Abstract
The development of clinically useful molecular diagnostics requires validation of clinical assay performance and achievement of clinical qualification in clinical trials. As discussed elsewhere in this Focus Section on Molecular Diagnostics, validation of assay performance must be rigorous especially when the assay will be used to guide treatment decisions. Here we review some of the problems or hurdles associated with assay development, especially for academic investigators. These include lack of expertise and resources for analytical validation, lack of experience in project design for a specific clinical use, lack of specimens from appropriate patient groups, and lack of access to Clinical Laboratory Improvement Act (CLIA) certified laboratories. In addition, financial support for assay validation has lagged behind financial support for marker discovery or drug development, even though the molecular diagnostic may be considered necessary for the successful use of the companion therapeutic. The National Cancer Institute supports a large number of clinical trials and a significant effort in drug development. In order to address some of these barriers for predictive and prognostic assays that will be used in clinical trials to select patients for a particular treatment, stratify patients into molecularly defined subgroups, or choose between treatments for molecularly defined tumors, the National Cancer Institute (NCI) has begun a pilot program designed to lessen barriers to the development of validated prognostic and predictive assays.
Keywords: prognostic assay, predictive assay, integral assay, assay validation, Clinical Assay Development Program
Introduction
Despite enthusiasm from patients, providers and clinicians for the use of predictive and prognostic biomarkers, to date few assays have met the standards needed to convince the clinical community that they can be used to make treatment decisions that will improve outcomes (1–5). In 2008, the President's Office of Science and Technology Policy (6) recommended that the National Institutes of Health (NIH) “develop a funding program for academic/industry collaborative projects addressing biomarker standardization, statistical methods, and other aspects of study design necessary for validating the clinical utility of molecular diagnostics based on genomic correlations with disease characteristics“ and that the Food and Drug Administration(FDA) develop “standards for study design and product performance with regard to regulatory review of new diagnostic products” as well as a “regulatory approach to co-development of diagnostics and therapeutics”. These statements were strongly supported by a combined FDA, American Association for Cancer Research (AACR) and National Cancer Institute (NCI) Cancer Biomarkers Collaborative (CBC) (7) that supported over 27 recommendations covering biospecimens, reference standards, analytical performance, standardization and harmonization, bioinformatics, collaboration and data sharing, regulatory issues, policy and education. But what are the major issues that prevent wider utilization of molecular diagnostics in cancer patients? Recent reviews have defined several issues relating to the complexity and heterogeneity of cancer, lack of appropriate tumor specimens, bias inherent in the assay platform or analysis, study design issues, issues related to analysis and interpretation of results, lack of appropriate controls or standards applicable to complex assays, and assay technical validation issues (8). Elsewhere in this Focus issue, Poste et al, as well as others (9–11) describe aspects of this problem by defining the different types of markers and their uses in clinical trials as well as the differences between the identification of markers during discovery and clinical assay development. In addition, Schilsky et al, and others (10–12) describe use of integral markers that are essential for the performance of a trial, such as identification of patients as candidates for a therapeutic, risk stratification, modification of the dose of a therapeutic or other aspect of medical decision-making. The use of such integral markers requires validation of the assay analytical performance so that the assay may be performed in a laboratory accredited under the Clinical Laboratory Improvement Act (CLIA) of 1988. This article reviews the background of analytical validation of a clinical assay, why the complete validation of most molecular diagnostic assays is currently not possible, and describes a new NCI pilot program designed to assist predictive and prognostic assays to move from research to clinical readiness.
Validation of Assay Performance – Introduction of “Fit-for-Purpose” and different classes of markers
Development of a prognostic or predictive assay that can guide treatment for cancer patients is usually an iterative process that begins with discovery of a molecular characteristic or signature and preliminary correlation with a clinical outcome. Subsequent steps involve optimization of the assay, including technical or analytical validation of the assay, clinical validation (clinical qualification), or evidence that the result of the analytically validated assay correlates with the clinical outcome of interest, and assessment of clinical utility for the intended use (does use of the assay result in better outcome than standard methods or treatment) (Fig 1).
Fig 1. Development of a prognostic or predictive assay for use in a clinical trial where clinical utility will be evaluated.
Figure depicts a potential path from discovery to clinical qualification of an assay,
The pharmaceutical industry has incorporated markers into its workflow for drug development (11, 12). However, in 2003, a workshop (13) of the American Association of Pharmaceutical Scientists, the Ligand Binding Assay Bioanalytical Focus Group and the U.S. Clinical Ligand Society, suggested that analytical validation of assays for markers should not follow the same process as bioanalytic assays commonly used by industry. Instead, the first step in assay development is to define the intended clinical use for the result of the assay. For predictive and prognostic assays, most clinicians consider the intended use to be how the marker assists medical decision-making in a specific clinical situation. The workshop suggested that, for the clinical laboratory scientist, a “useful starting point in determining the direction of a biomarker assay validation is to consider the assay to be used and the data type that the assay will generate (13).” The report defined several different types of assays depending on the type of data results. Definitive quantitative assays are those in which the result is a continuous number expressed using a definitive, approved or certified reference standard. An example is human insulin. Relative quantitative assays are those in which the output is a continuous number but the reference standard is not well characterized or “fully representative of the endogenous biomarker.” This category constitutes the great majority of clinical biomarkers where the reference standard is not approved or certified by a regulatory body but is added to the matrix of the sample in which the clinical assay is to be performed. This “spike in” can be affected by matrix components that interfere with detection of the analyte and, hence, lead to only relative accuracy. Quasi-quantitative assays were defined as those that also have an uncertified reference standard but are expressed in relation to a baseline characteristic of the sample. A fourth type of assay, very common in clinical practice, is the qualitative assay in which the results are presented in categorical terms – either as numbers (ordinal) or in nonnumeric (nominal) form. Immunohistochemistry results are qualitative assays usually expressed as low, medium or high or as 1+ to 3+. These assays generally lack calibrators but may have standards for the different categorical values that are usually not certified by a regulatory body. This report has come to be the foundation upon which the development of markers that are “fit-for-purpose” was initially based.
