Abstract
The current histoclinical breast cancer classification is simple but imprecise. Several molecular classifications of breast cancers based on expression profiling have been proposed as alternatives. However, their reliability and clinical utility have been repeatedly questioned, notably because most of them were derived from relatively small initial patient populations. We analyzed the transcriptomes of 537 breast tumors using three unsupervised classification methods. A core subset of 355 tumors was assigned to six clusters by all three methods. These six subgroups overlapped with previously defined molecular classes of breast cancer, but also showed important differences, notably the absence of an ERBB2 subgroup and the division of the large luminal ER+ group into four subgroups, two of them being highly proliferative. Of the six subgroups, four were ER+/PR+/AR+, one was ER−/PR−/AR+ and one was triple negative (AR−/ER−/PR−). ERBB2-amplified tumors were split between the ER−/PR−/AR+ subgroup and the highly proliferative ER+ LumC subgroup. Importantly, each of these six molecular subgroups showed specific copy-number alterations. Gene expression changes were correlated to specific signaling pathways. Each of these six subgroups showed very significant differences in tumor grade, metastatic sites, relapse-free survival or response to chemotherapy. All these findings were validated on large external datasets including more than 3000 tumors. Our data thus indicate that these six molecular subgroups represent well-defined clinico-biological entities of breast cancer. Their identification should facilitate the detection of novel prognostic factors or therapeutical targets in breast cancer.
Keywords: breast cancer; molecular classification; taxonomy; transcriptome; CGH-array, outcome
Introduction
Breast cancer is heterogeneous. Biological features have proven insufficient for a comprehensive description of the disease. Seminal work by Sorlie et al. (2003) has delineated five major molecular subtypes of breast cancer associated to different outcomes. This initial classification was reproduced in independent datasets (Bertucci et al., 2006) strongly suggesting the existence of distinct molecular entities in breast cancer. The Sorlie centroid approach has subsequently been redefined and adapted to more recent technological platforms (Hu et al., 2006; Parker et al., 2009).
However, criticisms have pointed to the instability of the defined subtypes (Kapp et al., 2006; Weigelt et al., 2010) and their dependence on the original set of samples or genes. Thus, although molecular classification brings interesting insights in breast cancer taxonomy, its implementation in the clinics is put in doubt because of insufficient reliability in single sample allocation (Weigelt et al., 2010). Rather, three broad classes of breast tumors drawn along their ER, PR and ERBB2/HER2 status are commonly used in the clinic. ER−/PR−/HER2− tumors were defined as triple negative, ER+/PR+/HER2− as luminal, and HER2+ tumors irrespective of their ER status form the third class (Foulkes et al., 2010). However, this simple classification is also criticized because of the biological heterogeneity within classes. In particular, the correspondence between the triple-negative group and basal-like breast tumors and the heterogeneity of the large ER+/PR+ group have been repeatedly questioned (Gusterson, 2009; Foulkes et al., 2010). This argues for a more elaborate stratification amenable to biological exploration and clinical choices.
This prompted us to construct a robust molecular classification on a large number of samples to reach high statistical power. To this aim, we produced transcriptomes of a series of 537 primary breast cancers and, using a semi-supervised analysis, revealed six stable molecular subgroups. A related classification rule was defined. Each of the six molecular subgroups showed distinct genomic changes, correlated with a specific set of signaling pathways and was associated with significant differences in tumor grade, metastatic sites and metastasis-free survival (MFS). We propose that this classification scheme could lay the bases of an operative tool to reliably classify breast cancers in more homogeneous molecular subgroups. This classification could be highly beneficial in future investigations aiming at identifying novel prognostic factors or therapeutical targets in breast cancer.
Results
Semi-supervised gene expression analysis identifies six prototypic molecular subtypes
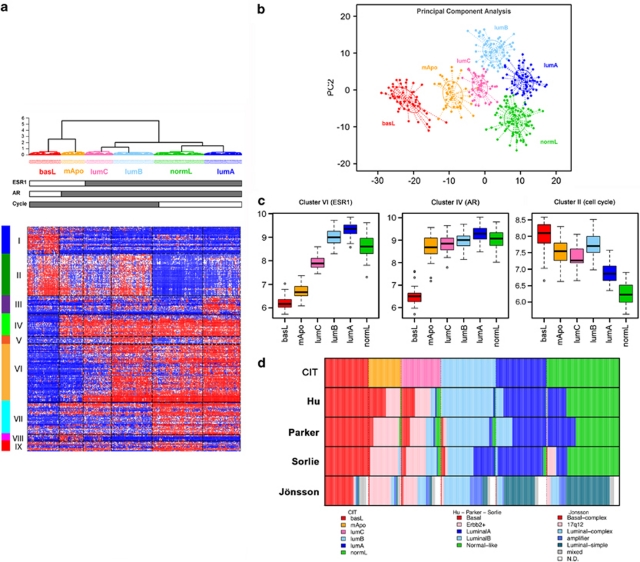
Our aim was to identify molecular subgroups representing homogeneous subsets of breast cancer. Our methodology is detailed in Supplementary Figure 1 and the Supplementary Methods section. Briefly, we produced a large dataset comprising 537 primary breast cancer transcriptomes on Affymetrix U133-Plus2.0 arrays to ensure proper statistical power. First, this tumor set was classified with three unsupervised methods (hierarchical clustering, Gaussian mixture models and k-means) in parallel. Of the 537 tumors, 355 yielded a consensus subgroup assignment (that is, were assigned to the same subclass) between all three methods. This subset was named coreset and was used for further analysis. Second, a minimal list of 256 discriminative genes with maximal intragroup homogeneity and intergroup heterogeneity was generated by analysis of variance (Supplementary Table 3). Hierarchical clustering based on this list delineated six homogeneous tumor subgroups, homogeneity being confirmed by the principal component analysis (Figure 1b). To allow the classification of independent sample profiles to one of the six subgroups we built a single sample predictor based on a distance-to-centroid approach (using the previously mentioned 256 genes; Supplementary Methods). The 182 tumors of the discovery set lying outside of the coreset were classified using this single sample predictor.
Figure 1.
Breast tumor classification according to the CIT classification into six subgroups of tumors. (a) Heatmap representing the expression of the 256 genes (nine clusters of genes represented by vertical color bars on the left of the heatmap) through the six groups. (b) Principal component analysis (PCA) of the samples of the coreset according to the 256 gene signature. The first principal component (PC1) represents the combined expression of the three transversal clusters (ER, AR and cell cycle), the second component (PC2) differentiates LumB and NormL. (c) Distribution of mean expression levels of the three transversal gene clusters (ER, AR and Cell Cycle) over the six main molecular subgroups. (d) Comparison of the CIT classification with those obtained using the Sorlie, Hu, Parker and Jönsson systems.
