Abstract
Background. Oseltamivir resistance among 2009 pandemic influenza A (H1N1) viruses (pH1N1) is rare. We investigated a cluster of oseltamivir-resistant pH1N1 infections in a hospital ward.
Methods. We reviewed patient records and infection control measures and interviewed health care personnel (HCP) and visitors. Oseltamivir-resistant pH1N1 infections were found with real-time reverse-transcription polymerase chain reaction and pyrosequencing for the H275Y neuraminidase (NA) mutation. We compared hemagglutinin (HA) sequences from clinical samples from the outbreak with those of other surveillance viruses.
Results. During the period 6–11 October 2009, 4 immunocompromised patients within a hematology-oncology ward exhibited symptoms of pH1N1 infection. The likely index patient became febrile 8 days after completing a course of oseltamivir; isolation was instituted 9 days after symptom onset. Three other case patients developed symptoms 1, 3, and 5 days after the index patient. Three case patients were located in adjacent rooms. HA and NA sequences from case patients were identical. Twelve HCP and 6 visitors reported influenza symptoms during the study period. No other pH1N1 isolates from the hospital or from throughout the state carried the H275Y mutation.
Conclusions. Geographic proximity, temporal clustering, presence of H275Y mutation, and viral sequence homology confirmed nosocomial transmission of oseltamivir-resistant pH1N1. Diagnostic vigilance and prompt isolation may prevent nosocomial transmission of influenza.
The 2009 pandemic influenza A (H1N1) virus (pH1N1) spread globally after being identified in April 2009 [1]. Oseltamivir, an oral neuraminidase (NA) inhibitor, is widely used for treatment and chemoprophylaxis of pH1N1 infection. Sporadic cases of oseltamivir-resistant pH1N1 have been identified [2], and only 2 clusters of possible person-to-person transmission of oseltamivir-resistant virus have been reported [3, 4]. Given the limited number of treatment options, the potential for widespread transmission of oseltamivir-resistant pH1N1 is a public health concern.
In September 2009, 4 patients were admitted to a hematology-oncology ward at Duke University Medical Center (DUMC). These patients were admitted for reasons unrelated to influenza infection. All 4 patients exhibited fever or respiratory symptoms while in the hospital and subsequently received a diagnosis of pH1N1 infection. All pH1N1 viral isolates from these patients carried the H275Y substitution in the NA gene, which is a mutation associated with oseltamivir resistance [5]. The hospital collaborated with the North Carolina Department of Health and Human Services (NC DHHS) and the Centers for Disease Control and Prevention (CDC) to investigate potential nosocomial transmission of oseltamivir-resistant pH1N1.
METHODS
Epidemiologic Investigation
We defined a case as laboratory-confirmed oseltamivir-resistant pH1N1 infection as demonstrated by presence of the H275Y mutation (through pyrosequencing) in any patient admitted to the hematology-oncology ward during the period 15 September–4 December 2009 (the study period).
We reviewed medical records of patients and collected information regarding reasons for admission, comorbidities, symptoms, patient location within the hospital, and antiviral use. We specifically reviewed patient records, examined work logs, and interviewed health care personnel (HCP), volunteers, and visitors to identify potential exposure to case patients. We defined a 7-day period before and including the date of symptom onset as the potential exposure period for each case patient (a continuous period of 29 September–11 October 2009) [6]. We defined date of symptom onset by new or unexplained fever or respiratory symptoms (cough, shortness of breath, sore throat, or inadequate oxygen saturation on oximetry).
We conducted a case-control study to identify clinical and demographic features associated with acquisition of oseltamivir-resistant pH1N1. We reviewed laboratory records during the study period and identified case patients as inpatients with confirmed oseltamivir-resistant pH1N1 infections, and control subjects were inpatients with wild-type pH1N1 infection.
A point-prevalence study was conducted to detect additional cases of oseltamivir-resistant pH1N1 infection. During the period 20–21 November 2009, nasopharyngeal specimens were collected from inpatients of the affected hematology-oncology ward and from patients who were hospitalized in the medical intensive care unit (ICU) at the same time as case patients. We also determined prevalence of oseltamivir-resistant pH1N1 by pyrosequencing pH1N1 specimens submitted to NC DHHS for routine influenza surveillance from counties that surround DUMC and in counties where the case patients lived during October 2009.
Additionally, we performed anonymous structured interviews with HCP, volunteers, and visitors who potentially had interacted with case patients to determine factors that might have facilitated nosocomial transmission of pH1N1.
Finally, we reviewed prevailing infection control procedures during the outbreak period and conducted observations for adherence to hand hygiene and isolation precautions. We reviewed data related to hand hygiene adherence independently recorded by auditors at DUMC.
The study was conducted under the auspices of outbreak investigation and protection of health of the community. An institutional review board approval was waived.
Laboratory Investigation
Confirmed pH1N1 isolates from 4 case patients underwent NA-275 pyrosequencing and conventional Sanger sequencing of HA and NA genes. Pyrosequencing was performed for viruses obtained from 73 other patients with pH1N1 infection diagnosed at the hospital. The HA regions of 11 of these 73 isolates were sequenced and used in phylogenetic analysis. An additional 145 unrelated surveillance specimens of pH1N1 from North Carolina were screened for the H275Y mutation.