Subsequently, Lee et al (14) expanded this report to describe in greater detail what was meant by “fit-for-purpose” which generally means insuring that the assay performance is within reasonable expectations for its intended use. Lee et al (14) outlined the steps necessary for an iterative approach for assay improvement that focuses on the intended use of the assay and method development and validation intended to meet that purpose. This paper was intended for assays for biomarkers as a component of drug development as opposed to the development of a clinical diagnostic. As a result, the intended use becomes critical and requires focusing on developing an assay that is validated within the matrix expected for the intended use as well considerations of analyte stability rather than the stability of the reagents used in the assay. Another major difference is that a diagnostic performed for medical decision-making needs to be performed in a CLIA-certified laboratory, following standards provided by the Clinical Laboratory Standards Institute. Lee et al (14) and others (15–19) also set out guidelines for the number of replicates needed for validation of the performance of molecular diagnostic assays as well as such considerations as linearity of assay response, dynamic range, limits of detection, analyte stability within the intended matrix and intra- and inter-laboratory coefficient of variability (Text Box). While several of these authors have prescribed how many replicates should be performed and the limits of allowable variability, these recommendations may need to be altered according to the type of assay and marker to be developed. Chau et al (16), in a prior FOCUS Section, provided specific recommendations that validation of quantitative assays include “at least five different concentrations of VS [validation samples] analyzed in duplicate on at least six different runs during the prestudy validation because quantitative biomarker assays often exhibit nonlinear calibration curves.” Reproducibility of an assay is generally measured by the per cent of coefficient of variation (% CV) which is defined as the standard deviation divided by the mean of the assay result expressed as a percent. As Chau et al (16) suggested, the % CV should be less 25% although many clinical laboratory scientists will only accept much lower % CV's. The reproducibility of such qualitative assays as immunohistochemistry is a distinct problem in that it is difficult to measure variation. For immunohistochemical assays, reproducibility is generally measured in terms of kappa statistic (20) as well as percent Agreement (21) between different observers. It is unclear what is an ideal level of agreement or concordance in such qualitative assays, although a level of agreement of 85% or better is considered to be acceptable.
An example illustrates this problem. The Eastern Cooperative Oncology Group (ECOG) led a randomized Phase III stage II colon carcinoma adjuvant trial in which the 18q Loss of Heterozygosity (LOH) assay was an integral marker used to assign adjuvant therapy. The assay was based on the original description of 18q LOH by Jen et al. that suggested loss of 18q/DCC was associated with a poor prognosis (22, 23) that might benefit from adjuvant therapy. Since the assay was not validated prior to beginning the trial, the Cancer Diagnosis Program performed an inter-laboratory validation study (24) that involved the reference laboratories for ECOG, the North Central Cooperative Treatment Group, and the Cancer and Leukemia Group B, all of whom participated in this and other related Phase III trials in colon carcinoma. All three laboratories used a single Standard Operating Procedure (SOP), common Taqman primers and probes for the polymerase chain reaction assays and similar ABI Prism equipment for this assay, which was performed in formalin-fixed paraffin embedded diagnostic samples - all elements considered to assure concordance among observers. LOH is reported as a categorical variable, and inter-observer analysis was performed with DNA extracts from 50 pairs of tumor (colon carcinomas) and normal tissue, tested by the three laboratories. The inter-observer agreement was only 73% among the three laboratories which was surprisingly low given the common DNA extracts as a starting point. Interestingly, a second phase of concordance testing was performed when one of the collaborators, Dr. Thibodeau, suggested that the concordance may be improved by the individual reference pathologists using their own macro-dissection techniques to prepare the DNA extracts. When this was done in a second set of over 100 samples, the inter-laboratory agreement improved to 92% (24). This experience suggests that even when almost all of the potential variables are standardized, there may still be variables that can affect the results, e.g. individual laboratory optimization of tumor content to increase enrichment of tumor DNA to improve detection of LOH.
Challenges for Clinical Assay Development and Acceptance by Clinicians and Investigators
Assay analytical validation involves assessing accuracy, precision, specificity (ability to detect the analyte in a complex matrix), and sensitivity. It is often difficult to “validate” novel biomarker assays because there is not a “gold standard” assay, and there may not be any approved reference standard that can be used to assist validation of the assay method. In addition, Chau et al. (16) remarked that only the definitive quantitative assay can be truly accurate because the only in this type of method can the exact amount of the endogenous marker be measured since there is a gold standard, certified quality control that is suited for the matrix of the clinical assay. All other categories of assays only estimate the accuracy of a marker with standards provided by the assay developer but not at a standard that is approved by either the United States Pharmacopeia (USP) or the FDA (25). A reference material is defined by NIST (26) as a material that is homogeneous and stable enough to be fit for use in a measurement process. A certified reference material is a type of “reference material characterized by a metrologically valid procedure for one or more specified properties, accompanied by a certificate that provides the value of the specified property, its associated uncertainty, and a statement of metrological traceability. A NIST standard reference material is a “certified reference material issued by NIST that also meets additional NIST-specific certification criteria and is issued with a certificate or certificate of analysis that reports the results of its characterizations and provides information regarding the appropriate use(s) of the material”. The USP and the National Formulary provide certified reference materials that meet the standards of the International Organization for Standardization (ISO) for reference materials in their ISO 17025 (27) and ISO Guide 34 (28) for Reference Material Producers. Standard or certified reference materials are critical for inter-laboratory harmonization efforts because they can serve as a common reference standard. Clearly, complex endogenous genes, proteins and lipids may undergo many alterations during malignant transformation and provide a far greater challenge to the development of certified reference materials than the development of certified reference materials for analytes such as sodium, other electrolytes and even enzymes for which clinical chemistry tests are routine and accepted. However, until that challenge is met, it will be difficult for both the clinical laboratory and the clinician to accept the accuracy of any laboratory developed test that is not approved or cleared by the FDA or accepted by major healthcare insurance payers.