The overall distribution of the six subgroups was determined by three large gene clusters shared by at least two subgroups. The first one (cluster-VI, Figures 1a and c, Supplementary Table 3) containing ESR1 and correlated genes, defined two ER-negative (ER−) and four ER-positive (ER+) subgroups (Figures 1a and c). The second gene cluster (cluster-IV) included the androgen receptor (AR) gene and encompassed five subgroups. Of the six subgroups, four were ER+/PR+/AR+, one was ER−/PR−/AR+ and one was triple negative (AR−/ER−/PR− Figures 1a and c). Interestingly, cluster-IV included transcription factors FOXA1, SPDEF and XBP1, which are usually associated to the ER-cluster (Bertucci et al., 2006). The third cluster (cluster-II) was predominantly composed of genes regulating DNA replication and cell cycle progression, thus defining elevated cell proliferation. This cluster encompassed both ER− and two ER+ subgroups (Figures 1a and c).
Each subgroup was defined by a specific gene cluster (Supplementary Figure 2) in which we found genes previously part of the Sorlie centroids. Hence, for simplicity we named our subgroups according to the Sorlie subtype (Sorlie et al., 2003). ER+ subgroups were split according to expression levels of the cell cycle cluster. Low proliferative ER+ subgroups were differentiated by clusters-III and IX (Figure 1a, Supplementary Figure 2), comprising respectively genes from the Sorlie luminal-A and normal-like centroids (Supplementary Table 3) and were, thus, designated LumA and NormL. The two high proliferation ER+ subgroups differed sharply in ER-cluster expression levels. The subgroup expressing highest levels of ER was named LumB. The other subgroup, positioned at the boundary between ER+ and ER− tumors, was designated LumC (Figures 1a–c). Noteworthy, 40% of LumC tumors overexpressed the ERBB2/HER2 gene.
Next was the AR+/ER−/PR− subgroup (Figure 1b), defined by cluster-VIII. The AR+/ER− status of this subgroup was reminiscent of the previously described ‘molecular-apocrine' subtype (Farmer et al., 2005) and we designated it mApo. Although ERBB2/HER2 was overexpressed by 72% of the tumors in this subgroup, cluster-VIII did not comprise genes co-amplified with ERBB2/HER2. In fact, ERBB2/HER2+ tumors distributed in mApo and LumC subgroups (Table 1). Finally, the AR−/ER−/PR− subgroup, defined by cluster-I, presented the greatest distance to all others (Figure 1). As it shared genes with the ‘basal-like' subtype, it was designated BasL (Supplementary Table 3).
Table 1. Molecular subgroups show differential correlation to breast cancer clinico-biological parameters and different sites of metastatic relapse.
|
CIT classification | |||||||
|---|---|---|---|---|---|---|---|
| Variable | pv | BasL | mApo | LumC | LumB | LumA | NormL |
| Total | 53 | 39 | 48 | 66 | 61 | 88 | |
| ER+ (IHC) | 1.00E–50 | 5 (10%) | 1 (3%) | 37 (84%) | 63 (98%) | 58 (97%) | 81 (93%) |
| ER− (IHC) | 46 (90%) | 35 (97%) | 7 (16%) | 1 (2%) | 2 (3%) | 6 (7%) | |
| ER+ (EXP) | 6.00E–68 | 3 (6%) | 2 (5%) | 48 (100%) | 66 (100%) | 61 (100%) | 87 (99%) |
| ER− (EXP) | 50 (94%) | 37 (95%) | 0 (0%) | 0 (0%) | 0 (0%) | 1 (1%) | |
| PR+ (IHC) | 2.00E–25 | 4 (8%) | 1 (3%) | 25 (54%) | 43 (67%) | 53 (88%) | 62 (71%) |
| PR− (IHC) | 48 (92%) | 34 (97%) | 21 (46%) | 21 (33%) | 7 (12%) | 25 (29%) | |
| PR+ (EXP) | 1.00E–37 | 5 (9%) | 5 (13%) | 32 (67%) | 47 (71%) | 58 (95%) | 85 (97%) |
| PR− (EXP) | 48 (91%) | 34 (87%) | 16 (33%) | 19 (29%) | 3 (5%) | 3 (3%) | |
| ERBB2+ (IHC) | 9.00E–19 | 3 (7%) | 19 (68%) | 10 (26%) | 5 (11%) | 0 (0%) | 0 (0%) |
| ERBB2− (IHC) | 43 (93%) | 9 (32%) | 28 (74%) | 41 (89%) | 37 (100%) | 74 (100%) | |
| ERBB2+ (EXP) | 4.00E–31 | 2 (4%) | 29 (74%) | 20 (42%) | 2 (3%) | 0 (0%) | 5 (6%) |
| ERBB2− (EXP) | 51 (96%) | 10 (26%) | 28 (58%) | 64 (97%) | 61 (100%) | 83 (94%) | |
| AR+ (EXP) | 2.00E–57 | 2 (4%) | 32 (82%) | 47 (98%) | 63 (95%) | 61 (100%) | 88 (100%) |
| AR− (EXP) | 51 (96%) | 7 (18%) | 1 (2%) | 3 (5%) | 0 (0%) | 0 (0%) | |
| P53mut | 1.00E–15 | 29 (83%) | 13 (72%) | 24 (69%) | 5 (16%) | 1 (4%) | 1 (5%) |
| P53wt | 6 (17%) | 5 (28%) | 11 (31%) | 27 (84%) | 27 (96%) | 21 (95%) | |
| Ductal | 0.05 | 51 (98%) | 32 (84%) | 39 (87%) | 54 (84%) | 50 (83%) | 61 (77%) |
| Lobular | 0.004 | 1 (2%) | 1 (3%) | 3 (7%) | 3 (5%) | 5 (8%) | 15 (19%) |
| Other | 0.1 | 0 (0%) | 5 (13%) | 3 (7%) | 7 (11%) | 5 (8%) | 3 (4%) |
| SBR Grade 1 | 8.00E–11 | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 7 (12%) | 23 (27%) |
| SBR Grade 2 | 2.00E–13 | 6 (11%) | 8 (21%) | 21 (47%) | 38 (58%) | 44 (77%) | 53 (62%) |
| SBR Grade 3 | 4.00E–26 | 47 (89%) | 30 (79%) | 24 (53%) | 28 (42%) | 6 (11%) | 9 (11%) |
| Age (median) | 4.00E–07 | 50 | 56 | 54 | 57 | 62 | 52 |
| MR 5year | 0.001 | 17 (36%) | 14 (38%) | 11 (34%) | 15 (26%) | 9 (20%) | 6 (8%) |
| MR 15year | 0.01 | 17 (36%) | 14 (38%) | 13 (41%) | 18 (32%) | 10 (22%) | 11 (15%) |
| Bones | 0.01 | 4 (24%) | 8 (57%) | 7 (54%) | 14 (78%) | 7 (70%) | 9 (82%) |
| Brain | 0.06 | 5 (29%) | 3 (21%) | 1 (8%) | 0 (0%) | 0 (0%) | 2 (18%) |
| Liver | 0.7 | 5 (29%) | 6 (43%) | 7 (54%) | 8 (44%) | 3 (30%) | 3 (27%) |
| Lung | 0.9 | 6 (35%) | 4 (29%) | 6 (46%) | 8 (44%) | 3 (30%) | 4 (36%) |
| Other | 0.1 | 4 (24%) | 1 (7%) | 7 (54%) | 8 (44%) | 3 (30%) | 3 (27%) |
Abbreviations: CIT, Cartes d'Identité des Tumeurs program; MR, metastasis relapse.
