Abstract
Progressive multifocal leukoencephalopathy (PML), a fatal demyelinating disease caused by JC virus (JCV) infection of oligodendrocytes, may develop in patients with immune disorders following reactivation of chronic benign infection. Mutations of JCV capsid viral protein 1 (VP1), the capsid protein involved in binding to sialic acid cell receptors, might favor PML onset.
Cerebrospinal fluid sequences from 37/40 PML patients contained one of several JCV VP1 amino acid mutations, which were also present in paired plasma but not urine sequences despite the same viral genetic background. VP1-derived virus-like particles (VLPs) carrying these mutations lost hemagglutination ability, showed different ganglioside specificity, and abolished binding to different peripheral cell types compared with wild-type VLPs. However, mutants still bound brain-derived cells, and binding was not affected by sialic acid removal by neuraminidase. JCV VP1 substitutions are acquired intrapatient and might favor JCV brain invasion through abrogation of sialic acid binding with peripheral cells, while maintaining sialic acid–independent binding with brain cells.
Progressive multifocal leukoencephalopathy (PML) is a rare demyelinating disease caused by the polyomavirus JC (JCV) infection of oligodendrocytes. It develops in the context of immune disorders such as human immunodeficiency virus (HIV) infection, hematological malignancies, transplantation, and treatments with immunomodulatory drugs. In most cases, PML is fatal within a few months from onset; there is no specific therapy, and reversion of the immune suppression, when feasible, remains the only proven approach for management of this disease [1, 2].
JCV establishes a chronic asymptomatic infection in the urinary tract in approximately 50% of the population and is excreted in urine in approximately one-third of healthy subjects [3, 4]. The mechanisms leading to PML are mostly unknown, yet involve JCV reactivation in the context of compromised immune control. The major capsid viral protein 1 (VP1) is likely a key player in pathogenesis, as it mediates immune responses, as well as cell attachment [5, 6] and viral cell entry [7] through sialic acid cell receptors. The VP1 gene is highly polymorphic; 13 distinct geographically associated subtypes have been identified [8]. However, there is no clear evidence of an association between any JCV subtype and PML.
We have recently examined all VP1 sequences deposited in GenBank (the National Institutes of Health genetic sequence database), and observed that a small number of mutations exclusively found in the brain or cerebrospinal fluid (CSF) of PML patients is positively selected. Three-dimensional modeling of VP1 molecular structure indicated that these substitutions are located at or close to binding sites for the sialic acid receptor [9]. Here, we undertook a detailed study of VP1 sequences derived from CSF, plasma, and urine samples of a large cohort of PML patients to assess whether positively selected mutations (defined as “PML-associated substitutions”) arise within the patient, and to characterize their spectrum and evolution during PML. To investigate their role in PML pathogenesis, we constructed mutated virus-like particles (VLPs) and studied their binding to different sialic acid–containing oligosaccharides as well as with various peripheral and central nervous system (CNS)-specific cells.
MATERIALS AND METHODS
Patients and Samples
This study was approved by the Ethical Committee of San Raffaele Hospital. To investigate the presence and evolution of PML-associated mutations in vivo, we examined CSF, and plasma and urine samples when available and positive for the presence of JCV DNA[10], from 84 PML patients followed at the Department of Infectious Diseases of San Raffaele Hospital between 1992 and 2009. Patient characteristics are shown in Table 1.
Table 1.
Patient Variables According to JCV-VP1 Amino Acid Substitution Position
| All | None | 50–61 | 122–134 | 265–271 | 283 | |
| No. of patients | 40 | 3 | 14 | 3 | 19 | 1 |
| HIV positive:HIV negative (no. of patients) | 30:10 | 3:0 | 12:2 | 3:0 | 12:7 | 0:1 |
| Year of observation, 1993–2000:2001–2009 (no. of patients) |
22:18 | 1:2 | 8:6 | 2:1 | 10:9 | 1:0 |
| Year of age (median, IQRa) |
41(34–54) | 42(36–48) | 39(36–47) | 38(35–42) | 49(34–6) | 8 |
| Females:males (no. of patients) | 16:24 | 0:3 | 4:10 | 2:1 | 10:9 | 0:1 |
| CSF JCV DNA log c/mL (median, IQR)b | 4.65(3.99–6.15) | 3.71(3.55–4.07) | 4.90(4.15–6.06) | 3.61(3.37–3.89) | 5.03(4.05–6.31) | 5.69 |
| Survivors:progressors (no. of patients) | 8:32 | 1:2 | 4:10 | 0:3 | 3:16 | 0:1 |
| Survivors:progressors, cART only (no. of patients) | 8:12 | 1:2 | 4:5 | 0:2 | 3:3 | 0:0 |
| CD4 cells/μL at PMLc (median, IQR) | 97(23–154) | 9(7–51) | 102(35–300) | 106(98–113) | 132(15–162) | NA |
| Plasma HIV-1 RNA log c/mL at PMLc (median, IQR) | 4.44(3.16–4.90) | 4.48(3.97–5.09) | 3.34(2.89–5.07) | 4.76(4.63–4.90) | 4.18(3.64–4.31) | NA |
| CD4 cells/μL at CSF samplingc (median, IQR) | 4.04(3.10–4.48) | 9(7–51) | 102(35–300) | 82(72–93) | 119(20–170) | NA |
| Plasma HIV-1 RNA log c/mL at CSF samplingc(median, IQR) | 4.04(3.10–4.48) | 4.48(3.97–5.09) | 3.34(2.63–5.04) | 2.87(2.28–3.45) | 4.18(3.64–4.31) | NA |
NOTE. JCV indicates JC virus; VP1, viral protein 1; HIV, human immunodeficiency virus; IQR, interquartile range; CSV, cerebrospinal fluid; cART, combination antiretroviral therapy; NA, not applicable.
1st and 3rd interquartile range.
No mutation and 122–134 substitutions vs. all the other substitutions, P = .0067.
In HIV-positive patients only.