The Global Initiative on Sharing Avian Influenza Data accession numbers for viruses from case patients are as follows: HA sequences from clinical specimens: EPI233164, EPI233171, EPI233179, and EPI233183; HA sequences of viral cultures: EPI233170, EPI233175, EPI233178, and EPI233181 and NA sequences of viral cultures: EPI233169, EPI233174, EPI233177, and EPI233182. Accession numbers of HA sequences from other patient specimens used in Figure 2 are pending.
Identification of 2009 Pandemic H1N1 Influenza.
The hospital microbiology laboratory used ProFlu+ (Gen-Probe) or direct fluorescent antibody staining to diagnose influenza A. The laboratory at NC DHHS performed real-time reverse-transcription polymerase chain reaction (rRT-PCR) assays to confirm pH1N1 in specimens from the hospital and in state-wide surveillance specimens.
rRT-PCR and Pyrosequencing.
Pyrosequencing was performed to detect the oseltamivir resistance marker, H275Y, on NA genes [7]. After RNA extraction, region encompassing codon 275 was amplified with primers H1N1pdm-N1-F780 and H1N1pdm-N1-R1273-biot. rRT-PCR products were subjected to pyrosequencing buffer washes and generated single-stranded DNA templates. H1N1pdm-N1-F804 was then used to sequence through the region of interest [7].
HA Amplification and Sanger Sequencing.
For full-length HA open reading frame amplification, F-1/R454 and F316/R-1778 were used to generate two overlapping amplicons. Superscript III One-Step RT-PCR System with Platinum Taq High Fidelity (Invitrogen) was used with the primers shown in Table 4.
Table 4.
Primers used for Amplification and Sequencing of the HA Gene of Pandemic Influenza A (H1N1) Virus (pH1N1) in this Study
| RT-PCR primers (20 μM) | Sequence |
| H1N1pdm-HA-F1 H1N1pdm-HA-R454 (Fragment 1) |
5'- ATGAAGGCAATACTAGTAG -3' 5'- CTGCCGTTACACCTTTG -3' |
| H1N1pdm-HA-F316 H1N1pdm-HA-R1778 (Fragment 2) |
5'- ACRTGTTACCCWGGRGATTTCA -3' 5'- TGTCAGTAGAAACAAGGGTGTTT -3' |
| Sequencing Primers (1 μM) | |
| H1N1pdm-HA-F1 H1N1pdm-HA-F316 H1N1pdm-HA-R454 H1N1pdm-HA-F727 H1N1pdm-HA-R928 H1N1pdm-HA-R1238 H1N1pdm-HA-R1505 H1N1pdm-HA-R1778 |
5'- ATGAAGGCAATACTAGTAG -3' 5'- ACRTGTTACCCWGGRGATTTCA -3' 5'- CTGCCGTTACACCTTTG -3' 5'- AGRATGRACTATTACTGGAC -3' 5'- GAAAKGGGAGRCTGGTGTTTA -3' 5'- TCTTTACCYACTRCTGTGAA -3' 5'- TCATAAGTYCCATTTYTGA -3' 5'- TGTCAGTAGAAACAAGGGTGTTT -3' |
NOTE. Primer numbers are based on full H1 hemagglutinin gene starting from the ATG with the signal peptide. RT-PCR, reverse-transcriptase polymerase chain reaction.
After RNA extraction, HA and NA genes of viral isolates from cell culture supernatants were amplified, as described elsewhere [8, 9]. Sequencing was performed using Big Dye Terminator chemistry, version 3.1 (Applied Biosystems). Sequencing reaction products were resolved with Applied Biosystems 3730 ABI sequencer, and sequences were generated with Sequencher, version 4.7 (Gene Codes).
Phylogenetic Analysis.
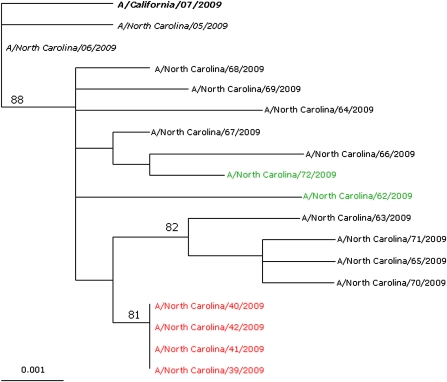
HA sequences from pH1N1 viruses were aligned, and phylogenetic trees were inferred using maximum likelihood method in the Genetic Algorithm for Rapid Likelihood Inference (GARLI .96b7) package using General Time Reversible (GTR) + I + γ4 substitution [10, 11]. Trees were visualized in TreeView, version 1.6.6 [12]. (Page, R. D. M. 1996. TREEVIEW: An application to display phylogenetic trees on personal computers. Computer Applications in the Biosciences 12: 357--358). Bootstrap values were determined for 1000 replicates, with topologies inferred using the neighbor-joining method [10]. Phylogenetic tree was rooted with the pandemic vaccine strain A/California/07/2009 (Figure 2).