Expression of ER, PR and ERBB2/HER2 were determined by immunohistochemistry as well as by RNA expression (for greater details see Supplementary Methods). TP53 mutation status was determined by the yeast functional assay (Supplementary Methods). P-values for qualitative variables (ER, PR, ERBB2/HER2, TP53 mutation, histological type, SBR grading) result from a Fisher exact test. P-values for quantitative variables (median age) result from an analysis of variance. MR was determined 5 and 15 years after surgery. Frequency of MR in a subgroup was calculated as the ratio of MR with the total number of MR. For each subgroup, percentages of MR in a given site are determined by the number of MR in this site over the whole number of MR in the subgroup. MR may occur at more than one site; hence, the sum of percentages may not equate 100.
Molecular subgroups show distinct clinical correlations, metastatic sites and outcomes
BasL and mApo at one end of the spectrum, and LumA and NormL at the other end showed an inverse balance between high-grade and ER/PR positivity (Table 1). TP53 mutation incidence reached 83% in the BasL subgroup and gradually went down to 4% in NormL and LumA tumors (Table 1). This distribution of high-grade/ER− versus low-grade/ER+ cancers was also coherent with the median age of onset: 50 and 62 for BasL and LumA patients, respectively. Correlation with histological type was observed as well. While the BasL subgroup was composed of 98% ductal carcinomas, NormL presented 19% of invasive lobular tumors, representing 53% of all lobular cancers in the dataset, in coherence with previous findings (Bertucci et al., 2008).
Molecular subgroups showed differences in sites of metastatic relapse. In line with previous studies (Smid et al., 2008), LumA and NormL predominantly metastasized to the bone and rarely or never to the brain, while BasL and mApo tumors metastasized to the brain and less to the bones (Table 1). ST6GALNAC5, COX2/PTGS2 and HBEGF, whose expression has recently been associated to brain metastasis (Bos et al., 2009), were increased in BasL (Supplementary Figure 3). Clear differences were also found in MFS (Figure 2). BasL and mApo subgroups showed earliest recurrence (18 to 60 months). LumA and NormL had the slowest course. Metastatic recurrence plateaued between 60 and 180 months in BasL and mApo, whereas it progressively increased after 60 months in ER+ subgroups. LumA and NormL tumors presented recurrences after 120 months post-surgery. Interestingly, patterns of recurrence (early versus late) matched cell cycle cluster expression levels in the different subgroups.
Figure 2.
Breast cancer molecular subgroups show distinctly different disease outcome. Kaplan–Meier curves shown in this figure represent disease-free survival with metastatic relapse as an end point. (a, b) show survival curves in the CIT and validation set, respectively. Abrupt breaks in some curves of (a) are related to small numbers of patients with long-term follow-up in these subgroups. These appear smoothed out in (b) because of greater numbers in the validation set.
Performance on external datasets
We applied our classification scheme to a large Affymetrix dataset comprising 2291 breast cancer transcriptomes we have collected from the literature (Supplementary Methods). The six molecular subgroups were perfectly reproduced, both in terms of distribution and clinical correlations and outcomes (Supplementary Table 4a, Figure 2b). To further ascertain its robustness, we tested our classification on three expression datasets from different technological platforms (Swegene, Qiagen/Operon, Eurofins MWG Operon, Roissy, France; and Agilent, Santa Clara, CA, USA). Our prediction rule being designed for Affymetrix datasets we had to adapt it to different technological contexts (Supplementary Methods). Overall molecular subgroups were reproduced on different platforms (Supplementary Table 4b and Supplementary Figure 4). Differences were noted according to the dataset, which may possibly be due to different tumor recruitment in each series. To test inter-platform reproducibility, we classified the GSE3155 dataset that was analyzed in parallel on two dual-color (Agilent and Stanford University, Palo Alto, CA, USA) and one uni-color (Applied Biosystems, Carlsbad, CA, USA) platforms (Supplementary Table 4c). Classification on both dual-color datasets showed a 90% overlap, suggesting a good inter-platform reproducibility. However, overlap dropped dramatically when dual and uni-color platforms were compared (48 and 52%). This indicates that classification rules need adaptation to technological specificities of each platform to perform optimally.
Comparison with other molecular classifiers
We next compared our classification with the Sorlie, Hu and Parker centroids (Sorlie et al., 2003; Hu et al., 2006; Parker et al., 2009). Variable overlaps were found for BasL, LumB, LumA and NormL subgroups (Figure 1d). However, significant differences were noted for the mApo and LumC subgroups, which not only overlapped at variable levels with the ERBB2 subtype, but also with basal-like, luminal-A and –B, and normal-like groups, depending on the classifier (Supplementary Table 5). Classification differences affected the distribution of bioclinical markers among molecular subgroups. Main differences were in the fraction of ER+/PR+ and AR+ tumors in basal-like subtypes and the distribution of ERBB2+ tumors (Supplementary Table 6). MFS curves showed better separation of good and bad outcome subgroups with our classification (the CIT classification) (Supplementary Figures 5 and 6).
Molecular subgroups show differential activation of signaling pathways
We selected 40 cancer relevant pathways from public databases and tested for specific enrichment in our molecular subgroups (Supplementary Methods). Genes specific for each subgroup were identified using four algorithms. Pathways were ordered for each subgroup on the mean rank of P-values across the four methods. As shown in Figure 3, each subgroup was associated to different up or downregulated signaling pathways. The upregulation of DNA replication and repair in BasL and LumB contrasted with its downregulation in NormL. The upregulation 4/5 immune system pathways in LumC was of further note. These data indicate that molecular subgroups relate to different signaling pathways and biological processes.
Figure 3.
Molecular subgroups show differential activation of major signaling pathways: correlations between a given pathway and a subgroup are indicated by color boxes. Red boxes show upregulation of the pathway, green downregulation. Up or downregulation was deduced using KEGGanim tool where relative expression measures are projected in the related KEGG pathway interaction graph. Pathways showing no clear direction of regulation were excluded.
Molecular subgroups show specific genomic anomalies
Of the 537 tumors profiled for RNA expression, 488 tumors were analyzed by array-CGH (comparative genome hybridization). A total of 21 regions of gain and 33 regions of loss were found in more than 30% of the tumors (Figure 4a, top panel). BasL and LumB showed extensive copy-number alterations (CNAs), whereas NormL and LumA were the least rearranged. Qualitative differences were also apparent (Figure 4a) and we searched for CNAs specifically associated to each subgroup. BasL and LumB tumors presented the greatest number of CNAs with 39 and 46 specific CNAs, respectively (Figure 4a, Supplementary Table 7). The number of specific events was lower in the other subgroups ranging from 2 to 8. Expectedly, amplifications at 17q12 were found in 70% of mApo tumors. LumA showed gains at 4q35 and 16p11-p13, whereas NormL tumors could be differentiated from LumA by frequency of gains at 9q33, 8p23, 16p13 and loss at 16q12.