Measurements
DNA was extracted from CSF, plasma, or urine samples using the QIAamp Blood Kit (Qiagen). JCV DNA was quantified by real-time polymerase chain reaction (PCR) [10]. VP1 was amplified either using primers flanking the whole VP1 gene, generating a fragment 2027 base pairs (bp) long (outer primers VP1-LF GCAGCCAGCTATGGCTTTAC and VP1-LR GCTGCCATTCATGAGAGGAT; inner primers VP1-SF CCTCAATGGATGTTGCCTTT and VP1-SR AAAACCAAAGACCCCT) or, when amplification with this method was not successful, by a seminested assay, generating a 1232-bp–long fragment [11] (see Supplementary Data for PCR conditions). Amplification products were purified by the Qiagen purification kit, and either cloned using a TOPO TA cloning kit (Invitrogen) or directly sequenced. Clones or purified PCR products were sequenced using a Model 3730XL DNA analyzer (Applied Biosystems). Following sequence translation, amino acid mutations were marked by comparison to a large selection of VP1 sequences from PML and non-PML cases obtained from GenBank [9]. Sequences were aligned using the ClustalW [12] multiple sequence alignment program (European Molecular Biology Laboratory–European Bioinformatics Institute [EMBL-EBI]), and the frequency of mutations in each sample was determined by the MargFreq program (freeware; http://sray.med.som.jhmi.edu/SCRoftware/margfreq/). Only mutations present in more than one clone for a sample were considered.
VLPs for all experiments were purified and tested for quality using electron microscopy as described [9].
Hemagglutination assay was performed as described [9].
For the ganglioside enzyme–linked immunosorbent assay (ELISA), specific gangliosides (EMD chemicals, Calbiochem; GM2, American Radiolabeled Chemicals, Inc.) were resuspended in methanol (MeOH) and coated onto a 96-well plate (Corning, 10 μg per well) overnight. After blocking with phosphate-buffered saline (PBS) containing Ca2+/Mg2, 1% bovine serum albumin (BSA, fraction V) (Sigma) and 0.1% Tween-20 VLPs were added to the wells at 30 μg/mL in the above buffer. After 90 min incubation at 4°C, the plates were washed and VLP binding was detected with anti-VP1 antibody PAB597 (courtesy Ed Harlow) and HRP–anti-mIgG (Jackson ImmunoResearch). Plates were developed with Turbo TMB-ELISA solution (Pierce Chemical Co.) and stopped with 2 N H3PO4; the plate was read on a spectrophotometer at 450 nm.
For flow cytometry analysis of VLP binding, 1–5 × 105 SVG-A cells (courtesy Walter Atwood), Jurkat cells (TIB-152, ATCC), primary human astrocytes (ScienCell), and renal proximal tubular epithelial cells (ScienCell; see Supplemental Material for culture conditions) were incubated with VLP (10–30 μg/mL) in FACS buffer (1% BSA, 2 mM NaN3 PBS with Ca2+/Mg2+) for 60 min at 4°C. Cells were washed with FACS buffer and further incubated with anti-VP1 antibody (PAB597) followed by incubation with AlexaFluor488–anti-mIgG (Jackson ImmunoResearch). After a brief fixation, cells were analyzed on a BD FACSCalibur system. In some cases, cells were pretreated with neuraminidase according to manufacturer’s recommendation (New England Biolabs). Appropriate controls and staining with lectins (peanut agglutinin [PNA], Sambucus nigra agglutinin [SNA], Maackia amurensis lectin II [MAL II]) were done as necessary. The degree of VLP binding was evaluated as the ratio of mean fluorescent intensity (MFI) of cells stained in the presence of VLPs to MFI of cells stained without VLPs (ie, background).
Statistics
Statistical analysis of VLP binding was done using Student t test, statistical analysis of PML patient variables was done by nonparametric tests for either continuous (Mann-Whitney test) or discrete (Fisher exact test) variables.
RESULTS
JCV VP1 Mutations in CSF and Plasma, But Not Urine of PML Patients
To investigate whether PML-associated mutations are specifically selected in CSF of PML patients, we analyzed JCV VP1 sequences from paired CSF and urine samples of 8 patients (Table 2, patients 1–8). In each patient, CSF carried 1 amino acid substitution that was not present in the urine-derived sequence. CSF and urine sequences were otherwise identical within individual patients and characterized by identical polymorphisms (ie, same virus subtype), as compared with the reference strain [8]. In all of 13 cases, plasma VP1 sequences (Table 2, patients 1–5, 9–16) were identical to the correspondent CSF-derived sequence, carrying the same PML-associated substitution. In both CSF and plasma, these substitutions were present in all or the vast majority (>95%) of the clones analyzed. These observations suggest that during PML, the virus circulating in CSF and blood has acquired specific VP1 changes relative to the parental urine virus within the patient, and constitutes the sole (or at least the predominant) viral population in these sites.
Table 2.
