: 10.1097/00005176-200405000-00001. [DOI] [PubMed] [Google Scholar]
- 101.Minami T, Tojo H, Shinomura Y, Matsuzawa Y, Okamoto M. Increased group II phospholipase A2 in colonic mucosa of patients with Crohn’s disease and ulcerative colitis. Gut. 1994;35(11):1593–1598. doi: 10.1136/gut.35.11.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yamashita S, Ogawa M, Abe T, et al. Group II phospholipase A2 in invasive gastric cancer cell line is induced by interleukin 6. Biochemical and Biophysical Research Communications. 1994;198(3):878–884. doi: 10.1006/bbrc.1994.1125. [DOI] [PubMed] [Google Scholar]
- 103.Ikegami T, Matsuzaki Y, Shoda J, Kano M, Hirabayashi N, Tanaka N. The chemopreventive role of ursodeoxycholic acid in azoxymethane-treated rats: suppressive effects on enhanced group II phospholipase A2 expression in colonic tissue. Cancer Letters. 1998;134(2):129–139. doi: 10.1016/s0304-3835(98)00248-1. [DOI] [PubMed] [Google Scholar]
- 104.Haapamäki MM, Grönroos JM, Nurmi H, Irjala K, Alanen KA, Nevalainen TJ. Phospholipase A2 in serum and colonic mucosa in ulcerative colitis. Scandinavian Journal of Clinical and Laboratory Investigation. 1999;59(4):279–288. doi: 10.1080/00365519950185643. [DOI] [PubMed] [Google Scholar]
- 105.Haapamäki MM, Grönroos JM, Nurmi H, Alanen K, Kallajoki M, Nevalainen TJ. Gene expression of group II phospholipase A2 in intestine in ulcerative colitis. Gut. 1997;40(1):95–101. doi: 10.1136/gut.40.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Nevalainen TJ, Haapamäki MM, Grönroos JM. Roles of secretory phospholipases A2 in inflammatory diseases and trauma. Biochimica et Biophysica Acta. 2000;1488(1-2):83–90. doi: 10.1016/s1388-1981(00)00112-8. [DOI] [PubMed] [Google Scholar]
- 107.Nevalainen TJ, Graham GG, Scott KF. Antibacterial actions of secreted phospholipases A2. Review. Biochimica et Biophysica Acta. 2008;1781(1-2):1–9. doi: 10.1016/j.bbalip.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 108.Kennedy BP, Soravia C, Moffat J, et al. Overexpression of the nonpancreatic secretory group II PLA2 messenger RNA and protein in colorectal adenomas from familial adenomatous polyposis patients. Cancer Research. 1998;58(3):500–503. [PubMed] [Google Scholar]
- 109.Tribler L, Jensen LT, Jørgensen K, et al. Increased expression and activity of group IIA and X secretory phospholipase A2 in peritumoral versus central colon carcinoma tissue. Anticancer Research. 2007;27(5):3179–3185. [PubMed] [Google Scholar]
- 110.Minami T, Shinomura Y, Miyagawa JI, Tojo H, Okamoto M, Matsuzawa Y. Immunohistochemical localization of group II phospholipase A2 in colonic mucosa of patients with inflammatory bowel disease. American Journal of Gastroenterology. 1997;92(2):289–292. [PubMed] [Google Scholar]
- 111.Femia AP, Luceri C, Toti S, Giannini A, Dolara P, Caderni G. Gene expression profile and genomic alterations in colonic tumours induced by 1,2-dimethylhydrazine (DMH) in rats. BMC Cancer. 2010;10, article 194 doi: 10.1186/1471-2407-10-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zallen G, Moore EE, Johnson JL, et al. New mechanisms by which secretory phospholipase A2 stimulates neutrophils to provoke the release of cytotoxic agents. Archives of Surgery. 1998;133(11):1229–1233. doi: 10.1001/archsurg.133.11.1229. [DOI] [PubMed] [Google Scholar]
- 113.Bingham CO, Frank Austen K. Phospholipase A2 enzymes in eicosanoid generation. Proceedings of the Association of American Physicians. 1999;111(6):516–524. doi: 10.1046/j.1525-1381.1999.99321.x. [DOI] [PubMed] [Google Scholar]
- 114.Marshall LA, Morgan DW. Modulation of arachidonic acid metabolism: focus on phospholipase A2(s) Drug News and Perspectives. 1998;11(2):82–91. doi: 10.1358/dnp.1998.11.2.863680. [DOI] [PubMed] [Google Scholar]
- 115.Droin N, Hendra JB, Ducoroy P, Solary E. Human defensins as cancer biomarkers and antitumour molecules. Journal of Proteomics. 2009;72(6):918–927. doi: 10.1016/j.jprot.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 116.Cunliffe RN, Mahida YR. Antimicrobial peptides in innate intestinal host defence. Gut. 2000;47(1):16–17. doi: 10.1136/gut.47.1.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kanmura S, Uto H, Numata M, et al. Human neutrophil peptides 1-3 are useful biomarkers in patients with active ulcerative colitis. Inflammatory Bowel Diseases. 2009;15(6):909–917. doi: 10.1002/ibd.20854. [DOI] [PubMed] [Google Scholar]