Figure 4.
Breast cancer molecular subgroups present different copy-number change (CNC) profiles. CNC profiles were established using genome-wide array-CGH on a 488 breast tumor dataset and subsequently stratified according to the CIT classification. Panel a shows frequency of gains (vertical bars going up) or losses (bars going down) at a given location on the genome. Graphs from top to bottom correspond to profiles of the whole CIT breast cancer set and each of the six molecular subgroups. Panel b represents regions of CNC correlating to a specific subgroup. Specific genomic regions for the whole CIT set are the ones for which the proportion of alterations (in gain or loss) exceeded 20%. Subgroup-specific regions are those that present significant increase in proportion (at a 0.1 FDR level) in a given subgroup tested against all others. Bars represent P-values after a standard logarithmic transformation.
CNAs were associated to large-scale gene expression modifications. A total of 786 genes comprised in intervals of gains or losses showed significantly modified expression levels. A number of regions of gains overexpressed genes encoding cell cycle and proliferation activators and, conversely, known tumor suppressors, pro-apoptotic or DNA damage checkpoint genes were found downregulated in regions of loss (Supplementary Table 7). These findings suggest that CNAs are part of a selective process associated with tumor progression, with differences from one subgroup to another. In that respect, 28 CNAs presented inverse patterns in different subgroups. These inverted patterns involved mainly BasL and LumB, but were also found in mApo and LumB or LumB and NormL (Supplementary Figure 7). Strikingly, they were associated to inverse expression of key cancer genes. These data support the notion that breast cancer subgroups arise along distinct genetic pathways.
Focal DNA amplification (defined as high-level gains occurring in regions not larger than 3 Mb) occurred significantly more frequently in LumB, mApo and LumC than in the other subgroups (Supplementary Table 8a). We further investigated the occurrence of focal CNAs and analyzed a subset of 72 tumors from the CIT discovery set with high-resolution Illumina 610K-SNP-arrays (Supplementary Table 8b). We detected 246 gains and 337 losses (mean size 132 and 161 kb, respectively). We noted that 53% of the gains were also detected in our BAC-array data, while the overlap was lower for losses (19%). However, gains showed modest copy-number increase and were infrequently recurrent. Losses showed greater recurrence but this corresponded mainly to probable CNVs (identical starts and ends).
We verified the overlap of our subgroups with the recently proposed CNA-based classification (Jönsson et al., 2010) and observed an overall coherence with our findings. Their CNA-based Basal-complex class overlapped with our BasL, 17q12 with part of our mApo and LumC, Luminal complex and amplifier with LumB and LumC, while the Luminal-simple corresponded globally to LumA and NormL (Figure 1d).
Fraction of non-tumor cells and distribution in molecular subgroups
The fraction of non-tumor cells is frequently discussed as a confounding factor in molecular analyses of breast cancer fostering the proposition that the normal-like group was a possible artifact (Prat et al., 2010). To get an objective estimate of the rate of non-diploid cells in our dataset and determine its distribution within molecular subgroups, we computed an estimate based on Illumina 610K-SNP data using a recent formula (Van Loo et al., 2010). Significant differences were seen among molecular subgroups (Supplementary Figure 8a) with, surprisingly, mApo showing the lowest rate of non-diploid cells. NormL ranked third and LumA and LumB presented the highest fraction of non-diploid cells. Our results agreed with recent data (Van Loo et al., 2010). However, a variable fraction of tumor cells may also be diploid, leading to an overestimation of normal cells. To assess this, a histological estimate of the non-tumor cell fraction was performed on the tumors analyzed with the Illumina 610K-SNP-arrays. This showed that SNP-based estimates of non-diploid cells were lower than pathological tumor cell content (Supplementary Figure 8b). Overall these data are coherent with the idea of NormL representing a bonafide breast cancer subgroup.
Breast cancer subgroups and mammary epithelial cell hierarchy
To test whether our subgroups relate to distinct cells of origins in the mammary gland, we took advantage of three published expression profiling datasets of sorted normal mammary epithelial cell subpopulations (Raouf et al., 2008; Lim et al., 2009; Pece et al., 2010). We inferred a signature that discriminated the mammary stem cell (MaSC) enriched, luminal progenitor (LPC), mature luminal (MLC) and stromal cell populations, and used this signature to classify our breast tumor expression data (Supplementary Methods). As shown in Figure 5, the principal component analysis ordered normal mammary epithelial cell fractions according to a differentiation gradient and breast tumors from BasL, mApo, LumC, LumB/NormL to LumA, suggesting a proximity of BasL and mApo with either MaSC or LPC, whereas ER+ subgroups showed a gradient between LPCs and MLCs. The correlation of BasL and mApo with least differentiated cells (MaSC or LPC) in the normal mammary gland was confirmed in a second analysis (Supplementary Table 9).
Figure 5.
Principal component analysis (PCA) of the CIT coreset expression profiles based on a meta-signature comparing normal mammary epithelial cell subpopulations. A 163 gene signature was produced by comparing different normal mammary cell contingents from three independent studies (GSE16997, GSE18931, GSE11395) and used in a PCA. Samples from the CIT coreset (panel a) and normal mammary gland samples (panel b) from GSE16997 were projected in the two first principal components in the upper and lower panel, respectively.
Prognostic significance of molecular subgroups
We next compared the prognostic significance in terms of metastatic relapse of our molecular subgroups to classical prognostic factors (ER, ERBB2/HER2, SBR grading and nodal involvement). As shown in Table 2, our classification signature performed better in both univariate and multivariate analyses than the classical prognostic factors, in both the discovery and validation sets. However, the absence of central pathology review in both datasets prevents us to draw firm conclusions on the independent prognostic power of our signature. In a comparative analysis with five expression signatures (Sorlie et al., 2003; van ‘t Veer et al., 2003; Hu et al., 2006; Sotiriou et al., 2006; Parker et al., 2009), our signature came second after the van't Veer signature in the discovery set and performed best in the validation set (Supplementary Table 10), demonstrating the important difference in terms of prognosis among molecular subgroups.
