Abstract
MicroRNA (miRNA) are small non-coding RNA molecules that posttranscriptionally effect mRNA stability and translation by targeting the 3′-untranslated region (3′-UTR) of various transcripts. Thus, dysregulation of miRNA affects a wide range of cellular processes such as cell proliferation and differentiation involved in organ remodeling processes. Divergent miRNA patterns were observed during chronic liver diseases of various etiologies. Chronic liver diseases result in uncontrolled scar formation ending up in liver fibrosis or even cirrhosis. Since it has been shown that miR-29 dysregulation is involved in synthesis of extracellular matrix proteins, miR-29 is of special interest. The importance of miR-29 in hepatic collagen homeostasis is underlined by in vivo data showing that experimental severe fibrosis is associated with a prominent miR-29 decrease. The loss of miR-29 is due to the response of hepatic stellate cells to exposure to the profibrogenic mediators TGF-β and PDGF-BB. Several putative binding sites for the Smad proteins and the Ap1 complex are located in the miR-29 promoter, which are suggested to mediate miR-29 decrease in fibrosis. Other miRNA are highly increased after profibrogenic stimulation, such as miR-21. miR-21 is transcriptionally upregulated in response to Smad-3 rather than Smad-2 activation after TGF-β stimulation. In addition, TGF-β promotes miR-21 expression by formation of a microprocessor complex containing Smad proteins. Elevated miR-21 may then act as a profibrogenic miRNA by its repression of the TGF-β inhibitory Smad-7 protein.
Keywords: microRNA, fibrosis, extracellular matrix, TGF-β, PDGF, Smad proteins, Ap1
Introduction
Liver fibrosis is characterized by excessive scarring, caused by chronic inflammatory processes during liver diseases of different origin. In response to chronic liver injuries, various cell types get activated and transdifferentiate into myofibroblastic cells that then participate in synthesis and reorganization of connective tissue (Hautekeete and Geerts, 1997; Friedman, 1999, 2008; Knittel et al., 1999). A main source of extracellular matrix (ECM) production are hepatic stellate cells (HSC) undergoing myofibroblastic transition (Hautekeete and Geerts, 1997; Friedman, 1999, 2008). Myofibroblastic differentiation and matrix accumulation of HSC is heavily triggered by profibrogenic mediators such as transforming growth factor β (TGF-β) and the β-isoform of platelet-derived growth factor (PDGF). Hence, in response to TGF-β, HSC take center stage during fibrosis by the enhanced synthesis of ECM proteins, in particular collagen I and II. Here, we emphasize the role of dysregulated microRNA (miRNA) during liver fibrosis, their function in ECM expression and in profibrogenic signaling in HSC.
Dysregulation of miRNA in Liver Fibrosis
The fibrotic remodeling process and the change of gene expression during liver fibrosis are associated with an altered pattern of miRNA. miRNA are small (∼19–24 nucleotides long) non-coding RNA molecules inhibiting posttranscriptionally gene expression. It is suggested that more than one-third of all human genes are regulated by miRNA (Krek et al., 2005; Lewis et al., 2005). The primary miRNA (pri-miRNA) molecules of more than 1000 bases in length are transcribed by RNA-Polymerase II. Cleavage of the pri-miRNA then generates the precursor miRNA (pre-miRNA). This pre-miRNA is exported into the cytoplasm and further processed by RNase III and Dicer resulting in the release of a mature miRNA (Ruby et al., 2007). After integration into the RNA-induced silencing complex (RISC), interaction of miRNA sequences with the untranslated region (UTR) of transcripts causes translational repression or transcript degradation (Bartel, 2009). Several algorithms predict that one miRNA might bind to a multitude of mRNA transcripts and, in turn, that one mRNA might be targeted by a widespread panel of miRNA species (Doench and Sharp, 2004; Lewis et al., 2005).
In liver, miR-122 is the most abundant miRNA, accounting for more than 70% of the total miRNA in hepatocytes (Jopling et al., 2005). miR-122 is involved in cholesterol synthesis and hepatitis C virus (HCV) replication (Jopling et al., 2005; Esau et al., 2006). After liver injury and fibrosis it is markedly decreased depending on severity of fibrosis (Cheung et al., 2008; Morita et al., 2011). Other miRNA species, that are also highly expressed in the healthy liver, e.g., miR-125b or miR-22, were shown to be additionally reduced in fibrotic liver biopsies of chronic hepatitis C patients. Furthermore, the members of the miR-29 family, as well as miR-194 and miR-150 are reported to be downregulated during fibrogenesis (Kwiecinski et al., 2011; Roderburg et al., 2011). Additionally, miR-19b is recently found to be diminished in fibrotic liver of rat and human (Table 1) (Lakner et al., 2012). Whereas a wide range of miRNA are reduced after fibrosis, only some miRNA like the members of the miR-199, the miR-200, and the miR-34 family are known to be increased (Alisi et al., 2010; Pogribny et al., 2010; Murakami et al., 2011). Though miR-21 upregulation was only analyzed on fibrotic liver biopsies of a limited number of HCV patients (Marquez et al., 2010), in many other organs miR-21 is one of the most important miRNA prevalently expressed after fibrosis induction (Thum et al., 2008; Liu et al., 2010; Zhu and Fan, 2011).