- 118.Cunliffe RN, Kamal M, Rose FRAJ, James PD, Mahida YR. Expression of antimicrobial neutrophil defensins in epithelial cells of active inflammatory bowel disease mucosa. Journal of Clinical Pathology. 2002;55(4):298–304. doi: 10.1136/jcp.55.4.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Rodríguez-García M, Oliva H, Climent N, García F, Gatell JM, Gallart T. Human immature monocyte-derived dendritic cells produce and secrete α-defensins 1-3. Journal of Leukocyte Biology. 2007;82(5):1143–1146. doi: 10.1189/jlb.0507295. [DOI] [PubMed] [Google Scholar]
- 120.Mothes H, Melle C, Ernst G, Kaufmann R, Von Eggeling F, Settmacher U. Human Neutrophil Peptides 1-3—early markers in development of colorectal adenomas and carcinomas. Disease Markers. 2008;25(2):123–129. doi: 10.1155/2008/693937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Albrethsen J, Bøgebo R, Gammeltoft S, Olsen J, Winther B, Raskov H. Upregulated expression of human neutrophil peptides 1,2 and 3 (HNP 1-3) in colon cancer serum and tumours: a biomarker study. BMC Cancer. 2005;5, article 8 doi: 10.1186/1471-2407-5-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Melle C, Ernst G, Schimmel B, et al. Discovery and identification of α-defensins as low abundant, tumor-derived serum markers in colorectal cancer. Gastroenterology. 2005;129(1):66–73. doi: 10.1053/j.gastro.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 123.Sorokin L. The impact of the extracellular matrix on inflammation. Nature Reviews Immunology. 2010;10(10):712–723. doi: 10.1038/nri2852. [DOI] [PubMed] [Google Scholar]
- 124.Ohtani H. Stromal reaction in cancer tissue: pathophysiologic significance of the expression of matrix-degrading enzymes in relation to matrix turnover and immune/inflammatory reactions. Pathology International. 1998;48(1):1–9. doi: 10.1111/j.1440-1827.1998.tb03820.x. [DOI] [PubMed] [Google Scholar]
- 125.Worthley DL, Giraud AS, Wang TC. The extracellular matrix in digestive cancer. Cancer Microenvironment. 2010;3(1):177–185. doi: 10.1007/s12307-010-0053-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Abelev GI, Lazarevich NL. Control of differentiation in progression of epithelial tumors. Advances in Cancer Research. 2006;95:61–113. doi: 10.1016/S0065-230X(06)95003-9. [DOI] [PubMed] [Google Scholar]
- 127.Kim SH, Turnbull J, Guimond S. Extracellular matrix and cell signalling: the dynamic cooperation of integrin, proteoglycan and growth factor receptor. Journal of Endocrinology. 2011;209(2):139–151. doi: 10.1530/JOE-10-0377. [DOI] [PubMed] [Google Scholar]
- 128.Groos S, Hünefeld G, Luciano L. Epithelial cell turnover–extracellular matrix relationship in the small intestine of human adults. Italian Journal of Anatomy and Embryology. 2001;106(2):353–361. [PubMed] [Google Scholar]
- 129.Simon-Assmann P, Kedinger M, Haffen K. Immunocytochemical localization of extracellular-matrix proteins in relation to rat intestinal morphogenesis. Differentiation. 1986;32(1):59–66. doi: 10.1111/j.1432-0436.1986.tb00556.x. [DOI] [PubMed] [Google Scholar]
- 130.Gagné D, Groulx JF, Benoit YD, et al. Integrin-linked kinase regulates migration and proliferation of human intestinal cells under a fibronectin-dependent mechanism. Journal of Cellular Physiology. 2010;222(2):387–400. doi: 10.1002/jcp.21963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Westcarr S, Farshori P, Wyche J, Anderson WA. Apoptosis and differentiation in the crypt-villus unit of the rat small intestine. Journal of Submicroscopic Cytology and Pathology. 1999;31(1):15–30. [PubMed] [Google Scholar]
- 132.Vachon PH, Simoneau A, Herring-Gillam FE, Beaulieu JF. Cellular fibronectin expression is down-regulated at the mRNA level in differentiating human intestinal epithelial cells. Experimental Cell Research. 1995;216(1):30–34. doi: 10.1006/excr.1995.1004. [DOI] [PubMed] [Google Scholar]
- 133.DeGiorgio-Miller AM, Treharne LJ, McAnulty RJ, Coleridge Smith PD, Laurent GJ, Herrick SE. Procollagen type I gene expression and cell proliferation are increased in lipodermatosclerosis. British Journal of Dermatology. 2005;152(2):242–249. doi: 10.1111/j.1365-2133.2004.06243.x. [DOI] [PubMed] [Google Scholar]