Figure 2.
Phylogenetic tree illustrating genetic relation between hemagglutinin (HA) of viruses from the outbreak and from specimens obtained in other areas of the hospital and for surveillance ( n = 17). Numbers indicate maximal likelihood value. Scale bar indicates nucleotide substitutions per site. The 2009 pandemic vaccine strain (bold italic), 4 identified oseltamivir-resistant strains from this outbreak (red), strains from two other patients from the same ward before and after the outbreak (green), and 2 strains collected early in the pandemic (italic) are shown. Other strains shown are 9 strains isolated from different localities in North Carolina.
Neuraminidase Inhibition Assay.
NA-Star neuraminidase inhibition (NI) assay (Applied Biosystems) was used to assess susceptibility to oseltamivir carboxylate, zanamivir, and peramivir [13]. We determined the concentration of NA inhibitors (NAIs) needed to inhibit 50% of NA enzyme. Statistical analyses were performed by using Fisher's exact test with SAS, version 9.2 (SAS).
RESULTS
Epidemiologic Investigation
Four cases of oseltamivir-resistant pH1N1 infection were identified within the hematology-oncology ward during the period 15 September–4 December 2009. Mean age of case patients was 57 years (range, 43–67 years) (Table 1). These 4 patients all had malignancies and were admitted for reasons unrelated to influenza infection.
Table 1.
Line Listing and Characteristics of 4 Case Patients with Oseltamivir-Resistant Pandemic Influenza A (H1N1) Virus (pH1N1) Infection during the Outbreak
| Patient | Date of influenza symptom onset (2009) | Underlying condition | Temperature >37.8°C | Cough | Sore throat | Dyspnea | Concurrent comorbidities | Prior oseltamivir use | No. of days between symptom onset and laboratory confirmation of pH1N1 |
| A | 6 Oct | AML | + | + | – | + | Escherichia coli bacteremia, neutropenia, colitis | + | 9 |
| B | 7 Oct | ALL | + | – | – | + | – | – | 7 |
| C | 9 Oct | MF | + | + | – | – | Line-related bacteremia, IFN | – | 13 |
| D | 11 Oct | Metastatic thymoma | – | – | – | + | Recent thoracotomy | – | 3 |
NOTE. ALL, acute lymphoblastic leukemia; AML, acute myeloid leukemia; IFN, interferon; MF, mycosis fungoides; pH1N1, 2009 pandemic influenza A (H1N1) virus.
All 4 case patients exhibited symptoms attributable to pH1N1 infection during the period 6 October–11 October 2009 (Figure 1). All had experienced dyspnea or cough before diagnosis, but none had sore throat (Table 1). Three case patients had experienced temperatures ≥37.8°C; however, alternate explanations for fever were present, including bacteremia, recent interferon therapy, and recent surgery. Mean time between admission to the ward and symptom onset was 15 days (range, 12–17 days); mean time from symptoms to first positive pH1N1 test result was 8 days (range, 2–13 days). For 1 patient, diagnosis of pH1N1 infection occurred posthumously.
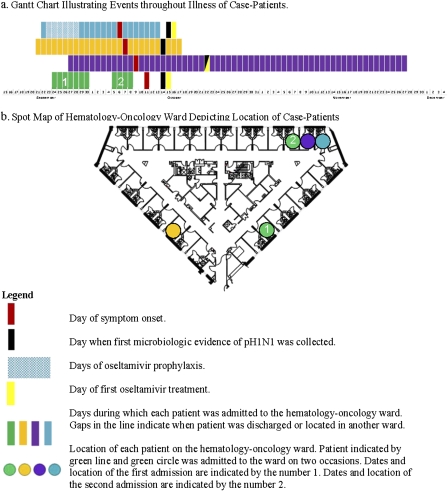
Figure 1.
A, Gantt chart illustrating key events in illness timeline. B, spot map illustrating the location of confirmed cases of oseltamivir-resistant pandemic influenza A (H1N1) virus (pH1N1) infection within the hematology-oncology ward during the outbreak. Each patient is represented by a color. Each colored line in the Gantt chart represents the date on which a confirmed case patient (A–D) was admitted to the affected ward. Each patient's room within the hematology-oncology ward is indicated by circles with corresponding colors. Bronchoalveolar lavage (BAL) specimens collected from patient A on 15 October 2009 and 27 October 2009, as well as their grown isolates, were tested. One BAL specimen collected from patient B on 14 October 2009 and its grown isolate were also tested. Two nasal wash specimens collected from patient C on 22 October 2009 and 29 October 2009, in addition to a grown isolate, were also tested. Two nasal swab specimens collected from patient D on 14 October and 23 October 2009 and a grown virus were tested.