Table 1.
Dysregulated miRNA during liver fibrosis and myofibroblastic HSC activation.
| Upregulated miRNA | Downregulated miRNA | Etiology | Reference | In HSC under fibrotic condition (Reference) |
|---|---|---|---|---|
| miR-122 | Human NAFLD, NAFLD (HFD rat, mice) | Alisi et al. (2010), Cheung et al. (2008) | Guo et al. (2009a), Ji et al. (2009)# | |
| miR-122 | Chronic HCV hepatitis | Morita et al. (2011) | ||
| miR-29a, -29b | NAFLD, chronic HCV hepatitis, CCl4 intoxication of mice | Kwiecinski et al. (2010), Ogawa et al. (2010), Roderburg et al. (2011) | Kwiecinski et al. (2011), Ogawa et al. (2010), Roderburg et al. (2011) | |
| miR-199a, -199a* miR-200a, -200b miR-34a, -34b |
CCl4 intoxication of mice, NAFLD (HFD rat, mice), chronic HCV hepatitis | Murakami et al. (2011) Alisi et al. (2010), Murakami et al. (2011), Pogribny et al. (2010) | p.o. | |
| miR-146b | Murakami et al. (2011), Pogribny et al. (2010) | p.o. | ||
| Murakami et al. (2011) | ||||
| miR-150, miR-194 | Venugopal et al. (2010) | |||
| miR-27a, -27b | NAFLD (HFD rat) | Alisi et al. (2010) | Ji et al. (2009) | |
| miR-15b, -16 | Guo et al. (2009b), Guo et al. (2009c) | |||
| miR-132 | Mann et al. (2010) | |||
| miR-335 | Chen et al. (2011) | |||
| miR-19b | Cholestasis (bile duct ligation) | Lakner et al. (2012) | Lakner et al. (2012) |
p.o., Personal observation; NAFLD, non-alcoholic fatty liver disease; HFD, high fat diet.
#The miR-122 decrease, observed in primary HSC during culture-induced myofibroblastic activation, might be due to the dilution of contaminating hepatocyte miRNA of the primary HSC isolate.
Recent reports have shown that altered miRNA levels are also associated with the phenotypical changes of HSC during the myofibroblastic transition process including the induction of ECM proteins (Guo et al., 2009a; Ji et al., 2009) (Table 1).
The Members of the miR-29 Family Repress Profibrogenic Gene Expression
The members of the miR-29 family are of particular interest because they are shown to inhibit ECM synthesis indicating an antifibrotic function. The miR-29 family consists of miR-29a, miR-29b, and miR-29c, differing only in two or three bases. miR-29a and miR-29b1 as well as miR-29c and miR-29b2, are encoded and transcribed in tandem by two genes located on chromosome 7 or chromosome 1, respectively (Mott et al., 2007; Wang et al., 2008). van Rooij et al. (2008) first reported the miR-29 function after myocardial infarction. The authors proved miR-29 mediated repression of elastin, collagen I and III synthesis in cardiac fibroblasts and its regulation by TGF-β. Accordingly, downregulation of miR-29 was suggested to enhance the fibrotic response after myocardial infarction, whereas overexpression of miR-29 in cardiac fibroblasts reduced collagen expression. Similarly, Ogawa et al. (2010) demonstrated that miR-29 inhibits the production of fibrillar collagen in HSC, suggesting also a function of miR-29 in liver fibrosis (Roderburg et al., 2011). Furthermore, highly conserved binding sites for the miR-29a and miR-29b are found in many 3′-UTR sequences of the various subunits of ECM proteins like collagen type V and XV subunits, laminin γ 1, and nidogen. The findings of Cushing et al. (2011) suggest that miR-29a and miR-29b are also involved in a widespread panel of other fibrosis associated genes including Adam metalloproteinases mainly type 12 and 9, ECM formatting components such as fibrillin-1 and follistatin-1, and different integrin chains. Recent work of Kwiecinski et al. (in review) showed that miR-29 not only targets gene expression of ECM associated proteins, but also the expression of profibrogenic growth factors (manuscript submitted). They found putative miR-29a and miR-29b binding sites in mRNA of IGF-I but most notably also in members of PDGF linked signaling pathways such as PDGF-β receptor, PDGF-C, and VEGF-A. Accordingly, miR-29 mediated inhibition of IGF-I and PDGF-C was proven in HSC. In addition, Sekiya et al. (2011) collect evidence that also the PDGF-β receptor is targeted by miR-29 regulation.