JCV VP1 Amino Acid Substitutions in Sequences Derived From Paired Urine, Cerebrospinal Fluid (CSF), and Plasma Samples From PML Patients
| Patient ID | Sample | JCV subtypea | PML-associated substitution | No. of clones with substitution/no. of clones analyzed | |
| 1 | 5067 | Urine | 1A | none | 0/11 |
| 5067 | CSF | 1A | H122R | 27/27 | |
| 5067 | Plasma | 1A | H122R | 28/28 | |
| 2 | 5167 | Urine | 2B-v69D,128A | none | 0/23 |
| 5167 | CSF | 2B-v69D,128A | S267Y | PCRb | |
| 5167 | Plasma | 2B-v69D,128A | S267Y | 42/44 | |
| 3 | 5166 | Urine | 1A | none | 0/29 |
| 5166 | CSF | 1A | S269F | 14/14 | |
| 5166 | Plasma | 1A | S269Fc | 18/21 | |
| 4 | 5174 | Urine | 1B | none | 0/19 |
| 5174 | CSF | 1B | S269F | 28/28 | |
| 5174 | Plasma | 1B | S269F | 37/38 | |
| 5 | 5180 | Urine | 1A | none | PCR |
| 5180 | CSF | 1A | S269F | PCR | |
| 5180 | Plasma | 1A | S269F | PCR | |
| 6 | 5211 | Urine | 2B | none | PCR |
| 5211 | CSF | 2B | none | PCR | |
| 5211 | Plasma | N | NA | NA | |
| 7 | 5087 | Urine | 1B | none | 0/11 |
| 5087 | CSF | 1B | L55F | 19/19 | |
| 5087 | Plasma | NA | NA | NA | |
| 8 | 5152 | Urine | 1A | none | 0/3 |
| 5152 | CSF | 1A | H122R; A2V | 44/46; 45/46 | |
| 5152 | Plasma | NA | NA | NA | |
| 9 | 5088 | Urine | NA | NA | NA |
| 5088 | CSF | 4-v164K | none | 0/3 | |
| 5088 | Plasma | 4-v164K | none | 0/24 | |
| 10 | 5092 | Urine | NA | NA | NA |
| 5092 | CSF | 1A | L55F | 20/20 | |
| 5092 | Plasma | 1A | L55F | PCR | |
| 11 | 5108 | Urine | NA | NA | NA |
| 5108 | CSF | 4-v128A,345K | L55F | 24/24 | |
| 5108 | Plasma | 4-v128A,345K | L55F | 30/30 | |
| 12 | 5201 | Urine | NA | NA | NA |
| 5201 | CSF | 1B | K60N | PCR | |
| 5201 | Plasma | 1B | K60N | PCR | |
| 13 | 5029 | Urine | NA | NA | NA |
| 5029 | CSF | 2B-v74T,345K | S269F | 21/22 | |
| 5029 | Plasma | 2B-v74T,345K | S269F | 19/19 | |
| 14 | 5060 | Urine | NA | NA | NA |
| 5060 | CSF | 2B | S269F | 23/23 | |
| 5060 | Plasma | 2B | S269F | 31/31 | |
| 15 | 5121 | Urine | NA | NA | NA |
| 5121 | CSF | 1B-v117T | S269F | 41/41 | |
| 5121 | Plasma | 1B-v117T | S269F | 12/12 | |
| 16 | 5207 | Urine | NA | NA | NA |
| 5207 | CSF | 1B | Q271H | PCR | |
| 5207 | Plasma | 1B | Q271H | PCR | |
| 17 | 5163 | Urine | 4-v164K | none | 0/24 |
| 5163 | CSF | NA | NA | NA | |
| 5163 | Plasma | 4-v164K | L55Fd | 32/32 |
NOTE. JCV indicates JC virus; VP1, viral protein 1; PML, progressive multifocal leukoencephalopathy; PCR, polymerase chain reaction; NA, sequence not available.
According to [8]. The subgroup, followed by “v” and position, means subgroup variant and polymorphism relative to that subtype.
PCR sequence information was obtained directly from a PCR product without cloning.
S267F also identified in 3/21 clones.
S267P also identified in 2/32 clones.
Characterization of JCV VP1 Mutations in CSF of PML Patients
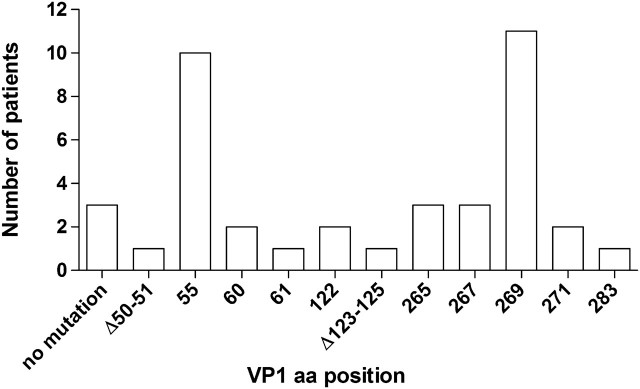
To define the type and frequency of VP1 PML-associated substitutions, we analyzed CSF-derived VP1 sequences in a larger group of patients. One main PML-associated mutation or deletion was identified in 37 of 40 patients (90%), always in all or most of the clones derived from each analyzed sample (Figure 1, Table 3). The most frequent changes involved amino acids 269 and 55, identified in 27% and 25% of the patients, respectively. In addition to the substitutions already reported for amino acids 55, 60, 61, 265, 267, 269, and 271 [9, 11], we newly identified several other PML-associated mutations or deletions involving nonpolymorphic (ie, JCV subtype–independent) VP1 codons (amino acid positions 50, 51, 122–125, and 283) and also located in likely critical VP1 binding sites.
Figure 1.
Frequencies of PML-associated substitutions identified in JCV VP1 from the CSF of PML patients. The number of PML patients harboring a JCV with particular VP1 mutation or deletion in the CSF is displayed. Mutations were identified by comparing the sequences to either VP1 sequences from matched urine samples (when available) or to 253 sequences isolated from the urine of non-PML individuals as reported in GenBank and assembled elsewhere [9]. The most dominant mutation is plotted for each patient (ie, mutations present in a low number of clones are not displayed) (for a comprehensive list, see Table 3).
NOTE: PML indicates progressive multifocal leukoencephalopathy; JCV, JC virus; VP1, viral protein 1; CSF, cerebrospinal fluid.
Table 3.