Table 2. Prognostic significance of the CIT classification.
| Variable |
Univariate analysis |
Multivariate analysis |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Value | HR | 95% CI | P-value modality | P-value model | n | HR | 95% IC | P-value modality | P-value model | n | |
| (a) Clinical parameters | |||||||||||
| CIT (ref=normL) | LumA | 1.66 | 0.84–3.30 | 0.15 | 1.8 × 10−5 | 426 | 1.66 | 0.78–3.52 | 2.5 × 10−4 | 6.4 × 10−6 | 371 |
| Other | 3.16 | 1.82–5.48 | 4.3 × 10−5 | 426 | 2.99 | 1.62–5.51 | |||||
| ER (ref=Pos) | Negative | 1.85 | 1.22–2.81 | 0.003 | 0.003 | 426 | 1.19 | 0.72–1.97 | 0.5 | ||
| ERBB2 (ref=Neg) | Positive | 1.18 | 0.74–1.9 | 0.49 | 0.49 | 426 | 0.89 | 0.52–1.5 | 0.66 | ||
| N (ref=0) | 1+ | 1.43 | 0.85–2.38 | 0.18 | 0.17 | 373 | 1.55 | 0.92–2.63 | 0.1 | ||
| T (ref=[0,1]) | >1 | 2.08 | 1.3–3.31 | 0.0021 | 0.0016 | 422 | 2.21 | 1.3–3.76 | 0.003 | ||
| SBR (ref=1) | 2 | 2.92 | 0.91–9.36 | 0.07 | 3 × 10−4 | 418 | |||||
| 3 | 5.19 | 1.63–16.53 | 0.005 | 418 | |||||||
| Chemotherapy adjuvant (ref=No) | Yes | 1.09 | 0.73–1.62 | 0.67 | 0.67 | 378 | |||||
| Hormononal adjuvant (ref=No) | Yes | 0.64 | 0.44–0.94 | 0.02 | 0.02 | 375 | |||||
| (b) Molecular signatures | |||||||||||
| CIT (ref=NormL) | LumA | 1.66 | 0.84–3.30 | 1.8 × 10−5 | 426 | 2.0 | 0.74–5.33 | 3.7 × 10−1 | 426 | ||
| Other | 3.16 | 1.82–5.48 | 426 | 1.8 | 0.63–5.06 | ||||||
| Sorlie (ref=NormL) | LumA | 1.37 | 0.74–2.52 | 2.4 × 10−3 | 426 | 1.9 | 0.7–5.03 | 5.0 × 10−1 | |||
| Other | 2.29 | 1.31–4.00 | 426 | 1.4 | 0.57–3.29 | ||||||
| Hu (ref=LumA) | NormL | 1.67 | 0.86–3.25 | 9.6 × 10−5 | 426 | 2.7 | 0.98–7.35 | 4.2 × 10−1 | |||
| Other | 2.88 | 1.69–4.93 | 426 | 1.6 | 0.72–3.73 | ||||||
| Parker (ref=LumA) | NormL | 1.43 | 0.74–2.75 | 3.5 × 10−3 | 426 | 1.25 | 0.49–3.18 | 3.5 × 10−1 | |||
| Other | 2.26 | 1.34–3.81 | 426 | 0.8 | 0.36–1.79 | ||||||
| GGI (ref=Low risk) | High risk | 2.51 | 1.60–3.93 | 3.4 × 10−5 | 426 | 1.0 | 0.43–2.47 | 8.0 × 10−1 | |||
| Van′t Veer (ref=Low risk) | High risk | 2.93 | 2.00–4.27 | 5.9 × 10−9 | 426 | 2.8 | 1.53–5.1 | 3.4 × 10−3 | |||
Abbreviations: CI, confidence-interval; CIT, our classification; HR, hazard ratio.
Relative risk was calculated taking metastatic relapse as an endpoint and compared with that of (a) clinical parameters and (b) of three molecular classifiers (Sorlie, Hu, Parker) and two prognostic signature (GGI, Van′t Veer). The dataset comprised 426 patients from the CIT discovery set for which MFS information was available. Complete clinical information was available in 371 cases explaining the smaller numbers in the multivariate analysis on prognostic factors. Prognostic significance was assessed by applying a Cox model. Columns refer to the HR, the 95% CI and the P-values for both univariate and multivariate models.
Molecular subgroups show differential response to chemotherapy
To test whether our classification could predict chemotherapy response, we analyzed three datasets of locally advanced breast cancers treated by neoadjuvant therapy followed by surgery and assessment of the pathological response. ER− breast cancers were overrepresented in the three cohorts, but our signature allowed the assignment of tumors to four subgroups after pooling LumB and LumC, as well as LumA and NormL to reach sufficient sample size by subgroup. Despite different chemotherapy protocols in individual cohorts, obvious differences in response were observed. BasL and mApo showed the best response rates with 50%, and 37% of complete response, respectively. ER+ subgroups showed 15% of complete response in LumB/LumC tumors and 0% in LumA/NormL (Table 3a). Prediction of complete pathological response of the CIT classification was then compared with that of ER status and SBR Grade in the three pooled datasets. Both in the univariate and multivariate analysis the CIT classification showed the strongest score (Table 3b).
Table 3. Differential response to chemotherapy according to molecular subgroups of the CIT classification.
| Treatment | n | Response | BasL | mApo | LumC/LumB | LumA/NormL | P-value | ||
|---|---|---|---|---|---|---|---|---|---|
| (a) Correlation between the molecular subgroups of the CIT classification and pathological complete response to chemotherapy | |||||||||
| Hess | T/FAC | 125 | pCR | 17 (68%) | 11 (32%) | 3 (7%) | 0 (0%) | 2.6 × 10−9 | |
| no pCR | 8 (32%) | 23 (68%) | 41 (93%) | 22 (100%) | |||||
| CIT | EC | 58 | pCR | 8 (53%) | 6 (46%) | 2 (7%) | 0 (0%) | 1.6 × 10−3 | |
| no pCR | 7 (47%) | 7 (54%) | 25 (93%) | 3 (100%) | |||||
| Bonnefoi | FEC | 66 | pCR | 16 (43%) | 7 (41%) | 5 (42%) | NS | ||
| no pCR | 21 (57%) | 10 (59%) | 7 (58%) | ||||||
| TET | 58 | pCR | 17 (45%) | 6 (35%) | 3 (100%) | 0.11 | |||
| no pCR | 21 (55%) | 11 (65%) | 0 (0%) | ||||||
| Total | 307 | pCR | 58 (50%) | 30 (37%) | 13 (15%) | 0 (0%) | 4.3 × 10−10 | ||
| no pCR | 57 (50%) | 51 (63%) | 73 (85%) | 25 (100%) | |||||
|
Univariate Analysis |
Multivariate Analysis |
||||||||
| |
Value |
n |
Odds ratio |
95% CI |
P-value |
n |
Odds ratio |
95% CI |
P-value |
| (b) Uni- and multivariate analyses of factors predictive for pathological complete response to chemotherapy in the three pooled datasets | |||||||||
| ER | ER− | 307 | 4.5 | 2.5–8.4 | 2.1 × 10−08 | 291 | 1.6 | 0.67–4.2 | 0.28 |
| Grade | Grade3 | 291 | 3.2 | 1.8–5.8 | 3.6 × 10−05 | 1.9 | 1–3.5 | 0.04 | |
| CIT molecular classification | BasL/mApo | 307 | 6.1 | 3.1–13 | 7.0 × 10−10 | 3.8 | 1.3–11 | 0.01 | |
Abbreviations: CIT, our classification; NS, not significant; pCR, pathological complete response.