As a consequence, miR-29 acts as antifibrogenic miRNA by two pathways (1) by inhibition of ECM formation and (2) interfering with the profibrogenic cell communication pathways via PDGF-B and PDGF-C signaling (Figure 1B). Interestingly, miR-29 expression is in turn highly regulated by PDGF-BB (Ogawa et al., 2010). Thus, the loss of miR-29 in myofibroblastic HSC during liver fibrosis is mainly due to profibrogenic stimulation by TGF-β and PDGF-BB (Ogawa et al., 2010; Kwiecinski et al., 2011).
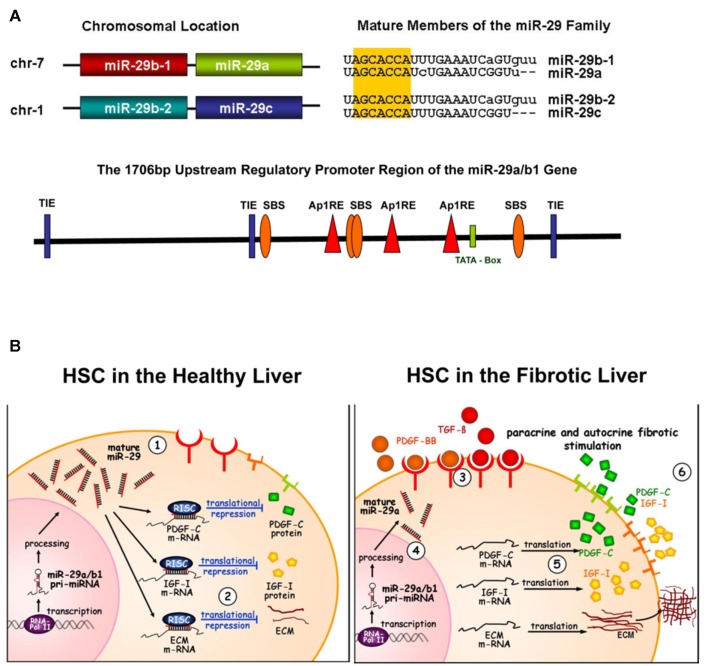
Figure 1.
miR-29 involved in profibrogenic signaling. (A) Genetic structure and promoter region of the miR-29 members. The members of the miR-29 family are encoded by two genes, located on chromosome 7 or 1, respectively. The pri-miR-29a and b1 as well as the miR-29c and b2 are each transcribed in tandem. The mature miR-29 sequences differ only in max. three bases, but sharing an identical seed region (yellow shadowed). The promoter region of the miR-29a/b1 gene contains several putative binding sites for the profibrogenic repression of miR-29: three Ap1 consensus sequences (Ap1-RE), three TGF-β inhibitory elements (TIE) and in addition the three putative Smad binding elements (SBE). These transcriptional binding sites and their interaction are suggested to mediate profibrogenic stimulation of PDGF-BB (i.p. of the Ap1 RE sites) and of TGF-β by Smad and Ap1 signaling. (B) miR-29 signaling in the healthy and the fibrotic liver. In the healthy liver, mir-29 is highly expressed in hepatic stellate cells (1) and is responsible for repression of extracellular matrix proteins (ECM) in particular collagens (2), but also of expression of growth factors such as PDGF-C and IGF-I. During fibrogenesis, profibrogenic growth factors are increased and stimulate hepatic stellate cells in a paracrine and autocrine manner (3). Profibrogenic stimulation of TGF-β and PDGF-BB results in miR-29 repression (4). The loss of miR-29 by TGF-β and PDGF-BB results in the abolished repression of profibrogenic expression of ECM, PDGF-C or IGF-I (5). The enhanced secretion of PDGF-C and IGF-I stimulates autocrinely stellate cells, leading to increased proliferation and ECM production (6).
Loss of miR-29 in the Fibrotic Liver is a Result of Profibrogenic Signaling
The decrease of miR-29 was observed after experimental fibrosis including liver intoxication in mice and cholestasis induced fibrosis after bile duct occlusion in rat. Furthermore, in liver biopsies of patients with chronic hepatitis C miR-29 levels are significantly reduced (Kwiecinski et al., 2011; Roderburg et al., 2011). These in vivo studies suggest that the loss of miR-29 during liver fibrogenesis leads to the abolishment of ECM and profibrogenic mediator repression. The findings on HSC provide additional evidence, that reduced miR-29 levels leading to profibrogenic gene expression are due to TGF-β or PDGF-BB exposure. The Smad-2/3 proteins act as the main signal transducers of TGF-β stimulation, initiating ECM synthesis and myofibroblastic transition (Dooley et al., 2000, 2001a). In renal fibrosis, silencing of the miR-29c/b2 gene is assumed to involve Smad-3 signaling (Qin et al., 2011). However, the analysis of promoter regions of both miR-29 genes, shown in Figure 1A, has just started by defining the transcriptional NF-KB and YY-NF-KB control, respectively (Wang et al., 2008; Mott et al., 2010), and did not yet focus on TGF-β signaling.