JCV VP1 Amino Acid Substitutions in Sequences Derived From Cerebrospinal Fluid (CSF) of PML Patients
| Patient IDa | Underlying disease | CSF JCV DNA log c/mL | JCV VP1 subtypeb | PML-associated substitution | No. of clones with substitution/no. of clones analyzed | |
| 1 | 5088 | HIV | 3.71 | 4v-164K | none | 0/3 |
| 2 | 5168 | HIV | 3.39 | 1B | none | PCRc |
| 3 | 5211 | HIV | 4.43 | 2B | none | PCR |
| 4 | 5162 (0) | HIV | 4.12 | 2B | 50-51del; D66G; N124S | 22/22; 3/22; 2/22 |
| 5162 (230) | 4.49 | 2B | 50-51 del | 44/44 | ||
| 5162 (719-r) | 3.68 | 2B | 50-51del;124-133 del | PCR | ||
| 5 | 5005 (0) | HIV | 3.49 | 1B | L55F | 9/9 |
| 5005 (17) | 4.87 | 1B | L55F | 23/23 | ||
| 5005 (92) | 3.47 | 1B | L55F | 48/48 | ||
| 6 | 5013 | HIV | 6.55 | 1A | L55F | 21/21 |
| 7 | 5031 | HIV | 6.12 | 3 | L55F; Q271H | 18/22; 4/22d |
| 8 | 5087 | HIV | 4.39 | 1B | L55F | 19/19 |
| 9 | 5092 | HIV | 5.6 | 1A | L55F | 20/20 |
| 10 | 5108 | HIV | 5.87 | 4-v128A,345K | L55F | 24/24 |
| 11 | 5135 | HIV | 3.94 | 1A-v75K,308R | L55F | PCR |
| 12 | 5177 | HIV | 6.51 | 1A | L55F | PCR |
| 13 | 5053 | HIV | 4.05 | 2B | L55F | PCR |
| 14 | 5175 | HIV | 4.26 | 2B-v264I,308R | K60M | PCR |
| 15 | 5169 (0) | HIV | 5.41 | 1B | S61L; 54-55 del | 15/22; 4/22d |
| 5169 (26) | 4.30 | 1B | S61L | PCR | ||
| 16 | 5067 | HIV | 4.18 | 1A | H122R | 27/27 |
| 17 | 5152 (0) | HIV | 3.61 | 1A | H122R; A2V | 44/46; 45/46 |
| 5152 (487) | 3.80 | 1A | H122R; A2V | 35/36; 35/36 | ||
| 18 | 5026 | HIV | 3.14 | 1B | 123-125 del | 8/8 |
| 19 | 5009 (0) | HIV | 2.75 | 4-v128A | N265D | 41/41 |
| 5009 (225-r) | 2.75 | 4-v128A | N265D | PCR | ||
| 20 | 5016 (0) | HIV | 6.24 | 4-v128A | N265D | 23/23 |
| 5016 (56) | 8.59 | 4-v128A | N265D | 48/48 | ||
| 21 | 5050 | HIV | 5.03 | 4 | Q271R | PCR |
| 22 | 5007 | HIV | 6.89 | 1B | S267F; S61L; Q271H | 15/22; 4/22; 2/22d |
| 23 | 5033 | HIV | 4.1 | 2B | S267F | 42/42 |
| 24 | 5029 | HIV | 6.78 | 2B-v74T,345K | S269F | 21/22 |
| 25 | 5174 | HIV | 7.62 | 1B | S269F | 28/28 |
| 26 | 5147 | HIV | 3.95 | 2B | S269F | PCR |
| 27 | 5040 (0) | HIV | 5.93 | 1A-v75K,128S | S269F | PCR |
| 5040 (3128-r) | 4.28 | 1A-v75K,128S | None | 0/14 | ||
| 28 | 5180 | HIV | 4.96 | 1A | S269F | PCR |
| 29 | 5058 | HIV | 4.53 | 1A-v75K | S269F | PCR |
| 30 | 5202 | HIV | 6.6 | Undet.e | S269Y | PCR |
| 31 | 5064 (0) | SLE | 4.31 | 2B | L55F | 47/47 |
| 5064 (66) | 2.98 | 2B | L55F | 23/23 | ||
| 32 | 5201 | NHL | 7 | 1B | K60N | PCR |
| 33 | 5041 | NHL | 5.54 | 2B | N265H | 20/20 |
| 34 | 5167 | ICL | 3.63 | 2B-v69D, 128A | S267Y | PCR |
| 35 | 5060 | NHL | 6.24 | 2B | S269F | 23/23 |
| 36 | 5121 | NHL | 3.62 | 1B-v117T | S269F | 41/41 |
| 37 | 5138 | NHL | 4.01 | 1B | S269F | 19/19 |
| 38 | 5166 | NHL | 4.78 | 1A | S269F | 14/14 |
| 39 | 5207 | NHL, SCT | 6.39 | 1B | Q271H | PCR |
| 40 | 5097 | Hyper IgE syndrome | 5.69 | 1B | V283I | PCR |
NOTE. JCV indicates JC virus; VP1, viral protein 1; PML, progressive multifocal leukoencephalopathy; HIV, human immunodeficiency virus; PCR, polymerase chain reaction; Undet., undetermined; SLE, systemic lupus erythematosus; NHL, non-Hodgkin lymphoma; ICL, Idiopathic CD4 lymphopenia; SCT, stem cell transplantation.
Number in parentheses indicate the number of days from first sample examined.
According to [8]. The subgroup followed by “v” and position indicates subgroup variant and polymorphism relative to that subtype.
PCR, sequence information was obtained directly from a PCR product without cloning.
Different mutations were observed in different clones.
Sequence available up to position 295.
We also investigated the dynamics of mutations in 8 PML patients with CSF collected over a timeframe of 17 days to 8.5 years (Table 3). PML-associated substitutions were maintained over time in all the progressive cases, irrespective of the JCV DNA level dynamics, and were even present during disease relapse in 2 of 3 patients months to years after first PML episode. By correlating PML-associated substitutions to clinical and laboratory variables (Table 1), the only significant finding was lower DNA levels in CSF of patients with no substitutions or substitutions/deletions involving amino acid positions 122–134, as compared with patients with all other substitutions.