Table 3a shows the correlation between pCR and CIT molecular subgroups. pCR and absence of response (no pCR) to chemotherapy were analyzed in three clinical trials (Hess et al., 2006, Bonnefoi et al., 2007, CIT set). Owing to the small number of data, four main subgroups and two intermediate subgroups were combined into two groups: (LumB; LumC; LumB/C) and (NormL; LumA; NormL/LumA). Treatment description: EC, six cycles of a dose-dense regimen of 75 mg/m2 epirubicin and 1200 mg/m2 cyclophosphamide, given every 14 days; T/FAC, 24 weeks of sequential paclitaxel and fluorouracil-doxorubicin-cyclophosphamide; FEC, fluorouracil, epirubicin, and cyclophosphamide for six cycles; TET, docetaxel for three cycles followed by epirubicin plus docetaxel for three cycles. Correlations were calculated using Fisher exact test. Table 3b shows uni- and multivariate analyses of factors predictive of pCR in the three pooled datasets. Univariate analysis was done using the Fisher exact test and multivariate analysis by logistic regression.
Discussion
Breast cancer heterogeneity, reflected in molecular subgroups, can be attributed to differences in molecular alterations, cellular origin or both. We present a classification of breast cancer into six molecular subgroups, which differed upon gene expression, genomic profiles, differentiation level and clinical features.
First, gene expression differences strongly suggested that they outlined distinct biological entities, reflecting initiating mutations and/or cell-of-origin. Specific sets of signaling pathways were associated to each subgroup.
The distribution of the six subgroups was determined by the combination of the expression of three large gene clusters organized around the (i) estrogen receptor, (ii) androgen receptor and (iii) cell cycle regulator genes. The ER cluster is well known as defining luminal breast tumors (Bertucci et al., 2006) and the expression of AR in breast cancer is long-known (Isola, 1993), but has been confounded with that of the ER cluster (Doane et al., 2006). Its combined expression with the ER cluster yields three broad classes determined by nuclear receptor expression; AR−/ER−/PR− (triple negative) corresponding to the BasL subgroup, AR+/ER−/PR− (mApo), AR+/ER+/PR+ (triple positive) including the four ER+ subgroups. The AR cluster comprises key genes previously associated to the ER cluster, such as the pioneer factor FOXA1, which recruits ER, AR and RAR/RXR (Carroll et al., 2006; Lupien et al., 2008).
The existence of an ER−/AR+ breast tumor subset (our mApo subgroup) has been proposed (Farmer et al., 2005; Doane et al., 2006), and its important overlap with ERBB2/HER2 amplification is intriguing, possibly reflecting cross-talks between the AR and ERBB2/HER2 pathways (Naderi & Hughes-Davies, 2008). However, it is notable that our classification did not define an ERBB2 subgroup. Instead, ERBB2-amplified cancers distributed in mApo (ER−) and LumC (ER+) subgroups. We found less expression differences between mApo/ERBB2+ and mApo/ERBB2− than between mApo and LumC tumors (Supplementary Figure 9). Interestingly, Staaf et al. (2010) showed that ER− and ER+ ERBB2-amplified tumors presented different 17q CNA patterns. These observations could have implications in the clinic as they indicate that ERBB2+ breast cancer correspond to a biologically heterogeneous group. Moreover, it seems important to distinguish ERBB2+ and mApo tumors, because the so-called triple-negative group comprises both BasL and ERBB2−/mApo tumors despite clear molecular and clinical differences.
Second, subgroups were also characterized by different patterns of genomic anomalies. These data were concordant with previous results (Chin et al., 2006; Natrajan et al., 2009) and the CGH classification recently proposed by Jönsson et al. (2010). Moreover, the existence of chromosomal regions showing inverse patterns (gain in one subgroup/loss in another) further supported the notion that these subgroups progress along distinct genetic routes, which possibly involve different mechanisms of genetic instability.
Third, our data indicated that subgroups differed in their differentiation level, pointing to possible differences in cell-of-origin. This was suggested by similarities between the transcriptome of distinct cellular contingents in the normal mammary gland and those of molecular subgroups. While BasL and mApo showed proximity to MaSC or luminal progenitors, ER+ subgroups formed a gradient between LPCs (LumC) and MLC cells (LumA). Our findings are consistent with recent work suggesting that LPCs were the cells of origin of basal cancer and Brca1 mammary tumors (Lim et al., 2009; Molyneux et al., 2010). These findings bring insight on the prevalence of Grade 3 tumors in BasL and mApo contrasting sharply with that of low-grade cancers in NormL and LumA. Our data thus suggest that breast cancer may arise from at least two distinct cell types and that the final phenotype will result from genetic and epigenetic changes occurring during cancer progression. This may also have some link with the striking gradient of TP53 mutations observed between BasL and NormL subgroups. The correlation with elevated expression of the cell-cycle cluster and increased genomic instability was also notable. Moreover, there is a striking parallel between the incidence of TP53 inactivation and the response rates of neoadjuvant chemotherapies. These data are in line with our previous observation proposing that TP53 is not the mediator of chemotherapy-induced cell death (Bertheau et al., 2007).
Fourth, molecular subgroups show striking differences with respect to metastatic relapse both in terms of kinetics and site of recurrence. While BasL and mApo tumors preferentially metastasized to the brain and rarely to the bone, ER+ subgroups exhibited an inverse pattern, strengthening previous studies (Smid et al., 2008). Our data suggest that these differences could be due to differential expression of key metastasis genes (Bos et al., 2009). Hence, metastasis to a specific organ can also be the result of a subgroup-specific gene program and coexist with the de novo acquisition of stochastic mutations, as recently shown by massively parallel sequencing work (Ding et al., 2010; Yachida et al., 2010). Outcomes of the different subgroups were very different as well. BasL and mApo showed earlier relapse, but a remarkably stable MFS for the next 100 months. In contrast, although all ER+ subgroups did better during the first years, a continuous incidence of late relapse was observed. LumB and LumC outcome progressively became worse than that of BasL or mApo. However, a number of recurrences occurring after 5 years in ER+ subgroups are probably linked to interruption of anti-estrogen treatments.
The status of the NormL subgroup is of particular interest because its existence has been put in doubt and attributed to an elevated content of normal cells (Prat et al., 2010). In line with recently published data (Van Loo et al., 2010), we showed that NormL tumors did not present a lower fraction of non-diploid cells than mApo or LumC. Furthermore, our data showed that 70% of NormL tumors showed loss at 16q, further supporting that this subgroup does not result from a co-cluterization of breast tumors presenting smaller fractions of tumor cells.
Our results are in favor of the existence of different breast cancer subtypes bearing distinct biologies and clinical courses. We propose that stratifying breast cancers according to such a classification could be highly beneficial when searching for new prognostic or response to treatment indicators. These would be subgroup specific instead of expressing the differences between highly and poorly proliferating tumors. Furthermore, such a classification, once adapted in a format compatible with clinical setting, could efficiently contribute to disease management. Indeed, the different subgroups outlined here occur in different age groups, metastasize to different organs and exhibit distinct survival kinetics. Similarly, the association with immune system activation pathways in LumC may be indicative for an anti-tumor immunity in this specific subgroup. All of these are clear indications that they represent distinct clinical and biological entities.