Interestingly, the promoter sequence of the miR-29a/b1 gene harbors several CAGA-boxes. CAGA-boxes function as Smad-3/4 protein binding elements (SBE) and Smad-3/4 binding is commonly followed by induction of gene transcription (Dennler et al., 1998). However, suppressive effects of Smad proteins are also possible, depending on their interaction with other regulatory factors like histone deacetylases (Liberati et al., 2001) or the activator protein 1 (Ap1; Hall et al., 2003). Thus, TGF-β induced Smad-3/4 signaling and the regulatory binding of the SBE may contribute to the transcriptional repression of the miR-29a/b1 gene. In addition, three consensus sequences for binding of the transcription factor Ap1 are located in the upstream region of the miR-29a/b1 gene. Ap1 is highly activated by the ras/raf pathway after TGF-β and PDGF-BB exposure (Angel and Karin, 1992; Zhang et al., 1998). Ap1 interaction with the Ap1 responsive elements (Ap1 RE) drives transcription of a broad spectrum of genes involved in the early gene response and acute inflammatory stimulation. However, Ap1 can also function as transcriptional suppressor by binding to TGF-β inhibitory elements (TIE), that are known to lead to gene silencing in the stromelysin (Kerr et al., 1990), the HGF, and the c-myc gene (Matrisian et al., 1992; Yagi et al., 2002). Thus, in future experiments it has to be proven if profibrogenic miR-29 repression is mediated by the consensus sequences of the TIE, the Ap1 RE, the SBE, or even by the interplay of different regulatory elements (Figure 1A).
Enhanced miR-21 Levels in Response to TGF-β by Transcriptional Induction and Promoted Microprocessing
miR-21 is an ubiquitously expressed miRNA, which is highly upregulated in the majority of cancer types. In cancer, miR-21 acts as an oncogenic miRNA (oncomiR) targeting PTEN and other tumor suppressor proteins (Krichevsky and Gabriely, 2009; Qi et al., 2009). Previous reports have demonstrated that miR-21 is also enhanced after initiation of myocardial, renal, or pulmonary fibrosis (Thum et al., 2008; Liu et al., 2010).
miR-21 enrichment during fibrosis is assumed to be based on profibrogenic stimulation by TGF-β, because Zhong et al. (2011) have demonstrated that Smad-3 transducing TGF-β signaling, is crucial for miR-21 transcription. Furthermore, the signal transducers of TGF-β, the receptor-regulated Smad-2 and Smad-3 (R-Smad), are able to interact with the p68 RNA helicase of the Drosha/PDCD4 microprocessor complex, promoting the restriction of pri-miR-21 into the precursor pre-miR-21 (Davis et al., 2008). Thus, the upregulation of miR-21 during fibrotic processes is based on two TGF-β stimulated mechanisms, that are mediated by the Smad proteins: (1) by transcriptional induction and (2) by enhanced miR-21 maturation (Figure 2).
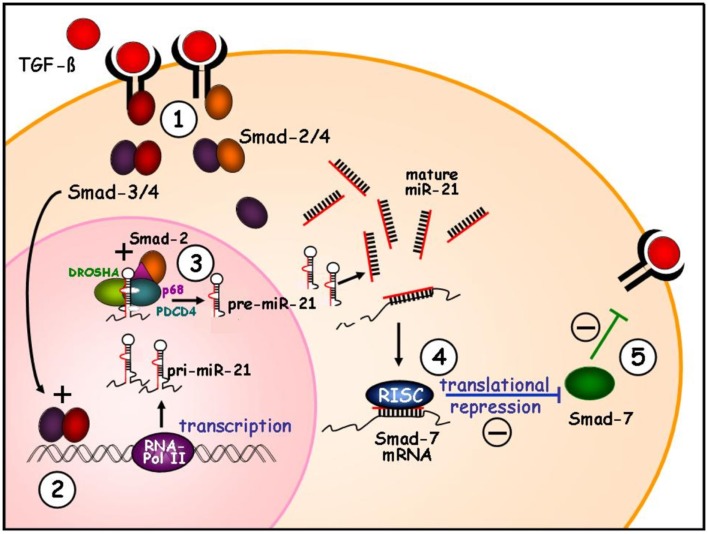
Figure 2.
Function of miR-21 in profibrogenic TGF-β signaling. During fibrosis TGF-β binds to the receptors leading to Smad-2 or Smad-3 phosphorylation, followed by aggregation with Smad-4 (1). The Smad-3/4 is shown to induce transcriptional induction of synthesis of pri-miR-21 (2). The further cleavage of pri-miRNA into hairpin-loop precursor miRNA is catalyzed by the PDCD4/Drosha complex. The Smad proteins can interact with the RNA helicase p68, which is integrated in the microprocessing complex. After TGF-β exposure, both the Smad-2 and Smad-3 protein is bound by p68 of the complex resulting in stabilization and high microprocessing efficiency (3). High levels of miR-21 target the Smad-7 mRNA and repress translation of the inhibitory Smad-7 (4). The decrease of the inhibitory Smad-7 protein in turn abolishes the negative feed-back mechanisms of TGF-β signaling (5) resulting in elevated profibrogenic TGF-β stimulation.