The Centers for Disease Control (CDC), through its initiation and support of the Get-RM project (29) has made reference materials that are publicly available through the Coriell Repository to support genetic tests. In addition to a number of reference materials for cystic fibrosis, Huntington disease, fragile × syndrome, and several genetic conditions with relatively high prevalence in the Ashkenazi Jewish population (30–33), it has also provided information for several pharmacogenetic markers, including members of the CYP450 gene family, VKORC1, and UGT1A1 (34). The GET-RM is now providing reference materials for both genomic and somatic mutations that occur in hematologic and solid malignancies (35, 36).
An example illustrates the importance of certified reference materials. Her2 testing in breast and other cancers may identify patients who could benefit from treatment that targets the gene (37). Her2 expression is identified by immunohistochemistry (IHC) for protein or Fluorescence In Situ Hybridization (FISH) for gene amplification. However, studies have shown considerable discordance in assay results even between CLIA-certified laboratories testing the same samples (38, 39). The College of American Pathology/American Society of Clinical Oncology issued guidelines in 2007 (40) that cover all aspects of pre-analytic and post-analytic procedures for the qualitative IHC and quasi-quantitative FISH assays and recommends proficiency testing twice a year with standard controls contained on each assay run. Since NIST does not provide a standard reference material for Her2 in its catalog (41) and USP does not have a Her2 analyte in its compendium (42), there is no certified traceable standard for the IHC and FISH assays. As a result, the accuracy of the tests can only be estimated, although the precision of the assay may be measured. Similar situations exist with other FDA approved, cleared or commonly accepted diagnostics, such as ALK translocation. This will become an increasing problem as clinical trials successfully demonstrate the clinical utility of an integral marker, because demand will increase that the marker be analyzed in community hospitals in the same manner as it was tested in the trial. Unfortunately, it is very difficult to assure that clinical laboratories will be able to provide the assay as it was performed in a clinical trial without standard or certified reference materials that trace back to specimens in the trial. The uncertainty caused in payers, clinicians and patients makes this an important issue. In future, with appropriate support from government agencies and the pharmaceutical industry, it may be possible to assure that samples from pivotal trials could be preserved to develop standard or certified reference materials for subsequent assay development.
Another major problem for scientists in academia or small business is the lack of funding to develop and validate clinical predictive and prognostic tests. The funding that supports development of new clinical and biological discoveries vastly exceeds that which supports clinical assay development and validation. In FY 2011 0.8% of the total NCI budget was spent on development of clinically useful assays as compared to 5.3% on identification of biomarkers (43). The actual difference between discovery and clinical assay development support may be considerably greater because it is difficult to determine what fraction of the molecular diagnostics portfolio actually is for development of assays within a CLIA-certified laboratory. The other major source of support for development and validation of clinical assays is diagnostics companies. These companies support the development of diagnostics for use in clinical cancer care, and/or collaborate with the Pharmaceutical industry to develop companion diagnostics. There has been a recent increase in this activity (44–47), but this source of support is generally not open to academic investigators or even many small biotechnology firms.
Several larger academic cancer centers e.g., M. D. Anderson Cancer Center, Memorial Sloan Kettering Cancer Center, Dana-Farber Cancer Institute with the Massachusetts General Hospital and the Brigham and Women's Hospital, Vanderbilt-Ingram Cancer Center, Moffitt Cancer Center and the Washington University Genomic Center, have recently begun to genotype patients who come to their institutions as a means of identifying patients who may be candidates for trials involving targeted therapy (48). These centers have resources to bring promising assays from the research laboratory to the CLIA certified laboratory. The costs for these efforts are shared between the institution and sponsors of the drugs that are under development. However, investigators at other institutions or small companies do not have access to these resources and find it difficult and bewildering if they want to convert their biologically important discovery into a clinically useful diagnostic. These investigators often have biologically important findings but may not be able to develop them into clinically useful tools unless they can turn to a clinical assay developer.
CADP – The NCI's Pilot Program Response to the Problems of Clinical Assay Development
In response to the need for technically validated assays to be incorporated in to NCI-supported clinical trials as integral assays, NCI developed The Clinical Assay Development Program (CADP) and the Molecular Characterization Clinical Assay Development Laboratory, The CADP is designed to overcome some of the hurdles faced by researchers in analytic and clinical validation of a biomarker assay. (See Table 1 for relevant definitions of terms.) Integral assays are those that must be performed on every patient entered in the trial in order for the trial to proceed (see Schilsky et al, in this Focus issue, (10)). These assays are used for patient selection (eligibility), stratification, or treatment assignment. It is especially important to perform analytical and clinical validation studies prior to using an assay in a clinical trial. (6–10). Whether or not an Investigational Device Exemption (IDE) from the FDA is needed depends on the risk the assay poses to the patient in the particular planned clinical use (see Meshinchi et al in this Focus issue, (49). Preparation for use of the assay either in a CLIA certified laboratory as a laboratory developed test or as an IDE, will require careful assessment of accuracy, precision, specificity, and sensitivity. The goals of the Clinical Assay Development Program (CADP) are to a) provide the needed services to evaluate, optimize and validate the analytical performance and clinical validity of promising assays that will be used in a clinical trial to demonstrate clinical utility; b) educate and engage researchers and others in a process of assay validation that leads to robust, clinically useful assays to guide cancer treatment and c) develop and evaluate standard operating procedures, controls and calibrators for clinical molecular characterization of malignancies. CADP is not a grants program but rather is a resource for investigators who want assistance converting their research assay into an assay that is performed in a CLIA-certified laboratory. Investigators apply to the program through an electronic submission process described on the CADP website (50). Investigators with markers that have a defined clinical use and biologic rationale, and a research assay that functions in human tissues but whose performance needs to be improved and validated for use in a clinical laboratory are excellent candidates for the CADP. Assays that are further along in development, but may need transfer to a CLIA environment, additional refinement or a platform transfer, are also suitable for CADP. If approved, investigators will be provided access to the NCI's full suite of assay development and validation resources. Appropriate attention to the validation of these assays will increase the probability for success of the clinical trials in which they are employed, resulting in improved patient management.