The likely index case patient (patient A) was well at admission for scheduled induction chemotherapy. She received oseltamivir (75 mg twice per day) for 7 days after exposure to a family member who had received a diagnosis of influenza (Figure 1a). No screening for influenza was performed prior to receipt of oseltamivir. Later, she was the first case patient to exhibit symptoms of influenza. Three other case patients did not receive oseltamivir before collection of respiratory specimens in which oseltamivir-resistant pH1N1 virus was detected.
Three of 4 case patients required transfer to the medical ICU for worsening respiratory status. Three patients were treated with oseltamivir at a dosage of 75 mg twice per day after pH1N1 infection was diagnosed or suspected. The dose of oseltamivir was increased to 150 mg twice per day for all 3 patients at various times during treatment. Repeat rRT-PCR testing of respiratory specimens from 3 case patients revealed persistent viral shedding for ≥7 days after initiation of oseltamivir therapy (median duration of documented virus detection, 16 days; range, 8–29 days). One patient received intravenous zanamivir after oseltamivir resistance was confirmed, but pH1N1 remained detectable for ≥13 days after initiation of intravenous zanamivir. Although all 4 case patients died, 2 clinically recovered from their influenza infections and were able to continue chemotherapy for underlying malignancies.
Case patients were admitted to the same ward during the period 21–27 September 2009 (Figure 1a). All 4 were patients within the ward during the same 8 days after 27 September. Three were patients within the ward continuously from 27 September through 13 October 13. One patient was admitted to the ward twice, first during the period 24 September–30 September and again from 5 October–8 October 2009.
The affected ward consists of 32 single-patient rooms with en-suite bathrooms. Three patients in the cluster were in close proximity within the ward (Figure 1b). Two patients resided in adjacent rooms during the period 27 September–13 October 2009. A third case patient was admitted to a room adjacent to these 2 case patients during a second hospitalization, 5–8 October 2009. The final case patient was located on the same ward from 21 September–17 October, but in a room that was distant from the other case patients. All case patients were ambulatory and periodically left the ward and interacted with staff, visitors, and other patients while outside of their rooms. The amount of interaction between case patients was unclear. Neutropenic patients were asked to wear a surgical mask while outside of their rooms; however, adherence to this policy was not monitored.
Our case-control study showed that features associated with being in the hematology-oncology ward were also associated with development of oseltamivir-resistant pH1N1 infection, including immunosuppression, malignancy, and recent chemotherapy (Table 2). Receipt of oseltamivir within 1 month of symptom onset occurred in 1 of 4 case patients and in none of the control subjects; however, this was not a statistically-significant association.
Table 2.
Case-Control Comparison of Patients with and without Oseltamivir-Resistant Pandemic Influenza A (H1N1) Virus (pH1N1) Infection during the Outbreak
| Factor | Case patients (n = 4) | Control subjects (n = 17) | P |
| Age, mean yeas | 57 | 48 | .35 |
| Age >25 years | 4 (100) | 16 (94) | >.99 |
| Age >45 years | 3 (75) | 11 (65) | >.99 |
| White race | 4 (100) | 12 (71) | .53 |
| Male sex | 1 (25) | 7 (41) | >.99 |
| Hematology-oncology ward | 4 (100) | 2 (12) | .003 |
| Medical ICU ward | 3 (75) | 7 (41) | .31 |
| Immunosuppressiona | 4 (100) | 5 (29) | .02 |
| Malignancy | 4 (100) | 3 (18) | .006 |
| Oseltamivir exposureb | 1 (25) | 0 | .19 |
| Any underlying illness | |||
| History of chemotherapyc | 4 (100) | 2 (12) | .003 |
| History of radiotherapyd | 1 (25) | 1 (6) | .35 |
| Current smoker | 1 (25) | 5 (29) | >.99 |
| History of COPD or emphysema | 2 (50) | 6 (35) | .62 |
| Alternate etiology for fever and respiratory symptoms | 4 (100) | 5 (29) | .02 |
| Fever at diagnosis | 3 (75) | 13 (76) | >.99 |
| Cough within 1 week of pH1N1 diagnosis | 2 (50) | 15 (88) | .15 |
| Cough at time of diagnosis | 1 (25) | 11 (65) | .12 |
| Sore throat | 0 (0) | 0 (0) | |
| ILI (fever with cough or sore throat) | 2 (50) | 9 (53) | >.99 |
| Myalgia | 0 | 2 (12) | >.99 |
| Dyspnea | 3 (75) | 12 (71) | >.99 |
| Oxygen desaturation | 3 (75) | 8 (47) | .59 |
| Radiograph changese | 2 (50) | 10 (59) | >.99 |
| ANC <1 at admission | 0 | 2 (12) | >.99 |
| WCC <3 at admission | 1 (25) | 1 (6) | .28 |
| ANC <1 at diagnosis | 1 (25) | 0 | .17 |
| WCC <3 at diagnosis | 2 (50) | 3 (18) | .23 |
NOTE. Data are no. (%) of subjects, unless otherwise indicated. Boldface type indicates statistical significance. ANC, absolute neutrophil count; COPD, chronic obstructive pulmonary disease; ICU, intensive care unit; ILI, influenza-like illness; WCC, white cell count.