A potential mechanism for the role of miR-21 in fibrosis is through regulating the protein synthesis of the inhibitory Smad-7 (Figure 2; Liu et al., 2010; Marquez et al., 2010). Smad-7 mediates a negative feed-back mechanism of TGF-β signaling. After TGF-β stimulation, Smad-7 transcription is driven by Smad-2/3 and Smad-4 (Stopa et al., 2000; Dooley et al., 2001b). Enhanced Smad-7 levels, in turn, are highly potent to antagonize TGF-β-mediated pathways (1) by blockade of the TGF-β type I receptor for further binding of the receptor Smads (Massague and Chen, 2000), and (2) by Smurf2 interaction, to form an E3 ubiquitin ligase that targets the TGF-β type I receptor for degradation (Kavsak et al., 2000).
Therefore, the inhibition of Smad-7 in response to miR-21 enhancement during fibrosis has to be considered as an important profibrogenic pathway, which might become an attractive target for future therapeutical approaches. In contrast to miR-21 that promotes TGF-β signaling, a recent study of Lakner et al. shows that miR-19b which is downregulated during fibrosis, inhibits TGF-β pathways. miR-19b represses fibrotic features after myofibroblastic activation of stellate cells by targeting the TGF-β II receptor and most notably Smad-3 (Lakner et al., 2012). Hence, upcoming knowledge of miRNA species, involved in profibrogenic signaling, will provide novel perspectives in understanding and probably also in treatment of chronic liver diseases.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Alisi A., Da Sacco L., Bruscalupi G., Piemonte F., Panera N., De Vito R., Leoni S., Bottazzo G. F., Masotti A., Nobili V. (2010). Mirnome analysis reveals novel molecular determinants in the pathogenesis of diet-induced nonalcoholic fatty liver disease. Lab. Invest. 91, 283–293 10.1038/labinvest.2010.166 [DOI] [PubMed] [Google Scholar]
- Angel P., Karin M. (1992). Specific members of the Jun protein family regulate collagenase expression in response to various extracellular stimuli. Matrix Suppl. 1, 156–164 [PubMed] [Google Scholar]
- Bartel D. P. (2009). MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233 10.1016/j.cell.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Wu C. Q., Zhang Z. Q., Yao D. K., Zhu L. (2011). Loss of expression of miR-335 is implicated in hepatic stellate cell migration and activation. Exp. Cell Res. 317, 1714–1725 10.1016/j.yexcr.2011.05.001 [DOI] [PubMed] [Google Scholar]
- Cheung O., Puri P., Eicken C., Contos M. J., Mirshahi F., Maher J. W., Kellum J. M., Min H., Luketic V. A., Sanyal A. J. (2008). Nonalcoholic steatohepatitis is associated with altered hepatic MicroRNA expression. Hepatology 48, 1810–1820 10.1002/hep.22569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cushing L., Kuang P. P., Qian J., Shao F., Wu J., Little F., Thannickal V. J., Cardoso W. V., Lu J. (2011). miR-29 is a major regulator of genes associated with pulmonary fibrosis. Am. J. Respir. Cell Mol. Biol. 45, 287–294 10.1165/rcmb.2010-0323OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis B. N., Hilyard A. C., Lagna G., Hata A. (2008). SMAD proteins control DROSHA-mediated microRNA maturation. Nature 454, 56–61 10.1038/nature07053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennler S., Itoh S., Vivien D., ten Dijke P., Huet S., Gauthier J. M. (1998). Direct binding of Smad3 and Smad4 to critical TGF beta-inducible elements in the promoter of human plasminogen activator inhibitor-type 1 gene. EMBO J. 17, 3091–3100 10.1093/emboj/17.11.3091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench J. G., Sharp P. A. (2004). Specificity of microRNA target selection in translational repression. Genes Dev. 18, 504–511 10.1101/gad.1184404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooley S., Delvoux B., Lahme B., Mangasser-Stephan K., Gressner A. M. (2000). Modulation of transforming growth factor beta response and signaling during transdifferentiation of rat hepatic stellate cells to myofibroblasts. Hepatology 31, 1094–1106 10.1053/he.2000.6126 [DOI] [PubMed] [Google Scholar]
- Dooley S., Delvoux B., Streckert M., Bonzel L., Stopa M., ten Dijke P., Gressner A. M. (2001a). Transforming growth factor beta signal transduction in hepatic stellate cells via Smad2/3 phosphorylation, a pathway that is abrogated during in vitro progression to myofibroblasts. TGFbeta signal transduction during transdifferentiation of hepatic stellate cells. FEBS Lett. 502, 4–10 10.1016/S0014-5793(01)02656-4 [DOI] [PubMed] [Google Scholar]