Table 1.
Definitions
| Term | Meaning |
|---|---|
| Analytical Validity | How accurately an assay detects the analyte of interest |
| Clinical Validity | How well the test relates to the clinical outcome of interest |
| Clinical Utility | Whether the results of the test provide information that can contribute to and improve current optimal management of the patient's disease |
| Integral marker (or assay) | Test(s) that must be performed in order for the trial to proceed. Integral studies are inherent to the design of the trial from the onset and must be performed in real time for the conduct of the trial. |
| Integrated marker (or assay) | Test(s) done on all patients, or a pre-defined subset of all patients, in a clinical trial, but will NOT be used to direct treatment for that clinical trial |
“Analytical Validity”, “Clinical Validity” and “Clinical Utility” defined as in reference 10
Components of the CADP
Clinical Assay Development Network (CADN)
The CADN has five academic and three commercial or reference CLIA laboratories to optimize and analytically validate complex assays. These laboratories were selected competitively for their proven ability to convert research assays to clinical assays. Thus, once an assay is selected for development by the CADP, each CADN laboratory bids on the ability to develop that assay. One or more laboratory(ies) are chosen, then the investigator/assay submitter works with the CADP management team and members of the CADN laboratory to develop the assay, which is then returned to the assay submitter. The CADN is intended to be very interactive with the original investigator and may continue to collaborate with the investigator once the assay is developed and validated. This may be very beneficial to the investigator who wants to use the assay in clinical trials but lacks the resources to do so directly in his or her institution. Available assay technologies in the CADN include immunohistochemistry, FISH, CISH, RT-PCR, genomic sequencing and LC-mass spectroscopy. Each assay to be developed will have a project management team, which includes the assay submitter. The project management team is responsible to devise a timeline, milestones, and go/no-go decision points for the project.
Specimen Retrieval System (SRS)
The Specimen Retrieval System is designed to provide needed specimens for analytic and clinical validation studies depending on the assay to be supported. Since it is anticipated that most tissue-based assays will use formalin-fixed paraffin embedded (FFPE) tissues because FFPE are the predominant form of diagnostic material currently available in US hospitals (51, 52), the CADP has contracted with the Health Maintenance Organization, Kaiser Permanente Northwest. This site was selected because of their participation in the Cancer Research Network of the Health Maintenance Organization Research Network. And can provide FFPE tissues from pathology archives that use standard handling and storage procedures. Tissues are obtained from patients undergoing surgery for malignancy with appropriate Institutional Review Board approval. In typical surgeries, more FFPE blocks are created than needed by institutions for diagnosis. The excess blocks are de-identified and provided to the CADP while the providing institutions retain the diagnostic blocks. These specimens are accompanied by clinical data that is routinely abstracted by Tumor Registries (part of the quality management system of the Commission on Cancer (53). These clinical data are derived from pathology reports, clinic visits, oncology visits, and pharmacy records. These records are de-identified by “scrubber” software which was derived from an earlier NCI-supported program for the retrieval of FFPE specimens from diagnostic archives for research purposes (54). This software has been modified by investigators from Harvard working with clinicians at Kaiser Permanente Northwest (55) and now is more than 99% effective in removing all personal health identifiers and information. Records are still manually checked so that when specimens are forwarded to the CADP the specimen and the clinical history are de-identified. This enables the CADP to access specimens that fit the clinical context for assay development and to access the clinical outcome information necessary to begin the process of qualifying assay results by correlation of assay results with clinical outcome. CADP may also contract with other sites as necessary for similar de-identified tissue if needed by a particular assay (e.g. frozen tissue, blood, plasma).
Other services
CADP provides advice on assay development, including potential development strategy, and focus on an appropriate and feasible intended use for the assay, through the involvement of the personnel in the Cancer Diagnosis Program. Statistical advice is also available through the NCI's Biomedical Research Branch. In addition, the potential applicant may be referred to other potential collaborators, with which they can negotiate their own collaboration (such as other academic investigators, Cooperative Groups, other tissue resources).
Procedures for access to the Clinical Assay Development Program
Academic researchers, commercial entities and government researchers may apply to the Program for needed validation services. Applications are received on the ProposalCentral application website. Applicants are asked to describe the potential clinical relevance of the proposed assay, must stipulate a single well defined intended clinical use, and must have a prototype assay that has been tested in relevant tumor tissue, as well as an estimate of the prevalence of the abnormality the assay will detect. Detailed data on additional previous work (if available), and relevant literature may be submitted as appendices. Assays that have progressed further toward validation are also encouraged. Applications are reviewed by a Special Emphasis Panel, constituted under the Federal Advisory Committee Act (FACA) composed of experts external to the NCI. Members of this committee include academic experts with disease, molecular biology and diagnostics expertise, members of industry with similar expertise, and patient advocates. Applications are evaluated on the basis of scientific rationale, feasibility, potential clinical impact and plans for assessment of clinical utility (see Table 2). Applications are then evaluated internally to assure that resources that are needed can indeed be obtained by NCI for the proposed assay or test. Unsuccessful applicants may resubmit the identical project once, and/or receive additional input on their efforts from Cancer Diagnosis Program staff. Applications are accepted three times a year.
