Abstract
Purpose: The PathVisio-Validator plugin aims to simplify the task of producing biological pathway diagrams that follow graphical standardized notations, such as Molecular Interaction Maps or the Systems Biology Graphical Notation. This plugin assists in the creation of pathway diagrams by ensuring correct usage of a notation, and thereby reducing ambiguity when diagrams are shared among biologists. Rulesets, needed in the validation process, can be generated for any graphical notation that a developer desires, using either Schematron or Groovy. The plugin also provides support for filtering validation results, validating on a subset of rules, and distinguishing errors and warnings.
Availability: The PathVisio-Validator plugin works with versions of PathVisio 2.0.11 and later on Windows, Mac OS X and Linux. The plugin along with the instructions, example rulesets for Groovy and Schematron, and Java source code can be downloaded at http://pathvisio.org/wiki/PathVisioValidatorHelp. The software is developed under the open-source Apache 2.0 License and is freely available for both commercial and academic use.
Contact: chandankmit@gmail.com; augustin@mail.nih.gov
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Biological researchers use pathway diagrams as the medium to relay information about interactions among biological entities. Graphical notations like the Molecular Interaction Map (MIM) notation and the Systems Biology Graphical Notation (SBGN) promote the creation of unambiguous biological pathway diagrams through the use of well-defined rules for syntax. The specifications for these notations are long and technical, and this is a barrier to the wider usage of these notations by researchers. This issue presents a clear need not only for tools that simplify the usage of these graphical notations, but also tools that guide users in their correct usage.
PathVisio is an open-source Java-based tool for creating and editing biological pathways and linking diagram elements to external bioinformatics databases (van Iersel et al., 2008). By itself, PathVisio does not compel users to create diagrams using any particular notation; support for specific notations is added through the use of plugins, such as the MIM plugin (Luna et al., 2011b) or the SBGN plugin, which is under development (http://www.pathvisio.org/wiki/SbgnPluginHelp). Given this open nature of PathVisio, we have developed a validation framework similar to what developers are accustomed to in source code editors that report syntax errors. This accommodates the validation of pathway diagrams against any notation that PathVisio can draw with just the click of a button.
There are several projects related to standardized formats for computational biology that have associated validators, including well-known formats like BioPAX (Demir et al., 2010) and SBML (Hucka et al., 2003); there is also validation capability in BCML, a SBGN-compliant format (Beltrame et al., 2011), and SBGN-ED (Czauderna et al., 2010), an SBGN diagram editor. A key difference between SBGN-ED and the plugin presented here is the extensible support for other notations in the latter, which is especially important for the PathVisio community since its users may be using various graphical notations for diverse purposes. A second difference is that rulesets (specifically, Schematron rulesets) created for use with this plugin can be reused in other software projects. The use of Schematron is a feature that this plugin shares with some generic XML editors with validation capability, such as XMLSpy (http://xml-tools.com/ValidatorBuddy.htm).
2 IMPLEMENTATION
The validation of pathways using the plugin is performed based on a ruleset (a collection of rules). The plugin allows users to use their own custom-created rulesets or standard rulesets by notation developers. Described below are the main features of the PathVisio-Validator plugin and its user interface:
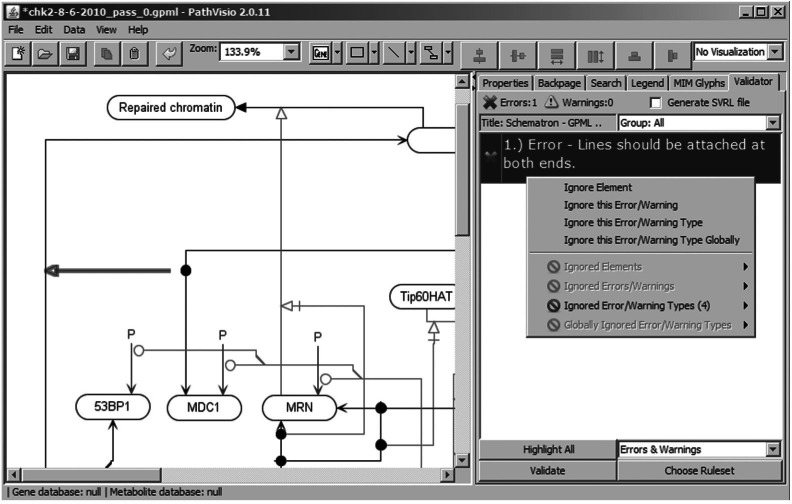
Plugin interface and current notation support: the interface for the plugin exists under the ‘Validator’ tab of PathVisio as shown in Figure 1. At the bottom of the tab are buttons for loading a ruleset and validating the current diagram. In order to validate a pathway diagram, the user simply needs to choose a ruleset and press the ‘Validate’ button. Validation messages for the current pathway diagram will then appear in the side panel as shown in Figure 1. These messages can be filtered to show only errors, only warnings, or both, using a drop-down menu. Users also have an option to select a specific rule group for validation. Using right-click options, users can ignore particular messages or types of messages. Currently, the plugin supports the MIM notation through a Schematron ruleset. We also provide a ruleset with rules used for the curation of diagrams on the WikiPathways database (http://wikipathways.org), a repository for biological pathway diagrams. SBGN support for PathVisio is currently in development as a separate project. Validation support of SBGN diagrams is expected through this plugin when that project is completed.