PML-Associated Mutations Decrease VP1 Hemagglutination Ability
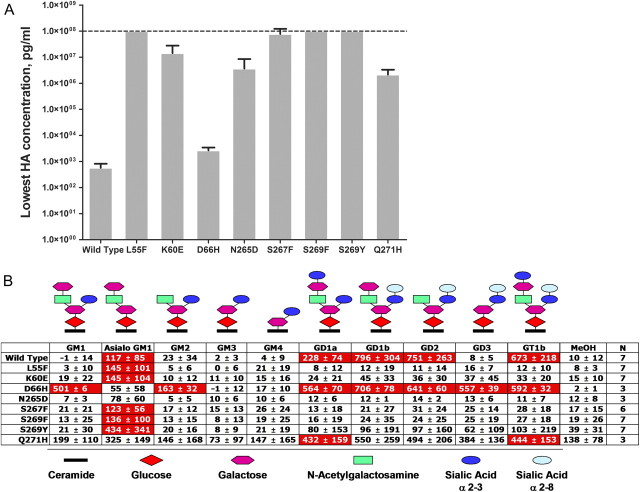
JCV is known to bind to sialic acid structures [5], thus we used red blood cell (RBC) agglutination as a model [13, 14] to study JCV VP1 interaction with sialylated receptors. We prepared VLPs that form complete viral capsid structures [9, 15] and are widely used as a model for polyomavirus interactions with their cellular receptors [15, 16, 17]. We introduced, on a backbone of a VLP from a single viral background (JCV type 3), a number of PML-associated mutations observed in vivo (ie, 55F, 60E, 265D, 267F, 269F, and 269Y), and additionally, the mutations 66H and 271H, which had been previously reported in PML brain-derived sequences [9]. While wild-type VLPs caused hemagglutination at concentrations as low as 760 pg/mL (Figure 2A), VLPs carrying the 55F, 267F, 269F, or 269Y mutations did not, even at the highest concentration of 100 μg/mL, which corresponds to >100,000-fold decrease in activity. Hemagglutination was still apparent with the 60E-, 265D-, and 271H-mutated VLPs, but only at high concentrations that corresponded to 200- to 25,000-fold losses in activity. Only binding of the 66H mutant was not strongly affected. The effect of VLP mutations on hemagglutination ability suggests that these may change cell tropism of JCV in vivo by abrogating the ability to bind cellular receptor(s).
Figure 2.
JCV VLPs carrying PML-associated mutations lose the ability to hemagglutinate red blood cells (RBCs) and bind sialylated gangliosides. (A) Ability of mutant VLPs to hemagglutinate red blood cells from different blood groups. RBCs were incubated with serial 2-fold dilutions of various VLPs, starting from 100 μg/mL. Minimum hemagglutination (HA) concentration is the lowest concentration of VLP that still agglutinated RBCs. HA results were examined by visual inspection. All HA reactions were conducted in duplicates. Mean ± SD for the minimum HA concentration is calculated based on the results obtained from hemagglutination of 4 to 7 different blood group donor RBCs (A, B, O, and AB). VLPs displaying the minimum HA concentration of 100 μg/mL (dotted line) did not cause any hemagglutination at this concentration (ie, highest VLP concentration tested). (B) Specificity of JC VLPs for sialylated gangliosides. VLPs binding to an array of gangliosides were detected with a 2-step process involving the detection of VLPs bound to a ganglioside with VP1-specific murine antibodies and anti-murine IgG HRP–labeled antibodies followed by development with TMB substrate. Numbers represent percent increase in the optical density obtained with the specific VLP relative to that obtained without VLP (ie, background) and were calculated as follows: % specific VLP binding = 100%*(OD450(with VLP)–OD450(without VLP))/OD450(without VLP). Statistically significant (P < .05) interaction between a VLP and ganglioside in comparison to the background interaction between the VLP and MeOH-treated well is denoted by a red cell background. Results are depicted as a mean ± SD binding for each VLP to each of the gangliosides or to the control for several independent experiments conducted; N denotes number of independent measurements for each VLP and all gangliosides. Schematic structure of ganglioside is shown to reveal core binding structure bound by various VLPs.
NOTE. JCV indicates JC virus; VLPs, virus-like particles; PML, progressive multifocal leukoencephalopathy; VP1, viral protein 1; TMB, tetramethylbenzidine.
PML-Associated Mutations Change Ganglioside Specificity of VP1
To further dissect VP1 receptor specificity, we investigated the binding of type-3 wild-type and mutated VLPs to different gangliosides. Gangliosides are a group of complex glycosphingolipids in which oligosaccharide chains containing one or more sialic acids (N-acetylneuraminic acid [NeuNAc]) are attached to a ceramide, which anchors the structure to the cellular membrane [18]. These molecules play an important role as cellular receptors and are abundant in the brain (for review, see [19]). Wild-type VLPs displayed the strongest binding to GD1b, GD2, and GT1a gangliosides (Figure 2B), which was consistent with previous observations for type 1A wild-type virus [16]. Binding to asialo-GM1 and GD1a was weaker but still more than 2- and 3-fold over background, respectively, while binding to other gangliosides was negligible. In contrast, the same mutated VLPs that lost the hemagglutination property (ie, mutants 55F, 60E, 265D, 267F, 269F, and 269Y), also lost binding to all sialylated gangliosides, while all but 265D exhibited binding to asialo-GM1 (Figure 2B). The 66H- and 271H-mutated VLPs still bound to sialylated structures but with different ganglioside specificities compared with nonmutated VLP, indicating changes of receptor specificity.