Table 2.
Evaluation Criteria for CADP applications
| Scientific Merit (30%) | Sound hypothesis that associates marker(s) for drug effects or state of disease with a known clinical outcome and is supported by current state of the field. Marker is expected to have clinical utility. |
|
| |
| Feasibility (30%) | Highly technically feasible approach — assay platform and specimen requirements should allow straightforward implementation for clinical use. |
|
| |
| Impact/ Clinical Need (30%) | If successful, the proposed diagnostic is likely to change clinical practice and have a major impact on outcome of treatment. |
|
| |
| Path to Clinical Implementation (10%) | The path to clinical implementation is clear. |
Molecular Characterization-Clinical Assay Development Laboratory (MC-CADL)
The MC-CADL is located at SAIC-Frederick under contract to the NCI. Established to provide a means of translating genome discoveries into clinical applications, the MC-CADL will focus on newer genomic technologies such as whole transcriptome gene expression and next generation sequencing. Importantly, the MC-CADL will apply these technologies using well defined, robust protocols and appropriate standards and controls to ensure reproducible results. The goal is to discern molecular properties of cancer that explain response, or its lack, to investigational targeted therapy. It will assist with early phases of assay development and transition to clinical laboratory readiness and support NCI-sponsored clinical trials within and outside the institute by developing standards and calibrators that can be used to compare assays between different laboratories or across platforms and can be shared with others. In this capacity the MC-CADL also seeks to work with the FDA, NIST and other regulatory bodies that may develop standard and certified reference materials.
The MC-CADL will use FFPE specimens, as these are the dominant form of tissue throughout the country and will facilitate adoption of developed assays. It will collaborate with the NCI's Office of Biorepositories and Biospecimen Research (OBBR) (56) and the Cancer Genome Atlas to study characteristics of specimens that affect the molecular characterization of malignancies particularly in massively parallel sequencing platforms. As part of the CADP, successful efforts from the MC-CADL can be transferred into the CADN and/or the MC-CADL could serve as a reference laboratory for the CADN and the investigator.
Conclusion
Many challenges remain to the adoption of molecular diagnostics into routine clinical practice. In addition to the performance of the assay, the association of the result with a clinical endpoint and the demonstration that such association is clinically useful, there are the other concerns highlighted by the OSTP Policy Report (6) and the CBC (7) that involve lack of appropriate biospecimens, standardization and harmonization, bioinformatics, collaboration and data sharing, regulatory issues, policy and education. It is difficult to address all of these issues at once. The CADP addresses several because it enables independent academic and small company investigators to develop clinically rigorous assays that then can enter clinical trials for clinical qualification and ultimately assessment of clinical utility. Members of the CADN may be available to the investigators and able to assist with supporting clinical assays in clinical trials in addition to assay validation assistance. A major unmet need recognized by the OSTP Policy Report (6) as well as the CBC (7) is the need for reference standards as a means to assess the accuracy of many molecular diagnostic assays. The MC-CADL is also beginning to address the reference issue through the creation of resources that may be able to be certified and distributed as standard or certified reference materials. Hopefully, as we move forward in the next few years certified reference materials will become available so that the accuracy of tests may actually be measured, and correlation of results across different laboratories will be facilitated.
A Few Typical Questions About Analytical Performance for Molecular Diagnostic Assays To Be Considered in Assay Development.
What is the dynamic range of the assay (Units)?
What are the usable limits of detection for the assay (Units)?
Is the assay Linear† in the usable range?
How stable is the analyte within its matrix? (< 7 days, 7–14 days, 15–30 days or longer and under what storage conditions)
What is the accuracy for detecting the analyte or alterations (mutations) in the analyte within the matrix?
What is the sensitivity and specificity of the assay?
What is the intra-lab reproducibility (% Coefficient of Variation, %CV)?
What is the inter-lab reproducibility (%CV, same specimens)?
Legend: This information for the analytical performance of a clinical assay is adapted from the templates for IHC and ISH available at the Cancer Diagnosis Program (http://cdp.cancer.gov/diagnostics/templates.htm) or Cancer Treatment and Evaluation Program (http://ctep.cancer.gov/protocolDevelopment/docs/IHC_TemplateHb.pdf and http://ctep.cancer.gov/protocolDevelopment/docs/ISH_Template_Ca.pdf) websites. The information in this table is not intended to be complete for analysis of analytical performance of all clinical assays but to provide a few characteristics that need to be considered during assay development.
† NCCLS Evaluation of the linearity of quantitative measurement procedures: a statistical approach. Approved guideline NCCLS document EP6-A (ISBN 1-56238-498-8). Wayne, PA: NCCLS; 2003:47pp. (Downloaded from: http://www.clsi.org/source/orders/free/ep6-a.pdf on Jan 14, 2012). Linearity is often important in Enzyme-Linked Immunosorbent Assays and other assays where a standard curve is made and the amount of analyte calculated from that standard curve.
%CV is equal to the standard deviation of the assay divided by the mean of the and expressed as a percent.