Defined as human immunodeficiency virus infection, corticosteroid therapy, or neutropenia.
Documented use of at least 1 dose of oseltamivir before pH1N1 diagnosis.
Any chemotherapy within the last 6 months.
Radiotherapy within the last 6 months.
Radiograph revealing new interstitial or bilateral infiltrates.
Two hundred forty-two HCP and volunteers had documented exposure to at least 1 case patient or were assigned to the ward during September and October. A total of 190 persons (79%) were interviewed during the period 24 November–2 December 2009. Three persons denied being on the affected ward; 3 declined interview; and 46 others could not be contacted. Of 107 HCP with documented exposure to the case patients, 97 (91%) were interviewed.
Twelve (6%) HCP reported influenza-like illness (ILI) (fever plus cough or sore throat), during the period 14 September–24 November 2009. Five of these HCP reported working ≥1 day while symptomatic. We were not able to confirm whether these HCP came into contact with case patients while they were symptomatic. Five HCP reported oseltamivir use (3 for prophylaxis, and 2 for treatment).
Vaccine against pH1N1 was in limited supply during the outbreak. A total of 108 (57%) of 190 HCP and volunteers who were interviewed reported having been vaccinated against pH1N1, and 87 (81%) of the interviewees provided a vaccination date, ranging from 3 September 2009 through 23 November 2009.
We also interviewed 30 of 51 visitors to at least 1 case patient (including the likely index patient) during the period 14 September–24 November 2009. These interviews occurred during the period 11–30 December 2009. Six visitors reported ILI during the period 14 September–24 November 2009. Three had used oseltamivir. No clinical specimens were obtained from visitors. One visitor with influenza A diagnosed by an externally performed rapid test reported visiting the likely index patient before she developed symptoms.
Certain enhanced infection-prevention measures were implemented to limit the transmission of pH1N1 before detection of this outbreak, including a hospital-wide recommendation that only adult family members or caregivers visit patients and a campaign to monitor and improve hand hygiene with use of independent auditors. Auditors performed 330 observations of hand hygiene throughout the affected ward from September through November 2009. Mean adherence to hand hygiene among HCP was 92%.
The hospital did not require preemptive isolation of patients while awaiting results of viral testing. However, clinicians could initiate precautions if clinical suspicion for influenza was high. All patients with confirmed pH1N1 infections were routinely placed on droplet precautions. Additionally, contact precautions were instituted for immunocompromised patients with confirmed pH1N1.
During 2.5 h of observation on the affected ward, 18 (100%) of the staff members complied with posted signage regarding isolation precautions, including correct use of personal protective equipment. One family member entered a contact precautions room wearing a gown but no gloves.
Air-pressure testing indicated positive air pressure relative to the corridor for the rooms of 3 case patients. One patient's room, located at a distance from those of the other 3 case patients (Figure 1), had mildly negative air pressure relative to the corridor.
Laboratory Investigation
Bronchoalveolar lavage (BAL) specimens from case patients A and D, nasal washes (NWs) from case patient B, and nasal swab (NS) specimens from case patient C were available for testing. rRT-PCR analysis confirmed that all were infected with pH1N1.
H275Y was found in viral isolates from various clinical specimens of the 4 case patients using pyrosequencing and was then confirmed by Sanger sequencing (see Figure 2 for details of the specimen types tested from each patient). Additional pyro- and Sanger sequencing performed on the propagated viruses showed the same results.
Mixtures of wild-type and mutant variants (275H/Y) were detected only in the BAL specimen from patient A, which was collected on 16 October 2009, and in its grown isolate. The BAL specimen had ∼75% of the H275Y variant and ∼25% of the wild-type H275. Upon propagation, the proportion of the H275Y variant in the isolate increased to ∼91%, whereas the wild-type variant ratio was reduced to 9%. The remaining samples (clinical specimen and grown viruses) from 4 case patients each contained 100% of the H275Y variant.
NA pyrosequencing allowed detection of both minor populations (9% and 25%) in the clinical specimen and virus isolate, respectively, whereas Sanger sequencing did not allow detection of any of the minor populations of the wild-type virus. These proportions of minor variants were below the reported limit of detection using Sanger sequencing [14, 15] and were indeed not detected.
Analysis from the NI assays is shown in Table 3. All 4 isolates from case patients had similar high 50% inhibitory concentrations (IC50) for oseltamivir, which are characteristic of oseltamivir-resistant viruses, whereas their IC50 values against zanamivir were comparable to those of the zanamivir-susceptible reference viruses and were within normal ranges. IC50 values against peramivir were 443–472-fold higher, compared with the average IC50 of the control oseltamivir-susceptible virus.
Table 3.