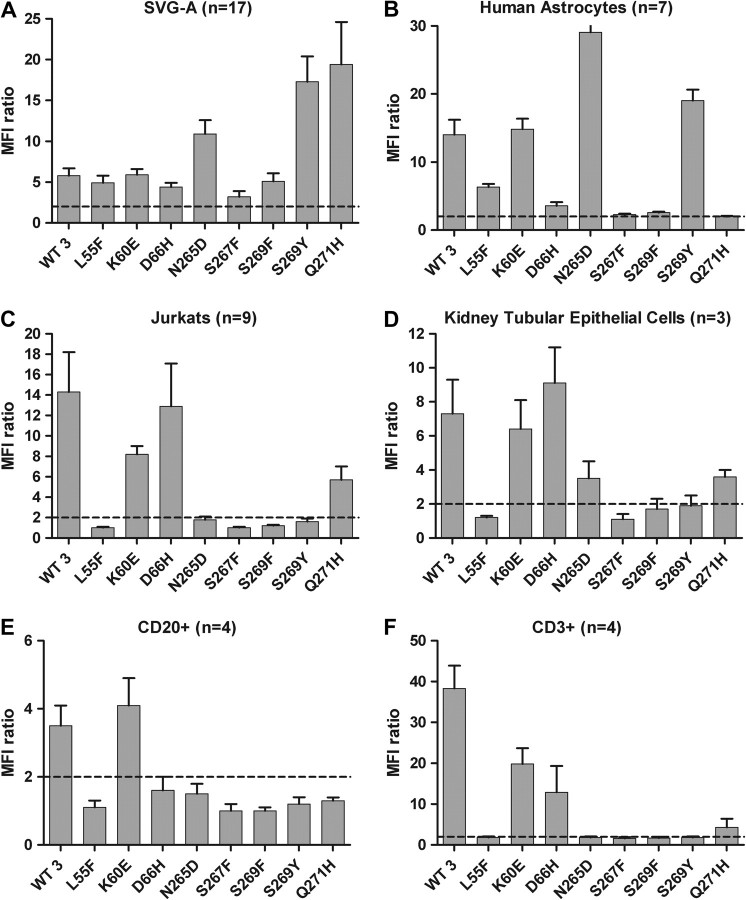
Binding of VLPs to Various Cellular Targets
By flow cytometric analysis (FACS), we measured binding of the above-described wild-type and mutant VLPs to the cell types reported to be important in the natural history of JCV infection: primary human renal proximal tubular epithelial cells, as target cells for asymptomatic viral persistence of nonmutated virus [20, 21]; primary human B lymphocytes, as the cell type suggested to be critical for viral spread from the periphery to the CNS [22, 23]; primary human T lymphocytes and the lymphocytic cell line Jurkat as additional peripheral blood cell types; and fetal astrocytes and the human glial cell line SVG-A [24, 25, 26], as model cells for CNS infection during PML in the lack of availability of purified primary human oligodendrocytes (ie, viral target in PML). Wild-type VLP strongly bound to all cell types, with an MFI >2-fold above background (Figure 3). However, the most frequent PML-associated mutations 55F, 267F, 269F, and 269Y abrogated binding to kidney and all lymphocytic cells, but not to CNS cells. VLPs carrying 60E or 66H still bound all cell types, while 265D or 271H VLPs bound kidney cells but not primary lymphocytes.
Figure 3.
JCV VLPs carrying PML-associated mutations bind differently to lymphoid vs CNS cells. (A) Glial cell line SVG-A, (B) primary human astrocytes, (C) Jurkat cells, (D) primary human kidney tubular epithelial cells (HRPTEC), or (E and F) peripheral blood mononuclear cells (PBMCs) were first incubated with different VLPs (as indicated on the x-axis) and then with anti-VP1 antibodies followed by staining with fluorescently labeled antibodies. Binding of VLPs to B (E) and T lymphocytes (F) was evaluated after costaining of PBMCs with anti-CD20 and anti-CD3 antibodies and gating on the corresponding population. The ratio of mean fluorescent intensity (MFI) of cells stained with anti-VP1 antibodies in the presence of VLPs relative to that of cells stained only with the detection antibodies in the absence of VLPs (background) is plotted for each VLP. Mean MFI ratio ± SD is calculated based on the results from several independent experiments (n = number of experiments). Dotted lines (MFI ratio = 2) represents a 2-fold increase in VLP binding over the background and deemed as being significant.
NOTE. JCV indicates JC virus; VLPs, virus-like particles; PML, progressive multifocal leukoencephalopathy; CSF, cerebrospinal fluid.
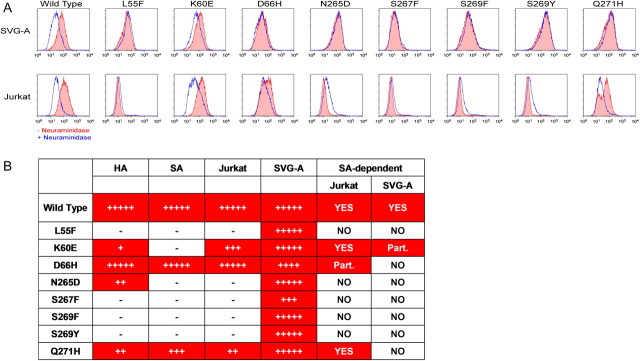
Mutant VLPs Bind Glial Cells in Neuraminidase Treatment–Insensitive Fashion
To test whether binding of mutated VLPs to CNS cells was still sialic acid–dependent, we treated either SVG-A or Jurkat cells (as controls) with neuraminidase to remove sialic acid from the cell surface prior to staining with various VLPs. Binding of wild-type VLPs was diminished after neuraminidase treatment (Figure 4A), indicating sialic acid–dependent binding. However, binding of all but the K60E mutant VLPs to SVG-A cells was not affected by the neuraminidase treatment, suggesting that binding of mutated virus to these CNS cells is independent of sialic acid moieties that are removed by this treatment and that wild-type VLP binding depends on. Interestingly, mutants that bound to Jurkat cells (ie, K60E, D66H, and Q271H) showed diminished binding to those cells after neuraminidase treatment (Figure 4A), indicating the sialic acid–dependent nature of those mutants binding to nonglial cells.
Figure 4.
Binding of JCV VLPs carrying PML-associated mutations to glial cells is neuraminidase treatment–insensitive. (A) SVG-A glial cells (top row) or Jurkat lymphoid cells (bottom row) were first either pretreated with neuraminidase (blue line) for 60 min at 37°C to remove all terminal α2–3–, α2–6–, and α2-8–linked sialic acid residues or mock-treated (red line) followed by incubation at 4°C with the indicated VLP and staining with the detection antibodies as described in the previous figure. One representative experiment out of 4 conducted is shown. Staining of the cells with Sambucus nigra agglutinin (SNA) lectin and Maackia amurensis lectin II (MAL II) proved neuraminidase treatment effectiveness in sialic acid removal (data not shown). (B) Sialic acid dependence as compared with VLP binding by various assays. The degree of binding was evaluated as a percent of wild-type VLP binding value in the assay (Figures 2 and 3) and were calculated as follows: 100%*(VLP VALUE – BASELINE VALUE)/(WT VALUE – BASELINE VALUE); hemagglutination assay values were LOG10 transformed first. Values were ranked accordingly: “–” < 10; 10 < “+” < 20; 20 < “++” < 40; 40 < “+++” < 60; 60 < “++++” < 80; and “+++++” > 80.