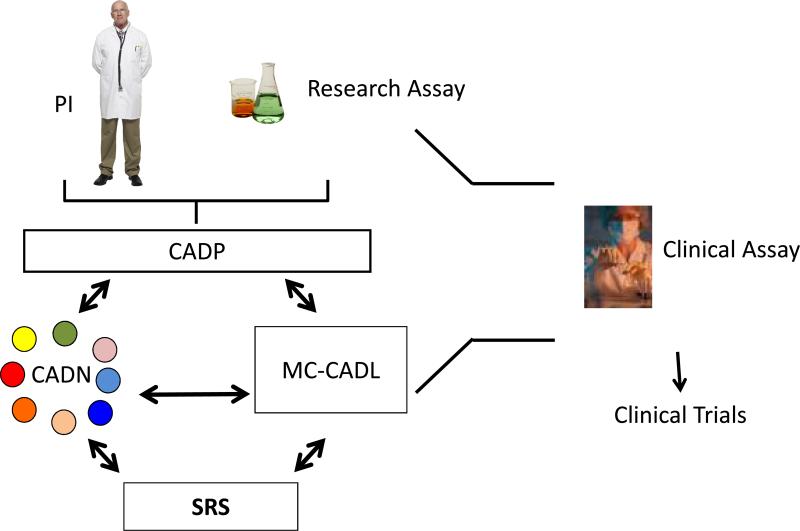
Fig 2. Overview of the workflow of the Clinical Assay Development Program.
The PI (assay submitter) with his or her research assay applies to the CADP (http://cadp.cancer.gov). If selected, the PI and the assay are assigned to one or more of the CADN laboratories (small colored circles) that work with the PI (submitter) and the CADP management team. Specimens are provided as needed and fit for purpose by the Specimen Retrieval System (SRS). If appropriate, the MC-CADL may assist with the development of the assay. When the assay is developed and its analytical performance is validated, the clinical assay is returned to the submitter from the CADP and is ready for use in clinical trials. It should be noted that it is possible for the CADN laboratories to assist in the performance of the assay in clinical trials, but this arrangement would be made by the assay submitter, not through CADP.
Footnotes
The opinions expressed in this manuscript are those of the authors and do not necessarily represent those of the National Cancer Institute, the National Institutes of Health or the Department of Health and Human Services.
References
- 1.Hayes DF, Bast RC, Desch CE, Fritsche H, Jr, Kemeny NE, Jessup JM, et al. Tumor marker utility grading system: a framework to evaluate clinical utility of tumor markers. J Natl Cancer Inst. 1996;88:1456–66. doi: 10.1093/jnci/88.20.1456. [DOI] [PubMed] [Google Scholar]
- 2.Locker GY, Hamilton S, Harris H, Jessup JM, Kemeny N, Macdonald JS, et al. ASCO 2006 update of recommendations for the use of tumor markers in gastrointestinal cancer. J Clin Oncol. 2006;24:5313–27. doi: 10.1200/JCO.2006.08.2644. [DOI] [PubMed] [Google Scholar]
- 3.Harris L, Fritsche H, Mellel R, Norton L, Ravdin P, Taube S, et al. American Society of Clinical Oncology 2007 update of recommendations for the use of tumor markers in breast cancer. J Clin Oncol. 2007;25:5287–312. doi: 10.1200/JCO.2007.14.2364. [DOI] [PubMed] [Google Scholar]
- 4.Allegra CJ, Jessup JM, Somerfield MR, Hamilton SR, Hammond EH, Hayes DF, et al. American Society of Clinical Oncology Provisional Clinical Opinion: Testing for KRAS gene mutations in patients with metastatic colorectal carcinoma to predict response to anti-epidermal growth factor receptor monoclonal antibody therapy. J Clin Oncol. 2009;27:2091–95. doi: 10.1200/JCO.2009.21.9170. [DOI] [PubMed] [Google Scholar]
- 5.Keedy VL, Temin S, Somerfield MR, Beasley MB, Johnson DH, McShane LM, et al. American Society of Clinical Oncology Provisional Clinical Opinion: Epidermal growth factor receptor (EGFR) mutation testing for patients with advanced non-small-cell lung cancer considering first-Line EGFR tyrosine kinase inhibitor therapy. J Clin Oncol. 2011;29:2121–27. doi: 10.1200/JCO.2010.31.8923. [DOI] [PubMed] [Google Scholar]
- 6.Priorities for Personalized Medicine. Report of the President's Council of Advisors on Science and Technology. 2008 September; Downloaded from http://www.whitehouse.gov/files/documents/ostp/PCAST/pcast_report_v2.pdf on December 12, 2011.