- Dooley S., Streckert M., Delvoux B., Gressner A. M. (2001b). Expression of Smads during in vitro transdifferentiation of hepatic stellate cells to myofibroblasts. Biochem. Biophys. Res. Commun. 283, 554–562 10.1006/bbrc.2001.4811 [DOI] [PubMed] [Google Scholar]
- Esau C., Davis S., Murray S. F., Yu X. X., Pandey S. K., Pear M., Watts L., Booten S. L., Graham M., McKay R., Subramaniam A., Propp S., Lollo B. A., Freier S., Bennett C. F., Bhanot S., Monia B. P. (2006). miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 3, 87–98 10.1016/j.cmet.2006.01.005 [DOI] [PubMed] [Google Scholar]
- Friedman S. L. (1999). Stellate cell activation in alcoholic fibrosis – an overview. Alcohol. Clin. Exp. Res. 23, 904–910 10.1097/00000374-199905000-00020 [DOI] [PubMed] [Google Scholar]
- Friedman S. L. (2008). Mechanisms of hepatic fibrogenesis. Gastroenterology 134, 1655–1669 10.1016/S0016-5085(08)63621-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C. J., Pan Q., Cheng T., Jiang B., Chen G. Y., Li D. G. (2009a). Changes in microRNAs associated with hepatic stellate cell activation status identify signaling pathways. FEBS J. 276, 5163–5176 10.1111/j.1742-4658.2008.06852.x [DOI] [PubMed] [Google Scholar]
- Guo C. J., Pan Q., Jiang B., Chen G. Y., Li D. G. (2009b). Effects of upregulated expression of microRNA-16 on biological properties of culture-activated hepatic stellate cells. Apoptosis 14, 1331–1340 10.1007/s10495-009-0401-3 [DOI] [PubMed] [Google Scholar]
- Guo C. J., Pan Q., Li D. G., Sun H., Liu B. W. (2009c). miR-15b and miR-16 are implicated in activation of the rat hepatic stellate cell: an essential role for apoptosis. J. Hepatol. 50, 766–778 10.1016/j.jhep.2008.11.025 [DOI] [PubMed] [Google Scholar]
- Hall M. C., Young D. A., Waters J. G., Rowan A. D., Chantry A., Edwards D. R., Clark I. M. (2003). The comparative role of activator protein 1 and Smad factors in the regulation of Timp-1 and MMP-1 gene expression by transforming growth factor-beta 1. J. Biol. Chem. 278, 10304–10313 10.1074/jbc.M212334200 [DOI] [PubMed] [Google Scholar]
- Hautekeete M. L., Geerts A. (1997). The hepatic stellate (Ito) cell: its role in human liver disease. Virchows Arch. 430, 195–207 10.1007/BF01324802 [DOI] [PubMed] [Google Scholar]
- Ji J., Zhang J., Huang G., Qian J., Wang X., Mei S. (2009). Over-expressed microRNA-27a and 27b influence fat accumulation and cell proliferation during rat hepatic stellate cell activation. FEBS Lett. 583, 759–766 10.1016/j.febslet.2009.01.034 [DOI] [PubMed] [Google Scholar]
- Jopling C. L., Yi M., Lancaster A. M., Lemon S. M., Sarnow P. (2005). Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science 309, 1577–1581 10.1126/science.1113329 [DOI] [PubMed] [Google Scholar]
- Kavsak P., Rasmussen R. K., Causing C. G., Bonni S., Zhu H., Thomsen G. H., Wrana J. L. (2000). Smad7 binds to Smurf2 to form an E3 ubiquitin ligase that targets the TGF beta receptor for degradation. Mol. Cell 6, 1365–1375 10.1016/S1097-2765(00)00134-9 [DOI] [PubMed] [Google Scholar]
- Kerr L. D., Miller D. B., Matrisian L. M. (1990). TGF-beta 1 inhibition of transin/stromelysin gene expression is mediated through a Fos binding sequence. Cell 61, 267–278 10.1016/0092-8674(90)90807-Q [DOI] [PubMed] [Google Scholar]
- Knittel T., Kobold D., Saile B., Grundmann A., Neubauer K., Piscaglia F., Ramadori G. (1999). Rat liver myofibroblasts and hepatic stellate cells: different cell populations of the fibroblast lineage with fibrogenic potential. Gastroenterology 117, 1205–1221 10.1016/S0016-5085(99)70407-5 [DOI] [PubMed] [Google Scholar]
- Krek A., Grun D., Poy M. N., Wolf R., Rosenberg L., Epstein E. J., MacMenamin P., da Piedade I., Gunsalus K. C., Stoffel M., Rajewsky N. (2005). Combinatorial microRNA target predictions. Nat. Genet. 37, 495–500 10.1038/ng1536 [DOI] [PubMed] [Google Scholar]