NOTE. JCV indicates JC virus; VLPs, virus-like particles; PML, progressive multifocal leukoencephalopathy; HA, hemagglutination; SA, sialic acid binding; HRPTEC, human renal proximal tubular epithelial cells; astrocytes, primary human astrocytes. Dependence on sialic acid binding is denoted as NO, no dependence; or Part., partial dependence. Positive values are denoted by red cell backgrounds.
In summary, the PML-associated mutations most frequently found in clinical samples largely abolished sialic acid–dependent JCV binding to different peripheral cells types, including RBCs, kidney tubular epithelial cells, and lymphoid cells. Binding of mutated virus to CNS-specific glial cells was unaffected, thus appearing largely sialic acid–independent (Figure 4B).
DISCUSSION
While immune dysfunction is a crucial prerequisite for the development of PML, the dramatic discrepancy between high JCV prevalence and low incidence of PML suggests that, in addition to immune dysfunction, there could be some unique viral characteristics that regulate the progression of asymptomatic infection to PML. In this work, we demonstrate that the part of the viral surface protein that is responsible for viral interactions with cellular receptors and hence viral infectivity acquires specific amino acid mutations in the patient en route from the urinary tract (site of asymptomatic infection) to the CNS (site of PML). Furthermore, we show that these PML-specific mutations change the ability of the viral capsid to bind to sialylated molecules and a variety of peripheral cell types, but retain the ability to bind CNS glial cells in a neuraminidase treatment–insensitive manner.
Through the analysis of urine and CSF samples taken from PML patients at the same time points, we showed that CSF VP1 sequences contained additional amino acid substitutions or deletion compared with sequences derived from paired urine samples of the same patients. Because background VP1 genotypes were the same in the CSF and urine samples from the same patients—whereas different patients carried different subtypes—our observations are parsimoniously explained by intrapatient origin of the substitutions.
In the larger group of PML patients, L55F and S269F were the most common mutations in CSF-derived sequences, both accounting for half of all observed. This observation correlates with the ability of both mutations to abrogate JCV hemagglutination and binding to peripheral cells and to sialic acid, suggesting that this loss of function is advantageous for the virus. Although quite distant in a linear structure, VP1 positions 55 and 269 colocalize on the 3-dimensional protein structure, recently described by Neu et al. [27], being part of the sialic acid binding site. All other VP1 mutations or deletions derived from the CSF of PML patients also colocalized to the sialic acid binding site, and most of those that we analyzed caused a loss of or dramatic change in sialic acid specificity of the virus. In addition, we did not observe any binding of 55F- and 269F-mutant VLPs to an a2, 6-sialylated oligosaccharide LSTc structure (Gorelik, unpublished observation, www.functionalglycomics.org), which was recently described to bind wild-type VLPs [27]. Although we did not characterize the function of the whole spectrum of mutations and deletions observed in vivo, the data from the examination of their vast majority provide support to the hypothesis that a change in VP1 specificity for sialic acids plays a crucial role in viral pathogenesis. These mutations are associated with some or complete loss of sialic acid binding capability, as apparently more mutations could be created to disrupt viral binding to sialic acid. However, we described a limited set of mutations that have originated from the viable and positively selected virus, thus they must provide some positive advantage to the virus and cannot be a simple loss of sialic acid binding function. Whether they provide additional viral function(s) (such as different receptor specificity) or are simply selected because they did not lead to a concurrent loss of some vital function for the virus would require further studies.
Finding the same sequences in CSF and plasma, both distinct from the urine sequences in PML, suggests that CSF or plasma virus populations originated from the other after a single viral escape event from the site of chronic infection in the urogenital tract. Whether this observation reflects subsequent continuous hematogenous spread to the CNS from a peripheral site or, rather, the release of CNS virus into the blood, could not be clarified in the present study. Determining the timing and tissue of origin of PML-associated mutations is central to better understand the role of these mutations in the etiology and pathogenesis of PML. Based on the current data, two main models can be hypothesized. According to a first model, mutations could occur during viral replication at a peripheral site and enable the virus to reach and replicate in the CNS. Although we have not observed PML-associated mutations in the urine of PML patients, it is possible that a mutation was originally acquired during viral replication in the urinary tract, and that the mutated virus had no competitive advantage over the resident nonmutated virus to become a detectable population at that site. Because most mutations make the virus lose specificity for widely occurring sialic acid–containing receptors, once the mutated virus is released into the blood circulation, it might escape being trapped by the multitude of sialo-containing oligosaccharide pseudoreceptors expressed on a great majority of peripheral cells (reviewed in [28, 29]). This same mechanism has been demonstrated to increase viral virulence of murine polyomavirus infection in mice. In that model, mutation at position 296, which is structurally orthologous to the critical JCV position 269, dramatically changed the viral specificity for sialic acids, affected the viral ability to bind target cells and RBCs, and also led to dramatic viral dissemination through the animal, resulting in a lethal outcome [30]. We do not know if VP1 mutations also provide any advantage for the virus to cross the blood-brain barrier (BBB), but it is possible that altered BBB permeability—not infrequent in patients with HIV infection [31, 32], who are at risk of developing PML—facilitates the virus spreading to the CNS. In such case, the required temporal constellation of events such as selection of mutations in the presence of BBB breaching might explain the rare occurrence of PML, even in patients with significant immune suppression. The observation that ∼10% of PML patients carry the virus without VP1 mutations is still consistent with the hypothesis that the mutations occur prior to viral entry into the CNS. Entrance of nonmutant virus into the CNS could be facilitated by brief high viremic titers, which might overwhelm the peripheral defense of viral pseudoreceptors, occurring simultaneously with some other predisposing event (eg, opening of the BBB).