Materials and methods
Patients and tumors
A total of 724 primary breast carcinomas were collected and analyzed for expression profiling on Affymetrix U133-Plus2.0 chips and a subset of 488 samples were analyzed by array-CGH. In addition, 58 fine-needle aspiration biopsies from patients undergoing neoadjuvant chemotherapy were analyzed by transcriptome and included in the response-to-chemotherapies set. Full description can be found in Supplementary Table 1 and Table 2. Mean follow-up time was of 65 months. Four RNA from normal human breast tissue were used as reference. Histological grade as well as ER, PR and HER2 levels determination are detailed in the Supplementary Methods.
Discovery and validation sets
Our 724 breast tumor transcriptome dataset was split in a CIT-discovery-set comprising 537 (75%) tumors of which 488 were analyzed by array-CGH and 187 (25%) cases were set apart for the validation-set. The Affymetrix validation set comprised the 187 samples from CIT and 2225 transcriptomes collected from GEO and array-express (Supplementary Table 2).
Expression profiling and data analysis
RNA profiling
Methods used for RNA purification, quality control, fluorescent probe production, hybridization and data processing were essentially as previously described (de Reynies et al., 2009).
Transcriptome analysis and molecular subgroup determination
Our rational was to ensure the greatest possible homogeneity to identified subgroups. Subgroup determination was based on the CIT-discovery-set including 537 transcriptomes and a clustering approach iterating unsupervised and supervised steps (Supplementary Figure 1, Supplementary Methods). Microarray data were first classified with a set of 244 most variant probe sets using in parallel hierarchical clustering, k-means and Gaussian mixture model. Tumors that were assigned to the same group by the three methods were kept, defining a coreset of 355 tumors. Based on this coreset most discriminative genes were selected by analysis of variance and ranked by random-forest, producing a 256 gene signature, leading to the identification of six homogeneous molecular subgroups. Validation datasets were independently classified in the CIT molecular subgroups by applying a classical distance-to-centroid approach, implemented in the citbcmst R package available at the following URL http://cran.r-project.org/web/packages/citbcmst/index.html and coming with a (Sweave) user documentation. The complete classification procedure is detailed in the Supplementary Methods.
Comparison with the Sorlie, Hu and Parker classifiers
Sorlie (Sorlie et al., 2003), Hu (Hu et al., 2006) and Parker (Parker et al., 2009) centroids were retrieved from http://genome-www.stanford.edu/breast_cancer/robustness/data/IntrinsicGeneList.txt, https://genome.unc.edu/pubsup/breastTumor/data/306genes-X-249samples-X-5subtypes+5centroids.xls and https://genome.unc.edu/pubsup/breastGEO/pam50_centroids.txt, respectively. To build the classifiers corresponding clone UniGene_IDs were mapped to Affymetrix (U133A or U133Plus2) probe sets. For Sorlie this was possible for 334 UniGene_IDs gene symbols, for Hu 232 UniGene_IDs and Parker all genes could be directly mapped.
Comparison with the Jönsson array-CGH-based classification
The 6 Jönsson centroids are relative to genomic regions determined with the GISTIC algorithm (Jönsson et al., 2010). Details are provided in the Supplementary Methods.
Cancer pathways analysis
Cancer relevant pathways were retrieved from KEGG (ftp://ftp.genome.ad.jp/pub/kegg/pathways/hsa), Biocarta (http://www.biocarta.com) and GO (http://www.geneontology.org/), and related genes were mapped to non-redundant HUGO Gene symbols. Four gene set analysis methods were used (Supplementary Methods), yielding P-values, which were transformed into ranks. Gene sets were ranked by order of interest according to the mean of the ranks across the four methods.
Array-CGH
Array-CGH was performed on a 4434 BAC-array with a median resolution of 0.6 Mb. DNA labeling, hybridization and data processing are as described in the Supplementary Methods.
Statistical tests
Clinical correlations were determined by χ2 for qualitative factors and analysis of variance for quantitative variables. Disease outcome was investigated with Kaplan–Meier curves using metastatic recurrence as an endpoint and subgroup for stratification. MFS was calculated from the date of diagnosis until first metastatic relapse. P-values at 60 and 180 months resulted from a log-rank test on Cox estimates. Benjamini and Hochberg method was applied for multiple-testing adjustment.
Acknowledgments
This work was supported by the Carte d'Identité des Tumeurs Program from the Ligue Nationale Contre le Cancer. We thank Pr Jacqueline Godet and Dr Jacqueline Métral for their constant support during the course of this work. The expertize of the personnel serving the RNA extraction and qualification (Saint-Louis Hospital, Paris), Affymetrix expression array (IGBMC, Strasbourg), CIT database and array-CGH (IRCM, Montpellier) CIT platforms is gratefully acknowledged. We further thank Dr Stan du Manoir for critical reading of the manuscript, Vessela N Kristensen and Peter Van Loo for making their data available to us.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc)
Supplementary Material
References
- Bertheau P, Turpin E, Rickman DS, Espie M, de Reynies A, Feugeas JP, et al. Exquisite sensitivity of TP53 mutant and basal breast cancers to a dose-dense epirubicin-cyclophosphamide regimen. PLoS Med. 2007;4:e90. doi: 10.1371/journal.pmed.0040090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertucci F, Finetti P, Cervera N, Maraninchi D, Viens P. Gene expression profiling and clinical outcome in breast cancer. Omics. 2006;10:429–443. doi: 10.1089/omi.2006.10.429. [DOI] [PubMed] [Google Scholar]
- Bertucci F, Orsetti B, Negre V, Finetti P, Rouge C, Ahomadegbe JC, et al. Lobular and ductal carcinomas of the breast have distinct genomic and expression profiles. Oncogene. 2008;27:5359–5372. doi: 10.1038/onc.2008.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnefoi H, Potti A, Delorenzi M, Mauriac L, Campone M, Tubiana-Hulin M, et al. Validation of gene signatures that predict the response of breast cancer to neoadjuvant chemotherapy: a substudy of the EORTC 10994/BIG 00-01 clinical trial. Lancet Oncol. 2007;8:1071–1078. doi: 10.1016/S1470-2045(07)70345-5. [DOI] [PubMed] [Google Scholar]
- Bos PD, Zhang XH, Nadal C, Shu W, Gomis RR, Nguyen DX, et al. Genes that mediate breast cancer metastasis to the brain. Nature. 2009;459:1005–1009. doi: 10.1038/nature08021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll JS, Meyer CA, Song J, Li W, Geistlinger TR, Eeckhoute J, et al. Genome-wide analysis of estrogen receptor binding sites. Nat Genet. 2006;38:1289–1297. doi: 10.1038/ng1901. [DOI] [PubMed] [Google Scholar]