Results of Neuraminidase Inhibition and Pyrosequencing Assay Testing of the Viral Isolates Obtained from the 4 Case Patients with Pandemic Influenza A (H1N1) Virus (pH1N1) Infection during the Outbreak
| Isolatea | Pyrosequencingb | IC50, nM |
||
| Oseltamivir | Zanamivir | Peramivir | ||
| A/North Carolina/39/2009 | H275Y | 70.96 | 0.31 | 8.51 |
| A/North Carolina/40/2009 | H275Y | 75.10 | 0.33 | 7.68 |
| A/North Carolina/41/2009 | H275Y | 72.83 | 0.33 | 8.75 |
| A/North Carolina/42/2009 | H275Y | 75.61 | 0.34 | 8.43 |
| Reference virusesc | ||||
| A/California/07/2009 (WT) | 275H | 0.16 | 0.21 | 0.13 |
| A/Texas/48/2009 (Mutant) | H275Y | 76.29 | 0.39 | 11.29 |
NOTE. H275Y is a mutation associated with oseltamivir resistance. IC50 values are the average of 2–4 experiments. IC50, the concentration of a drug needed to inhibit enzyme activity by 50%; WT, wild-type pH1N1.
Isolate pyrosequencing results are consistent with those determined using the clinical specimens.
Pyrosequencing and sequencing were performed on all positive specimens from each case patient, collected at different times.
Established oseltamivir-susceptible and oseltamivir-resistant control viruses.
Pyrosequencing analysis of clinical specimens of 13 pH1N1 viruses collected from other patients admitted to DUMC, as well as 145 additional pH1N1viruses submitted to public health surveillance from 15 September–20 November, revealed that none carried the H275Y mutation.
Additionally, no new cases of oseltamivir-resistant pH1N1 were detected among the 73 inpatients admitted to the affected ward and to the medical ICU during the same period when the 3 confirmed case patients were treated in the ICU.
Sequence analysis of HA genes revealed complete homology among the oseltamivir-resistant pH1N1 viruses from case patients (Figure 2). The viruses from case patients were also found to be more closely related to other local NC isolates, compared with existing pH1N1 virus sequences available at the time in GenBank (Figures 2 and S1).
DISCUSSION
Our investigation confirmed that a cluster of oseltamivir-resistant pH1N1 infections occurred among 4 immunocompromised patients within a hematology-oncology ward. Four factors provided evidence of nosocomial transmission of resistant virus—(1) temporal overlap of inpatient stay within the ward, (2) geographic proximity of patients, (3) presence of the H275Y resistance mutation in viral specimens obtained before oseltamivir use, and (4) complete homology of the HA and NA genes on sequencing.
We were unable to establish whether HCP or visitors contributed to viral transmission between patients. No evidence indicated that this variant of oseltamivir-resistant pH1N1 circulated in other parts of the hospital or in surrounding communities in NC. Interestingly, lack of documented transmission of oseltamivir-resistant pH1N1 virus to other patients in or around the ward could suggest that immunocompromised patients may be more at risk of acquiring infection.
Resistance to oseltamivir remains rare among pH1N1 strains; <1.2% of all pH1N1 isolates tested in the United States have been categorized as oseltamivir resistant [2]. Oseltamivir resistance in pH1N1 influenza has been described among immunocompromised patients and patients with receipt of oseltamivir for post-exposure prophylaxis or for therapy [2, 4, 14, 16]. Oseltamivir resistance probably emerged in this cluster when a single immunocompromised patient received oseltamivir after exposure to a close contact with influenza. In our case, transmission of oseltamivir-resistant virus from healthy HCP or visitors to case patients is unlikely, because there is no evidence that this variant of oseltamivir-resistant pH1N1 was circulating in other parts of the hospital or in surrounding communities in NC. Neither theory can be proven; the source of oseltamivir-resistant pH1N1 in this outbreak remains unknown.
Genetic analysis of NA genes of viruses from case patients indicated that they were 100% identical at both the protein and nucleotide levels. The isolate A/North Carolina/39/2009 had a mix of nucleotides A and G at position 807, which resulted in a mixture of amino-acids methionine and isoleucine at residue 269 (there is no known significance to this change). Comparison of the NA sequences from viruses from this outbreak with those from 2 viruses (A/North Carolina/05/2009 and A/North Carolina/06/2009) collected earlier in the pandemic and the reference vaccine strain virus A/California/07/2009 revealed that the outbreak strain differed from these viruses by changes at V106I and N248D. These mutations are not unique to the outbreak stain and are present in a large number of already circulating pH1N1 viruses [8, 15], and compensatory mutations that could have contributed to the transmission of the virus were not detected. Further NA sequencing and analysis of a larger number of pH1N1 viruses would be required to determine the closest ancestor of the outbreak viruses' NA gene.
This is, to our knowledge, the first report of nosocomial transmission of oseltamivir-resistant pH1N1 virus. Three mechanisms of viral transmission are possible. Drug-resistant virus might have been transmitted between patients by HCP or visitors. Six visitors and 12 HCP reported respiratory illness; however, none of these HCP or visitors was specifically tested for pH1N1. Alternately, drug-resistant virus might have been transmitted directly from one patient to another before diagnosis and implementation of pH1N1 isolation precautions. All case patients were ambulatory and had the opportunity to interact directly before diagnosis. Finally, drug-resistant pH1N1 might have been transmitted by a combination of direct exposures between case patients and by HCP or visitors as vectors. Results of air pressure testing indicated that airborne transmission was unlikely; however, we were unable to examine the role of fomites and environmental surfaces in viral transmission.