Fig. 1.
A pathway diagram being validated using the PathVisio-Validator plugin.
Schematron and Groovy ruleset support: the plugin supports rulesets written either in Schematron or Groovy; tutorials are provided on the project website.
Schematron (http://www.schematron.com) is an XML validation language that uses XSL Transformations (XSLT). The result of validation is a simple XML formatted report using the Schematron Validation Report Language. This validation report is parsed and error or warning messages for the current diagram are displayed in the ‘Validator’ tab as shown in Figure 1. Schematron rulesets are reusable and can facilitate the addition of validation support in other software projects where an XSLT processor is available. For instance, the ruleset provided for MIM was originally described for use as part of an automated command-line pipeline (Luna et al., 2011a). More information on the process used with the Schematron rulesets is on the project website.
Groovy (http://groovy.codehaus.org) is a scripting language with syntax similar to Java. Creating rulesets in Groovy is easy for users with a basic understanding of Java. Groovy rules are basically a collection of methods with specific signature for them to be recognized as valid rules. These are run directly against the internal memory representation of the diagram in PathVisio, producing results, which are then parsed and displayed on the plugin's panel. Validation against a Groovy ruleset, consumes less time than its Schematron counterpart, since it does not require any XSLT to take place. However, the downside to this approach is that the Groovy rules are tied to the internal workings of PathVisio. An example rule checking for unattached lines in pathway diagrams is provided in the Supplementary Material in both Schematron and Groovy.
Support for validation based on rule groups: validation rules in rulesets may be put into groups. Thus, a user can validate a diagram with all the rules in a ruleset or only those under a specific group, thereby making it more selective. Schematron supports rule groups using the ‘phase’ XML tag with one or more ‘active’ rules as child XML nodes. Groovy supports rule groups through a specific method that defines rule groups and their associated rules.
3 CONCLUSION
The PathVisio-Validator plugin has been developed to enhance the quality of pathways created in PathVisio by assisting biologists in drawing pathways according to specific graphical notations. Validation of this nature allows researchers to be more confident that their diagrams will not be ambiguous to readers. The extensible nature of the rulesets should allow it to adapt to preferences of PathVisio users, and the support for different ruleset formats (i.e. Schematron and Groovy) should allow users to create rulesets based on their priorities. Moreover, this plugin will also encourage the users of PathVisio, especially the beginners to create pathways based on specific notations, such as MIM, helping to promote the adoption of such standards. In the future, we plan to integrate this validation framework into WikiPathways to provide automated diagram validation (Pico et al., 2008), so that all uploaded/updated pathways are validated against common rules. Presented here is a narrow use case of the validation of graphical notations, but the ideas presented here can be extended to other biological standards written in XML-based formats. These ideas are of use in the wider standards community in providing validation support in multiple programming languages.
Supplementary Material
4 ACKNOWLEDGEMENTS
We thank the PathVisio community, Thomas Kelder and Margot Sunshine for their useful feedback, and Alexander Pico for coordinating the Google Summer of Code project.
Funding: Google Summer of Code program, Intramural Research Program of the National Institutes of Health, Center for Cancer Research, National Cancer Institute; Netherlands Consortium for Systems Biology (NCSB), which is part of the Netherlands Genomics Initiative/Netherlands Organization for Scientific Research, in part.
Conflict of Interest: none declared.
REFERENCES
- Beltrame L., et al. The Biological Connection Markup Language: a SBGN-compliant format for visualization, filtering and analysis of biological pathways. Bioinformatics. 2011;27:2127–2133. doi: 10.1093/bioinformatics/btr339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czauderna T., et al. Editing, validating and translating of SBGN maps. Bioinformatics. 2010;26:2340–2341. doi: 10.1093/bioinformatics/btq407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demir E., et al. The BioPAX community standard for pathway data sharing. Nature Biotechnology. 2010;28:935–942. doi: 10.1038/nbt.1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hucka M., et al. The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics. 2003;19:524–531. doi: 10.1093/bioinformatics/btg015. [DOI] [PubMed] [Google Scholar]
- Luna A., et al. A formal MIM specification and tools for the common exchange of MIM diagrams: an XML-based format, an API, and a validation method. BMC Bioinformatics. 2011a;12:167. doi: 10.1186/1471-2105-12-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna A., et al. PathVisio-MIM: PathVisio plugin for creating and editing Molecular Interaction Maps (MIMs) Bioinformatics. 2011b;27:2165–2166. doi: 10.1093/bioinformatics/btr336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pico A.R., et al. WikiPathways: pathway editing for the people. PLoS Biol. 2008;6:4. doi: 10.1371/journal.pbio.0060184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Iersel M.P., et al. Presenting and exploring biological pathways with PathVisio. BMC Bioinformatics. 2008;9:399. doi: 10.1186/1471-2105-9-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.