- 7.Khleif SN, Doroshow JH, Hait WN. AACR-FDA-NCI Cancer Biomarkers Collaborative. AACR-FDA-NCI Cancer Biomarkers Collaborative consensus report: advancing the use of biomarkers in cancer drug development. Clin Cancer Res. 2010;16:3299–318. doi: 10.1158/1078-0432.CCR-10-0880. [DOI] [PubMed] [Google Scholar]
- 8.Coyle VM, Johnston PG. Genomic markers for decision making: what is preventing us from using markers? Nat Rev Clin Oncol. 2010;7:90–97. doi: 10.1038/nrclinonc.2009.214. [DOI] [PubMed] [Google Scholar]
- 9.Poste G, Carbone DP, Parkinson DR, Verweij J, Hewitt S, Jessup JM. Leveling the playing Field: bringing development of biomarkers and molecular diagnostics up to the standards for drug development. Clin Cancer Res. 2012;18 doi: 10.1158/1078-0432.CCR-11-2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schilsky RL, Doroshow JH, LeBlanc M, Conley BA. Development and use of integral assays in clinical trials. Clin Cancer Res. 2012;18 doi: 10.1158/1078-0432.CCR-11-2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carden CP, Sarker D, Postel-Vinay S, Yap TA, Attard G, Banerji U, et al. Can molecular biomarker-based patient selection in Phase I trials accelerate anticancer drug development? Drug Discov Today. 2010;15:88–97. doi: 10.1016/j.drudis.2009.11.006. [DOI] [PubMed] [Google Scholar]
- 12.Valentin MA, Ma S, Zhao A, Legay F, Avrameas A. Validation of immunoassay for protein biomarkers: bioanalytical study plan implementation to support pre-clinical and clinical studies. J Pharm Biomed Anal. 2011;55:869–77. doi: 10.1016/j.jpba.2011.03.033. [DOI] [PubMed] [Google Scholar]
- 13.Lee JW, Weiner RS, Sailstad JM, Bowsher RR, Knuth DW, O'Brien PJ, et al. Method validation and measurement of biomarkers in nonclinical and clinical samples in drug development: a conference report. Pharm Res. 2005;22:499–511. doi: 10.1007/s11095-005-2495-9. [DOI] [PubMed] [Google Scholar]
- 14.Lee JW, Devanarayan V, Barrett YC, Weiner R, Allinson J, Fountain S, et al. Fit-for-purpose method development and validation for successful biomarker measurement. Pharm Res. 2006;23:312–28. doi: 10.1007/s11095-005-9045-3. [DOI] [PubMed] [Google Scholar]
- 15.Cummings J, Raynaud F, Jones L, Sugar R, Dive C. Fit-for-purpose biomarker method validation for application in clinical trials of anticancer drugs. Br J Cancer. 2010;103:1313–17. doi: 10.1038/sj.bjc.6605910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chau CH, Rixe O, McLeod H, Figg WD. Validation of analytic methods for biomarkers used in drug development. Clin Cancer Res. 2008;14:5967–76. doi: 10.1158/1078-0432.CCR-07-4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wagner JA, Williams SA, Webster CJ. Biomarkers and surrogate end points for fit-for-purpose development and regulatory evaluation of new drugs. Clin Pharmacol Ther. 2007;81:104–7. doi: 10.1038/sj.clpt.6100017. [DOI] [PubMed] [Google Scholar]
- 18.Williams SA, Slavin DE, Wagner JA, Webster CJ. A cost-effectiveness approach to the qualification and acceptance of biomarkers. Nat Rev Drug Discov. 2006;5:897–902. doi: 10.1038/nrd2174. [DOI] [PubMed] [Google Scholar]
- 19.Lathia CD, Amakye D, Dai W, Girman C, Madani S, Mayne J, et al. The value, qualification, and regulatory use of surrogate end points in drug development. Clin Pharmacol Ther. 2009;86:32–43. doi: 10.1038/clpt.2009.69. [DOI] [PubMed] [Google Scholar]
- 20.Mengel M, von Wasielewski R, Wiese B, Rudiger T, Muller-Hermelink HK, Kreipe H. Inter-laboratory and inter-observer reproducibility of immunohistochemical assessment of the Ki-67 labelling index in a large multi-centre trial. J Pathol. 2002;198:292–9. doi: 10.1002/path.1218. [DOI] [PubMed] [Google Scholar]
- 21.Garg K, Broaddus RR, Soslow RA, Urbauer DL, Levine DA, Djordjevic B. Pathologic scoring of PTEN immunohistochemistry in endometrial carcinoma is highly reproducible. Int J Gynecol Pathol. 2012;31:48–56. doi: 10.1097/PGP.0b013e3182230d00. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jen J, Kim H, Piantadosi S, Liu ZF, Levitt RC, Sistonen P, et al. Allelic loss of chromosome 18q and prognosis in colorectal cancer. N Engl J Med. 1994;331:213–21. doi: 10.1056/NEJM199407283310401. [DOI] [PubMed] [Google Scholar]
- 23.Shibata D, Reale MA, Lavin P, Silverman M, Fearon ER, Steele G, Jr, et al. The DCC protein and prognosis in colorectal cancer. N Engl J Med. 1996;335:1727–32. doi: 10.1056/NEJM199612053352303. [DOI] [PubMed] [Google Scholar]
- 24.Jessup JM, Dobbin K, Hamilton S, Thibodeau S, Redston M, Taube S, et al. Interlaboratory assay reproducibility study for loss of heterozygosity on chromosome 18 (18q LOH) in colon cancer. J Clin Oncol. 2009;27(suppl):15s. abstr 4052. [Google Scholar]
- 25.FDA Device Advice. website: http://www.fda.gov/MedicalDevices/DeviceRegulationandGuidance/default.htm.
- 26. http://www.nist.gov/srm/definitions.cfm.
- 27. http://www.iso.org/iso/Catalogue_detail?csnumber=39883.
- 28. http://www.iso.org/iso/iso_catalogue/catalogue_tc/catalogue_detail.htm?csnumber=50174.
- 29. http://wwwn.cdc.gov/dls/genetics/rmmaterials.
- 30.Pratt VM, Caggana M, Bridges C, Buller AM, DiAntonio L, Highsmith, et al. Development of genomic reference materials for cystic fibrosis genetic testing. J Mol Diagn. 2009;11:186–93. doi: 10.2353/jmoldx.2009.080149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kalman L, Johnson MA, Beck J, Berry-Kravis E, Buller A, Casey B, et al. Development of genomic reference materials for Huntington disease genetic testing. Gen Med. 2007;9:719–23. doi: 10.1097/gim.0b013e318156e8c1. [DOI] [PubMed] [Google Scholar]
- 32.Amos Wilson J, Pratt VM, Phansalkar A, Muralidharan K, Highsmith WE, et al. Fragile Xperts Working Group of AMP: Consensus characterization of 16 FMR1 reference materials: a consortium study. J Mol Diagn. 2008;10:2–12. doi: 10.2353/jmoldx.2008.070105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kalman L, Amos Wilson J, Buller A, Dixon J, Edelmann L, Geller L, et al. Development of genomic DNA reference materials for genetic testing of disorders common in people of Ashkenazi Jewish descent. J Mol Diagn. 2009;11:530–36. doi: 10.2353/jmoldx.2009.090050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pratt VM, Zehnbauer B, Wilson JA, Baak R, Babic N, Bettinotti M, et al. Characterization of 107 genomic DNA reference materials for CYP2D6, CYP2C19, CYP2C9, VKORC1, and UGT1A1: a GeT-RM and Association for Molecular Pathology collaborative project. J Mol Diagn. 2010;12:835–46. doi: 10.2353/jmoldx.2010.100090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barker SD, Bale S, Booker J, Buller A, Das S, Friedman K, et al. Development and characterization of reference materials for MTHFR, SERPINA1, RET, BRCA1, and BRCA2 genetic testing. J Mol Diagn. 2009;11:553–61. doi: 10.2353/jmoldx.2009.090078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. http://www.cdc.gov/dls/genetics/rmmaterials/default.aspx.