Patients infected with pH1N1 typically present with fever and respiratory symptoms, although some patients have milder symptoms [17, 18]. Additionally, diagnosing influenza among immunocompromised patients is difficult. Recognition of pH1N1 infection among immunocompromised patients can be confounded by attenuated symptoms or by the presence of other potential causes of fever or respiratory symptoms [19]. As a result, early diagnosis of influenza infection and timely implementation of isolation precautions might not occur. Fever and dyspnea occurred in 3 patients in this cluster, but these signs were initially attributed to etiologies other than pH1N1 infection.
Moreover, immunocompromised patients might have fluctuations in the intensity of influenza symptoms. For instance, 2 case patients experienced intermittent fevers, and another case patient reported spontaneous improvement of cough. Such fluctuation of symptoms may be attributable to altered inflammatory response in immunocompromised hosts, leading to difficulty with diagnosis and incorrect interpretation as spontaneous improvement or as response to therapeutic interventions.
Our findings underscore the need for vigilance for pH1N1 infection and other respiratory viral infections in the health care setting, especially among immunocompromised patients. Moreover, influenza and other respiratory viruses should always be considered in the differential diagnosis for fever or respiratory symptoms in any hospitalized patient when the prevalence of such respiratory viral infections is high in the community. Finally, if antiviral drugs are considered for prophylaxis or therapy of influenza infection, clinicians should consult up-to-date guidelines [20]. Despite antiviral prophylaxis, patients could still develop influenza as a result of failure of chemoprophylaxis [21]. Symptoms of fever or respiratory illness after oseltamivir prophylaxis should raise suspicion of influenza infection and prompt testing for influenza infection and oseltamivir resistance. Although no “rapid assay kits” are currently available, antiviral resistance can be determined by referring viruses to laboratories to identify markers of NAI resistance.
Transmission of seasonal influenza is well-described in hospitalized settings. However, isolation precautions and hand hygiene can reduce transmission of influenza infections in hospitals [22]. We suspect case patients were not placed on isolation precautions because the perceived risk for pH1N1 infection was low, even when the patient received oseltamivir chemoprophylaxis or underwent testing to rule out pH1N1 infection. Such delays in initiating isolation precautions could have contributed to transmission of pH1N1. Indeed, guidelines now recommend placing patients with suspected influenza on droplet precautions [23]. Finally, vaccines can effectively prevent influenza, and infection control programs should incorporate influenza vaccination for personnel in all health care settings.
This investigation had limitations. First, date of illness onset among case patients was difficult to determine because of concurrent medical problems that mimicked symptoms of influenza infection. Additionally, only a limited number of eligible patients were available for our case-control study. The retrospective nature of the investigation limited the ability to test HCP, visitors, or caregivers for pH1N1. Data obtained through interviews were subject to recall bias, social desirability bias, and underreporting. All case patients had died or were terminally ill at the time that the investigation began; we were unable to confirm their interactions with other patients, HCP, or visitors. Finally, although homologous HA and NA genes suggested transmission of a single strain of virus, confirmation would require full genome sequencing.
Results of our investigation indicate that, first, clinicians should include influenza in the differential diagnosis of any patient with fevers or any respiratory illness. Second, infections with pH1N1 or other influenza viruses can occur even in patients who have received antiviral prophylaxis or treatment. Finally, obtaining respiratory specimens and performing viral testing can facilitate early identification of cases. However, initiation of treatment and implementation of isolation precautions should not be delayed while awaiting the results of laboratory tests.
Supplementary Data
Supplementary data are available at http://www.oxfordjournals.org/our_journals/jid/online.