- 134.Gradl D, Kühl M, Wedlich D. The Wnt/Wg signal transducer β-catenin controls fibronectin expression. Molecular and Cellular Biology. 1999;19(8):5576–5587. doi: 10.1128/mcb.19.8.5576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Takayasu H, Horie H, Hiyama E, et al. Frequent deletions and mutations of the β-catenin gene are associated with overexpression of cyclin D1 and fibronectin and poorly differentiated histology in childhood hepatoblastoma. Clinical Cancer Research. 2001;7(4):901–908. [PubMed] [Google Scholar]
- 136.Kolachala VL, Bajaj R, Wang L, et al. Epithelial-derived fibronectin expression, signaling, and function in intestinal inflammation. Journal of Biological Chemistry. 2007;282(45):32965–32973. doi: 10.1074/jbc.M704388200. [DOI] [PubMed] [Google Scholar]
- 137.Walia B, Castaneda FE, Wang L, et al. Polarized fibronectin secretion induced by adenosine regulates bacterial-epithelial interaction in human intestinal epithelial cells. Biochemical Journal. 2004;382(2):589–596. doi: 10.1042/BJ20040021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Graham LL, Friel T, Woodman RL. Fibronectin enhances Campylobacter fetus interaction with extracellular matrix components and INT 407 cells. Canadian Journal of Microbiology. 2008;54(1):37–47. doi: 10.1139/w07-115. [DOI] [PubMed] [Google Scholar]
- 139.Isenmann R, Schwarz M, Rozdzinski E, et al. Interaction of fibronectin and aggregation substance promotes adherence of Enterococcus faecalis to human colon. Digestive Diseases and Sciences. 2002;47(2):462–468. doi: 10.1023/a:1013763331708. [DOI] [PubMed] [Google Scholar]
- 140.Konkel ME, Christensen JE, Keech AM, Monteville MR, Klena JD, Garvis SG. Identification of a fibronectin-binding domain within the Campylobacter jejuni CadF protein. Molecular Microbiology. 2005;57(4):1022–1035. doi: 10.1111/j.1365-2958.2005.04744.x. [DOI] [PubMed] [Google Scholar]
- 141.Jaluria P, Konstantopoulos K, Betenbaugh M, Shiloach J. A perspective on microarrays: current applications, pitfalls, and potential uses. Microbial Cell Factories. 2007;6, article 4 doi: 10.1186/1475-2859-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Terzić J, Grivennikov S, Karin E, Karin M. Inflammation and colon cancer. Gastroenterology. 2010;138(6):2101–2114.e5. doi: 10.1053/j.gastro.2010.01.058. [DOI] [PubMed] [Google Scholar]
- 143.Klaunig JE, Wang Z, Pu X, Zhou S. Oxidative stress and oxidative damage in chemical carcinogenesis. Toxicology and Applied Pharmacology. 2011;254(2):86–99. doi: 10.1016/j.taap.2009.11.028. [DOI] [PubMed] [Google Scholar]
- 144.Meira LB, Bugni JM, Green SL, et al. DNA damage induced by chronic inflammation contributes to colon carcinogenesis in mice. Journal of Clinical Investigation. 2008;118(7):2516–2525. doi: 10.1172/JCI35073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Din FVN, Theodoratou E, Farrington SM, et al. Effect of aspirin and NSAIDs on risk and survival from colorectal cancer. Gut. 2010;59(12):1670–1679. doi: 10.1136/gut.2009.203000. [DOI] [PubMed] [Google Scholar]
- 146.Harris RE. Cyclooxygenase-2 (cox-2) blockade in the chemoprevention of cancers of the colon, breast, prostate, and lung. Inflammopharmacology. 2009;17(2):55–67. doi: 10.1007/s10787-009-8049-8. [DOI] [PubMed] [Google Scholar]
- 147.Abulafi AM, Williams NS. Local recurrence of colorectal cancer: the problem, mechanisms, management and adjuvant therapy. British Journal of Surgery. 1994;81(1):7–19. doi: 10.1002/bjs.1800810106. [DOI] [PubMed] [Google Scholar]
- 148.Ushijima T. Epigenetic field for cancerization. Journal of Biochemistry and Molecular Biology. 2007;40(2):142–150. doi: 10.5483/bmbrep.2007.40.2.142. [DOI] [PubMed] [Google Scholar]
- 149.Braakhuis BJM, Tabor MP, Kummer JA, Leemans CR, Brakenhoff RH. A genetic explanation of slaughter’s concept of field cancerization: evidence and clinical implications. Cancer Research. 2003;63(8):1727–1730. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The sequences of the primer pairs used to confirm gene changes with qRT-PCR. Gene specific primers for cell division cycle 2 (Cdc2), cyclin B1 (Ccnb1), topoisomerase II alpha (Top2A), group IIA secretory phospholipase A2 (Pla2g2a), fibronectin 1 (Fn1), collagen, type I, alpha 1 (Col1a1), rat neutrophil (NP) defensin 3 (RatNP-3), aquaporin 8 (Aqp8), and 3-hydroxy-3-methylglutaryl-Coenzyme A synthase 2 (Hmgcs2) were designed with the Primer Express 2.0 program (Applied Biosystems, Foster City, CA). β-actin was used as an internal control.