- Krichevsky A. M., Gabriely G. (2009). miR-21: a small multi-faceted RNA. J. Cell. Mol. Med. 13, 39–53 10.1111/j.1582-4934.2008.00556.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiecinski M., Elfimova N., Noetel A., Schievenbusch S., Strack I., Toex U., Drebber U., Steffen H. M., Dienes H. P., Odenthal M. (2010). Mir-29, inhibiting synthesis of profibrogenic mediators, is released into the blood stream after chronic hepatitis C infection, indicating progression of fibrosis. Hepatology 52, 446A–447A 10.1016/S0168-8278(10)61155-5 [DOI] [Google Scholar]
- Kwiecinski M., Noetel A., Elfimova N., Trebicka J., Schievenbusch S., Strack I., Molnar L., von Brandenstein M., Töx U., Nischt R., Coutelle O., Dienes H., Odenthal M. (2011). Hepatocyte growth factor (HGF) inhibits collagen synsthesis in hepatic stellate cells by miRNA-29 induction. PLoS ONE 6, e24568. 10.1371/journal.pone.0024568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakner A. M., Steuerwald N. M., Walling T. L., Ghosh S., Li T., McKillop I. H., Russo M. W., Bonkovsky H. L., Schrum L. W. (2012). Inhibitory effects of microRNA 19b in hepatic stellate cell-mediated fibrogenesis. Hepatology (in press). 10.1002/hep.25613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis B. P., Burge C. B., Bartel D. P. (2005). Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120, 15–20 10.1016/j.cell.2004.12.035 [DOI] [PubMed] [Google Scholar]
- Liberati N. T., Moniwa M., Borton A. J., Davie J. R., Wang X. F. (2001). An essential role for Mad homology domain 1 in the association of Smad3 with histone deacetylase activity*. J. Biol. Chem. 276, 22595–22603 10.1074/jbc.M010778200 [DOI] [PubMed] [Google Scholar]
- Liu G., Friggeri A., Yang Y., Milosevic J., Ding Q., Thannickal V. J., Kaminski N., Abraham E. (2010). miR-21 mediates fibrogenic activation of pulmonary fibroblasts and lung fibrosis. J. Exp. Med. 207, 1589–1597 10.1084/jem.20100035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann J., Chu D. C., Maxwell A., Oakley F., Zhu N. L., Tsukamoto H., Mann D. A. (2010). MeCP2 controls an epigenetic pathway that promotes myofibroblast transdifferentiation and fibrosis. Gastroenterology 138, 705–714 10.1016/S0016-5085(10)63444-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquez R. T., Bandyopadhyay S., Wendlandt E. B., Keck K., Hoffer B. A., Icardi M. S., Christensen R. N., Schmidt W. N., McCaffrey A. P. (2010). Correlation between microRNA expression levels and clinical parameters associated with chronic hepatitis C viral infection in humans. Lab. Invest. 90, 1727–1736 10.1038/labinvest.2010.126 [DOI] [PubMed] [Google Scholar]
- Massague J., Chen Y. G. (2000). Controlling TGF-beta signaling. Genes Dev. 14, 627–644 [PubMed] [Google Scholar]
- Matrisian L. M., Ganser G. L., Kerr L. D., Pelton R. W., Wood L. D. (1992). Negative regulation of gene expression by TGF-beta. Mol. Reprod. Dev. 32, 111–120 10.1002/mrd.1080320206 [DOI] [PubMed] [Google Scholar]
- Morita K., Taketomi A., Shirabe K., Umeda K., Kayashima H., Ninomiya M., Uchiyama H., Soejima Y., Maehara Y. (2011). Clinical significance and potential of hepatic microRNA-122 expression in hepatitis C. Liver Int. 31, 474–484 10.1111/j.1478-3231.2010.02433.x [DOI] [PubMed] [Google Scholar]
- Mott J. L., Kobayashi S., Bronk S. F., Gores G. J. (2007). mir-29 regulates Mcl-1 protein expression and apoptosis. Oncogene 26, 6133–6140 10.1038/sj.onc.1210436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mott J. L., Kurita S., Cazanave S. C., Bronk S. F., Werneburg N. W., Fernandez-Zapico M. E. (2010). Transcriptional suppression of mir-29b-1/mir-29a promoter by c-Myc, hedgehog, and NF-kappaB. J. Cell. Biochem. 10, 155–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami Y., Toyoda H., Tanaka M., Kuroda M., Harada Y., Matsuda F., Tajima A., Kosaka N., Ochiya T., Shimotohno K. (2011). The progression of liver fibrosis is related with overexpression of the miR-199 and 200 families. PLoS ONE 6, e16081. 10.1371/journal.pone.0016081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa T., Iizuka M., Sekiya Y., Yoshizato K., Ikeda K., Kawada N. (2010). Suppression of type I collagen production by microRNA-29b in cultured human stellate cells. Biochem. Biophys. Res. Commun. 391, 316–321 10.1016/j.bbrc.2009.11.056 [DOI] [PubMed] [Google Scholar]