Acknowledgments
We thank the Duke University Medical Center, Division of Medical Oncology; the Duke Program for Infection Prevention and Healthcare Epidemiology; Freda Kohn, MT (ASCP) SM, Nancy G. Henshaw, MPH PhD, and Sylvia F. Costa, MD, of Duke Clinical Microbiology; Monte D. Brown, MD, and Jessica R. Thompson, MHA, of the Duke University Health System H1N1 Influenza Taskforce; Emilie G. Lamb, MPH, of the North Carolina Department of Health and Human Services; and John G. Bartlett, MD, the Johns Hopkins University Bloomberg School of Public Health Faculty Advisor for LFC. The findings and conclusions of this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
References
- 1.Centers for Disease Control Prevention. Swine influenza A (H1N1) infection in two children–Southern California, March-April 2009. MMWR Morb Mortal Wkly Rep. 2009;58:400–2. [PubMed] [Google Scholar]
- 2.FluView—A weekly influenza surveillance report prepared by the influenza Division. http://www.cdc.gov/flu/weekly/. Accessed 23 October 2010 . [Google Scholar]
- 3.Le QM, Wertheim HF, Tran ND, van Doorn HR, Nguyen TH, Horby P. A community cluster of oseltamivir-resistant cases of 2009 H1N1 influenza. N Engl J Med. 2009;362:86–7. doi: 10.1056/NEJMc0910448. [DOI] [PubMed] [Google Scholar]
- 4.Centers for Disease Control Prevention. Oseltamivir-resistant 2009 pandemic influenza A (H1N1) virus infection in two summer campers receiving prophylaxis–North Carolina, 2009. MMWR Morb Mortal Wkly Rep. 2009;58:969–72. [PubMed] [Google Scholar]
- 5.Gubareva LV, Kaiser L, Matrosovich MN, Soo-Hoo Y, Hayden FG. Selection of influenza virus mutants in experimentally infected volunteers treated with oseltamivir. J Infect Dis. 2001;183:523–31. doi: 10.1086/318537. [DOI] [PubMed] [Google Scholar]
- 6.Dawood FS, Jain S, Finelli L, et al. Emergence of a novel swine-origin influenza A (H1N1) virus in humans. N Engl J Med. 2009;360:2605–15. doi: 10.1056/NEJMoa0903810. [DOI] [PubMed] [Google Scholar]
- 7.Deyde VM, Sheu TG, Trujillo AA, et al. Detection of molecular markers of drug resistance in 2009 pandemic influenza A (H1N1) viruses by pyrosequencing. Antimicrob Agents Chemother. 2010;54:1102–10. doi: 10.1128/AAC.01417-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garten RJ, Davis CT, Russell CA, et al. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science. 2009;325:197–201. doi: 10.1126/science.1176225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.World Health Organization. Sequencing primers and protocol. http://www.who.int/csr/resources/publications/swineflu/sequencing_primers/en/index.html. Accessed 22 October 2010 . [Google Scholar]
- 10.Deyde V, Garten R, Sheu T, et al. Genomic events underlying the changes in adamantane resistance among influenza A(H3N2) viruses during 2006–2008. Influenza Other Respi Viruses. 2009;3:297–314. doi: 10.1111/j.1750-2659.2009.00103.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zwickl DJ. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. Austin, TX: The University of Texas; 2006. Ph.D. dissertation. [Google Scholar]
- 12.Page RDM. TREEVIEW: An application to display phylogenetic trees on personal computers. Computer Applications in the Biosciences. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 13.Sheu TG, Deyde VM, Okomo-Adhiambo M, et al. Surveillance for neuraminidase inhibitor resistance among human influenza A and B viruses circulating worldwide from 2004 to 2008. Antimicrob Agents Chemother. 2008;52:3284–92. doi: 10.1128/AAC.00555-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Centers for Disease Control Prevention. Oseltamivir-resistant novel influenza A (H1N1) virus infection in two immunosuppressed patients—Seattle, Washington, 2009. MMWR Morb Mortal Wkly Rep. 2009;58:893–6. [PubMed] [Google Scholar]
- 15.Itoh Y, Shinya K, Kiso M, et al. In vitro and in vivo characterization of new swine-origin H1N1 influenza viruses. Nature. 2009;460:1021–5. doi: 10.1038/nature08260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baz M, Abed Y, Papenburg J, Bouhy X, Hamelin ME, Boivin G. Emergence of oseltamivir-resistant pandemic H1N1 virus during prophylaxis. N Engl J Med. 2009;361:2296–7. doi: 10.1056/NEJMc0910060. [DOI] [PubMed] [Google Scholar]
- 17.Ciblak MA, Albayrak N, Odabas Y, et al. Cases of influenza A(H1N1)v reported in Turkey, May–July 2009. Euro Surveill. 2009;14:pii:19304. [PubMed] [Google Scholar]
- 18.Yang J, Yang F, Huang F, Wang J, Jin Q. Subclinical infection with the novel Influenza A (H1N1) virus. Clin Infect Dis. 2009;49:1622–3. doi: 10.1086/644775. [DOI] [PubMed] [Google Scholar]
- 19.Redelman-Sidi G, Sepkowitz KA, Huang CK, et al. 2009 H1N1 influenza infection in cancer patients and hematopoietic stem cell transplant recipients. J Infect. 2010;60:257–63. doi: 10.1016/j.jinf.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 20.Centers for Disease Control, Prevention. Treatment (Antiviral drugs) http://www.cdc.gov/flu/professionals/antivirals/. Accessed 23 October 2010 . [Google Scholar]
- 21.Hayden FG, Belshe R, Villanueva C, et al. Management of influenza in households: a prospective, randomized comparison of oseltamivir treatment with or without postexposure prophylaxis. J Infect Dis. 2004;189:440–9. doi: 10.1086/381128. [DOI] [PubMed] [Google Scholar]
- 22.Daniels TL, Talbot TR. Unmasking the confusion of respiratory protection to prevent influenza-like illness in crowded community settings. J Infect Dis. 2010;201:483–5. doi: 10.1086/650395. [DOI] [PubMed] [Google Scholar]
- 23.Centers for Disease Control and Prevention. Prevention strategies for seasonal influenza in healthcare settings. http://www.cdc.gov/flu/professionals/infectioncontrol/healthcaresettings.htm. Accessed 23 October 2010 . [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.