- 37.Slamon DJ, Leyland-Jones B, Shak S, Fuchs H, Paton V, Bajamonde A, et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med. 2001;344:783–92. doi: 10.1056/NEJM200103153441101. [DOI] [PubMed] [Google Scholar]
- 38.Paik S, Bryant J, Tan-Chiu E, Romond E, Hiller W, Park K, et al. Real-world performance of HER2 testing—National Surgical Adjuvant Breast and Bowel Project experience. J Natl Cancer Inst. 2002;94(11):852–54. doi: 10.1093/jnci/94.11.852. [DOI] [PubMed] [Google Scholar]
- 39.Roche PC, Suman VJ, Jenkins RB, Davidson NE, Martino S, Kaufman PA, et al. Concordance between local and central laboratory HER2 testing in the breast intergroup trial N9831. J Natl Cancer Inst. 2002;94(11):855–57. doi: 10.1093/jnci/94.11.855. [DOI] [PubMed] [Google Scholar]
- 40.Wolff AC, Hammond ME, Schwartz JN, Hagerty KL, Allred DC, Cote RJ, et al. American Society of Clinical Oncology/College of American Pathologists. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. Arch Pathol Lab Med. 2007;131:18–43. doi: 10.5858/2007-131-18-ASOCCO. [DOI] [PubMed] [Google Scholar]
- 41.NIST catalog. http://www.nist.gov/srm/upload/SRM-2011-Cat-v2_All.pdf.
- 42.USP. website for CRM: http://www.usp.org/referenceStandards/
- 43.This is based on a Query, View and Report (QVR, http://era.nih.gov/nih_and_grantor_agencies/other/query_view_and_report.cfm) search on 12/12/2011 for active grants on QVR using the terms `assay development' and `molecular diagnostics' compared to grants found with `biomarkers' alone. The total budget for NCI from public sources for FY2011 was given as slightly less than $5.3 billion (http://www.cancer.gov/aboutnci/servingpeople/nci-budget-information).
- 44.Wilson C, Schulz S, Waldman SA. Biomarker development, commercialization, and regulation: Individualization of medicine lost in translation. Clin Pharmacol Ther. 2007;81:153–55. doi: 10.1038/sj.clpt.6100088. [DOI] [PubMed] [Google Scholar]
- 45.Oakman C, Bessi S, Zafarana E, Galardi F, Biganzoli L, Di Leo A. Recent advances in systemic therapy: new diagnostics and biological predictors of outcome in early breast cancer. Breast Cancer Res. 2009;11:205. doi: 10.1186/bcr2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. http://www.fda.gov/MedicalDevices/ProductsandMedicalProcedures/DeviceApprovalsandClearances/Recently-ApprovedDevices/ucm268836.htm.
- 47. http://www.fda.gov/MedicalDevices/ProductsandMedicalProcedures/DeviceApprovalsandClearances/Recently-ApprovedDevices/ucm270832.htm.
- 48.Kaiser J. Looking for a target on every tumor. Science. 2009;326:218–20. doi: 10.1126/science.326_218. [DOI] [PubMed] [Google Scholar]
- 49.Meshinchi S, Hunger SP, Aplenc R, Adamson PC, Jessup JM. Lessons learned from the Investigational Device Exemption (IDE) review of Children's Oncology Group Trial AAML1031. Clin Cancer Res. 2012;18 doi: 10.1158/1078-0432.CCR-11-2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. http://cadp.cancer.gov/
- 51.Signoretti S, Bratslavsky G, Waldman FM, Reuter VE, Haaga J, Merino M, et al. Tissue-based research in kidney cancer: current challenges and future directions. Clin Cancer Res. 2008;14:3699–705. doi: 10.1158/1078-0432.CCR-07-4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lim MD, Dickherber A, Compton CC. Before you analyze a human specimen, think quality, variability, and bias. Anal Chem. 2011;83:8–13. doi: 10.1021/ac1018974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stewart AK, McNamara E, Gay EG, Banasiak J, Winchester DP. The Rapid Quality Reporting System--a new quality of care tool for CoC-accredited cancer programs. J Registry Manag. 2011;38:61–3. [PubMed] [Google Scholar]
- 54.Patel AA, Gupta D, Seligson D, Hattab EM, Balis UJ, Ulbright TM, et al. Shared Pathology Informatics Network.. Availability and quality of paraffin blocks identified in pathology archives: a multi-institutional study by the Shared Pathology Informatics Network (SPIN) BMC Cancer. 2007;28:7–37. doi: 10.1186/1471-2407-7-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Glass A, Taube S, McMurry A, Eddy C, La Chance PA, McShane L, et al. C-C4-02: Using a natural language processor to remove all elements of personal health information (PHI) to de-identify clinical annotations for the Specimen Retrieval System (SRS) Clin Med Res. 2011;9:170. [Google Scholar]
- 56. http://biospecimens.cancer.gov/default.asp.