- Pogribny I. P., Starlard-Davenport A., Tryndyak V. P., Han T., Ross S. A., Rusyn I., Beland F. A. (2010). Difference in expression of hepatic microRNAs miR-29c, miR-34a, miR-155, and miR-200b is associated with strain-specific susceptibility to dietary nonalcoholic steatohepatitis in mice. Lab. Invest. 90, 1437–1446 10.1038/labinvest.2010.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi L., Bart J., Tan L. P., Platteel I., Sluis T., Huitema S., Harms G., Fu L., Hollema H., Berg A. (2009). Expression of miR-21 and its targets (PTEN, PDCD4, TM1) in flat epithelial atypia of the breast in relation to ductal carcinoma in situ and invasive carcinoma. BMC Cancer 9, 163. 10.1186/1471-2407-9-163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin W., Chung A. C., Huang X. R., Meng X. M., Hui D. S., Yu C. M., Sung J. J., Lan H. Y. (2011). TGF-beta/Smad3 signaling promotes renal fibrosis by inhibiting miR-29. J. Am. Soc. Nephrol. 22, 1462–1474 10.1681/ASN.2010090967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roderburg C., Urban G. W., Bettermann K., Vucur M., Zimmermann H., Schmidt S., Janssen J., Koppe C., Knolle P., Castoldi M., Tacke F., Trautwein C., Luedde T. (2011). Micro-RNA profiling reveals a role for miR-29 in human and murine liver fibrosis. Hepatology 53, 209–218 10.1002/hep.23922 [DOI] [PubMed] [Google Scholar]
- Ruby J. G., Jan C. H., Bartel D. P. (2007). Intronic microRNA precursors that bypass Drosha processing. Nature 448, 83–86 10.1038/nature05983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiya Y., Ogawa T., Yoshizato K., Ikeda K., Kawada N. (2011). Suppression of hepatic stellate cell activation by microRNA-29b. Biochem. Biophys. Res. Commun. 412, 74–79 10.1016/j.bbrc.2011.07.041 [DOI] [PubMed] [Google Scholar]
- Stopa M., Anhuf D., Terstegen L., Gatsios P., Gressner A. M., Dooley S. (2000). Participation of Smad2, Smad3, and Smad4 in transforming growth factor beta (TGF-beta)-induced activation of Smad7. THE TGF-beta response element of the promoter requires functional Smad binding element and E-box sequences for transcriptional regulation. J. Biol. Chem. 275, 29308–29317 10.1074/jbc.M003282200 [DOI] [PubMed] [Google Scholar]
- Thum T., Gross C., Fiedler J., Fischer T., Kissler S., Bussen M., Galuppo P., Just S., Rottbauer W., Frantz S., Castoldi M., Soutschek J., Koteliansky V., Rosenwald A., Basson M. A., Licht J. D., Pena J. T., Rouhanifard S. H., Muckenthaler M. U., Tuschl T., Martin G. R., Bauersachs J., Engelhardt S. (2008). MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature 456, 980–984 10.1038/nature07511 [DOI] [PubMed] [Google Scholar]
- van Rooij E., Sutherland L. B., Thatcher J. E., DiMaio J. M., Naseem R. H., Marshall W. S., Hill J. A., Olson E. N. (2008). Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc. Natl. Acad. Sci. U.S.A. 105, 13027–13032 10.1073/pnas.0805038105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venugopal S. K., Jiang J., Kim T. H., Li Y., Wang S. S., Torok N. J., Wu J., Zern M. A. (2010). Liver fibrosis causes downregulation of miRNA-150 and miRNA-194 in hepatic stellate cells, and their overexpression causes decreased stellate cell activation. Am. J. Physiol. Gastrointest. Liver Physiol. 298, G101–G106 10.1152/ajpgi.00220.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Garzon R., Sun H., Ladner K. J., Singh R., Dahlman J., Cheng A., Hall B. M., Qualman S. J., Chandler D. S., Croce C. M., Guttridge D. C. (2008). NF-kappaB-YY1-miR-29 regulatory circuitry in skeletal myogenesis and rhabdomyosarcoma. Cancer Cell 14, 369–381 10.1016/j.ccr.2008.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi K., Furuhashi M., Aoki H., Goto D., Kuwano H., Sugamura K., Miyazono K., Kato M. (2002). c-myc is a downstream target of the Smad pathway. J. Biol. Chem. 277, 854–861 10.1074/jbc.M104170200 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Feng X. H., Derynck R. (1998). Smad3 and Smad4 cooperate with c-Jun/c-Fos to mediate TGF-beta-induced transcription. Nature 394, 909–913 10.1038/29814 [DOI] [PubMed] [Google Scholar]
- Zhong X., Chung A. C., Chen H. Y., Meng X. M., Lan H. Y. (2011). Smad3-mediated upregulation of miR-21 promotes renal fibrosis. J. Am. Soc. Nephrol. 22, 1668–1681 10.1681/ASN.2010111168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H., Fan G. C. (2011). Role of microRNAs in the reperfused myocardium towards post-infarct remodelling. Cardiovasc. Res. [Epub ahead of print]. 10.1093/cvr/cvr291 [DOI] [PMC free article] [PubMed] [Google Scholar]