- Chin K, DeVries S, Fridlyand J, Spellman PT, Roydasgupta R, Kuo W-L, et al. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell. 2006;10:529–541. doi: 10.1016/j.ccr.2006.10.009. [DOI] [PubMed] [Google Scholar]
- de Reynies A, Assie G, Rickman DS, Tissier F, Groussin L, Rene-Corail F, et al. Gene expression profiling reveals a new classification of adrenocortical tumors and identifies molecular predictors of malignancy and survival. J Clin Oncol. 2009;27:1108–1115. doi: 10.1200/JCO.2008.18.5678. [DOI] [PubMed] [Google Scholar]
- Ding L, Ellis MJ, Li S, Larson DE, Chen K, Wallis JW, et al. Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature. 2010;464:999–1005. doi: 10.1038/nature08989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doane AS, Danso M, Lal P, Donaton M, Zhang L, Hudis C. An estrogen receptor-negative breast cancer subset characterized by a hormonally regulated transcriptional program and response to androgen. Oncogene. 2006;25:3994–4008. doi: 10.1038/sj.onc.1209415. [DOI] [PubMed] [Google Scholar]
- Farmer P, Bonnefoi H, Becette V, Tubiana-Hulin M, Fumoleau P, Larsimont D, et al. Identification of molecular apocrine breast tumours by microarray analysis. Oncogene. 2005;24:4660–4671. doi: 10.1038/sj.onc.1208561. [DOI] [PubMed] [Google Scholar]
- Foulkes WD, Smith IE, Reis-Filho JS. Triple-negative breast cancer. N Engl J Med. 2010;363:1938–1948. doi: 10.1056/NEJMra1001389. [DOI] [PubMed] [Google Scholar]
- Gusterson B. Do ‘basal-like' breast cancers really exist. Nat Rev Cancer. 2009;9:128–134. doi: 10.1038/nrc2571. [DOI] [PubMed] [Google Scholar]
- Hess KR, Anderson K, Symmans WE, Valero V, Ibrahim N, Mejia JA, et al. Pharmacogenomic predictor of sensitivity to preoperative chemotherapy with paclitaxel and fluorouracil, doxorubicin, and cyclophosphamide in breast cancer. J Clin Oncol. 2006;24:4236–4244. doi: 10.1200/JCO.2006.05.6861. [DOI] [PubMed] [Google Scholar]
- Hu Z, Fan C, Oh DS, Marron JS, He X, Qaqish BF, et al. The molecular portraits of breast tumors are conserved across microarray platforms. BMC Genomics. 2006;7:96. doi: 10.1186/1471-2164-7-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isola JJ. Immunohistochemical demonstration of androgen receptor in breast cancer and its relationship to other prognostic factors. J Pathol. 1993;170:31–35. doi: 10.1002/path.1711700106. [DOI] [PubMed] [Google Scholar]
- Jönsson G, Staaf J, Vallon-Christersson J, Ringnér M, Holm K, Hegardt C, et al. Genomic subtypes of breast cancer identified by array-comparative genomic hybridization display distinct molecular and clinical characteristics. Breast Cancer Res. 2010;12:R42. doi: 10.1186/bcr2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapp AV, Jeffrey SS, Langerod A, Borresen-Dale AL, Han W, Noh DY, et al. Discovery and validation of breast cancer subtypes. BMC Genomics. 2006;7:231. doi: 10.1186/1471-2164-7-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim E, Vaillant F, Wu D, Forrest NC, Pal B, Hart AH, et al. Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat Med. 2009;15:907–913. doi: 10.1038/nm.2000. [DOI] [PubMed] [Google Scholar]
- Lupien M, Eeckhoute J, Meyer CA, Wang Q, Zhang Y, Li W, et al. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell. 2008;132:958–970. doi: 10.1016/j.cell.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molyneux G, Geyer FC, Magnay F-A, McCarthy A, Kendrick H, Natrajan R, et al. BRCA1 basal-like breast cancers originate from luminal epithelial progenitors and not from basal stem cells. Cell Stem Cell. 2010;7:403–417. doi: 10.1016/j.stem.2010.07.010. [DOI] [PubMed] [Google Scholar]
- Naderi A, Hughes-Davies L. A functionally significant cross-talk between androgen receptor and ErbB2 pathways in estrogen receptor negative breast cancer. Neoplasia. 2008;10:542–548. doi: 10.1593/neo.08274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natrajan R, Lambros MB, Rodríguez-Pinilla SM, Moreno-Bueno G, Tan DSP, Marchió C, et al. Tiling path genomic profiling of grade 3 invasive ductal breast cancers. Clin Cancer Res. 2009;15:2711–2722. doi: 10.1158/1078-0432.CCR-08-1878. [DOI] [PubMed] [Google Scholar]
- Parker JS, Mullins M, Cheang MCU, Leung S, Voduc D, Vickery T, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27:1160–1167. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pece S, Tosoni D, Confalonieri S, Mazzarol G, Vecchi M, Ronzoni S, et al. Biological and molecular heterogeneity of breast cancers correlates with their cancer stem cell content. Cell. 2010;140:62–73. doi: 10.1016/j.cell.2009.12.007. [DOI] [PubMed] [Google Scholar]
- Prat A, Parker JS, Karginova O, Fan C, Livasy C, Herschkowitz JI, et al. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 2010;12:R68. doi: 10.1186/bcr2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raouf A, Zhao Y, To K, Stingl J, Delaney A, Barbara M, et al. Transcriptome analysis of the normal human mammary cell commitment and differentiation process. Cell Stem Cell. 2008;3:109–118. doi: 10.1016/j.stem.2008.05.018. [DOI] [PubMed] [Google Scholar]
- Smid M, Wang Y, Zhang Y, Sieuwerts AM, Yu J, Klijn JG, et al. Subtypes of breast cancer show preferential site of relapse. Cancer Res. 2008;68:3108–3114. doi: 10.1158/0008-5472.CAN-07-5644. [DOI] [PubMed] [Google Scholar]
- Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci USA. 2003;100:8418–8423. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotiriou C, Wirapati P, Loi S, Harris A, Fox S, Smeds J, et al. Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J Natl Cancer Inst. 2006;98:262–272. doi: 10.1093/jnci/djj052. [DOI] [PubMed] [Google Scholar]
- Staaf J, Jönsson G, Ringnér M, Vallon-Christersson J, Grabau D, Arason A, et al. High-resolution genomic and expression analyses of copy number alterations in HER2-amplified breast cancer. Breast Cancer Res. 2010;12:R25. doi: 10.1186/bcr2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van ‘t Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Bernards R. Expression profiling predicts outcome in breast cancer. Breast Cancer Res. 2003;5:57–58. doi: 10.1186/bcr562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Loo P, Nordgard SH, Lingjærde OC, Russnes HG, Rye IH, Sun W, et al. Allele-specific copy number analysis of tumors. Proc Natl Acad Sci USA. 2010;107:16910–16915. doi: 10.1073/pnas.1009843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigelt B, Mackay A, A'Hern R, Natrajan R, Tan DS, Dowsett M, et al. Breast cancer molecular profiling with single sample predictors: a retrospective analysis. Lancet Oncol. 2010;11:339–349. doi: 10.1016/S1470-2045(10)70008-5. [DOI] [PubMed] [Google Scholar]
- Yachida S, Jones S, Bozic I, Antal T, Leary R, Fu B, et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 2010;467:1114–1117. doi: 10.1038/nature09515. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.