Still, we cannot rule out the alternative hypothesis that the virus always enters the CNS in the nonmutant form, and VP1 mutations originate and are positively selected within the CNS. Of note, the JCV replication level in CSF was lower in patients with nonmutated virus compared with the other patients, leading to the speculation that sialic acid independent cell entry, observed for mutated viruses, leads to more efficient viral replication in the CNS than sialic acid–dependent cell entry, expected for nonmutated virus.
Variation of the JCV noncoding regulatory region (NCCR) has also been associated with changes in viral tropism (reviewed in [22]). NCCR contains binding sites for host cell transcription factors, and rearrangements of this region (ie, patient-specific DNA duplications and deletions of variable length) are consistently found in CSF- and brain-derived sequences of PML patients, as opposed to the archetypal form of the NCCR that is normally shed in urine. Thus, NCCR rearrangements could confer a change of JCV properties in concert with the selection of VP1 mutations. These changes might, on one hand (ie, via NCCR rearrangements) decrease viral fitness for peripheral cells while conferring optimal viral fit with the transcriptional machinery of newly infected CNS cells and, on the other hand (ie, via VP1 mutations) favor CNS infection through peripheral escape and increased binding ability to CNS cells through sialic acid–independent mechanisms. Indeed, murine polyomavirus virulence is increased by the presence of VP1 mutations independently of NCCR changes [30] and NCCR can undergo further changes once the polyomavirus is forced to replicate in a new cell type [7], supporting the hypothesis that VP1 mutations might even occur first and NCCR rearrangements follow. Studies involving analysis of sequential samples would be required to determine the exact sequence of JCV mutation events, although these are hampered by the unavailability of CSF specimens and undetectability of JCV in plasma (our unpublished observation) before PML is clinically manifest.
In conclusion, our in vivo and in vitro observations on the presence and functional role of VP1 mutations in PML enhance our understanding of PML etiopathogenesis by revealing an aspect of viral–host interactions essential for disease development and progression. Our data also show that polyomaviruses may use a common mechanism involving capsid protein mutations to change host cell distribution and hence their virulence. As polyomaviruses begin to play a more prominent role in human diseases due to the development of new immunoregulatory drugs, these findings can pave the way for development of new diagnostic options and treatment paradigms.
Supplementary Data
Supplementary data are available at The Journal of Infectious Diseases online.
Funding
This work was supported by Biogen Idec and Italian National Health Ministry, AIDS Projects grant numbers 50F.11 and 30F.8.
Supplementary Material
Acknowledgments
The authors would like to thank Drs Rich Tizard and Huo Li for help and advice with sequencing and sequence annotation. The authors are grateful to Dr Susan Goelz for many helpful discussions.
References
- 1.Major EO. Reemergence of PML in natalizumab-treated patients—new cases, same concerns. N Engl J Med. 2009;361:1041–3. doi: 10.1056/NEJMp0906248. [DOI] [PubMed] [Google Scholar]
- 2.Cinque P, Koralnik IJ, Gerevini S, Miro JM, Price RW. Progressive multifocal leukoencephalopathy in HIV-1 infection. Lancet Infect Dis. 2009;9:625–36. doi: 10.1016/S1473-3099(09)70226-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Egli A, Infanti L, Dumoulin A, et al. Prevalence of polyomavirus BK and JC infection and replication in 400 healthy blood donors. J Infect Dis. 2009;199:837–46. doi: 10.1086/597126. [DOI] [PubMed] [Google Scholar]
- 4.Kean JM, Rao S, Wang M, Garcea RL. Seroepidemiology of human polyomaviruses. PLoS Pathog. 2009;5:e1000363. doi: 10.1371/journal.ppat.1000363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu CK, Wei G, Atwood WJ. Infection of glial cells by the human polyomavirus JC is mediated by an N-linked glycoprotein containing terminal alpha(2-6)-linked sialic acids. J Virol. 1998;72:4643–9. doi: 10.1128/jvi.72.6.4643-4649.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Neu U, Stehle T, Atwood WJ. The Polyomaviridae: contributions of virus structure to our understanding of virus receptors and infectious entry. Virology. 2009;384:389–99. doi: 10.1016/j.virol.2008.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen BJ, Atwood WJ. Construction of a novel JCV/SV40 hybrid virus (JCVS) reveals a role for the JCV capsid in viral tropism. Virology. 2002;300:282–90. doi: 10.1006/viro.2002.1522. [DOI] [PubMed] [Google Scholar]
- 8.Cubitt CL, Cui X, Agostini HT, et al. Predicted amino acid sequences for 100 JCV strains. J Neurovirol. 2001;7:339–44. doi: 10.1080/13550280152537201. [DOI] [PubMed] [Google Scholar]
- 9.Sunyaev SR, Lugovskoy A, Simon K, Gorelik L. Adaptive mutations in the JC virus protein capsid are associated with progressive multifocal leukoencephalopathy (PML) PLoS Genet. 2009;5:e1000368. doi: 10.1371/journal.pgen.1000368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bossolasco S, Calori G, Moretti F, et al. Prognostic significance of JC virus DNA levels in cerebrospinal fluid of patients with HIV-associated progressive multifocal leukoencephalopathy. Clin Infect Dis. 2005;40:738–44. doi: 10.1086/427698. [DOI] [PubMed] [Google Scholar]
- 11.Zheng HY, Takasaka T, Noda K, et al. New sequence polymorphisms in the outer loops of the JC polyomavirus major capsid protein (VP1) possibly associated with progressive multifocal leukoencephalopathy. J Gen Virol. 2005;86:2035–45. doi: 10.1099/vir.0.80863-0. [DOI] [PubMed] [Google Scholar]
- 12.Larkin MA, Blackshields G, Brown NP, et al. ClustalW and ClustalX version 2.0. Bioinformatics. 2007;23:2947–8. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 13.Isaacs A, Donald HB. Particle counts of haemagglutinating viruses. J Gen Microbiol. 1955;12:241–7. doi: 10.1099/00221287-12-2-241. [DOI] [PubMed] [Google Scholar]
- 14.Chapagain ML, Nguyen T, Bui T, Verma S, Nerurkar VR. Comparison of real-time PCR and hemagglutination assay for quantitation of human polyomavirus JC. Virol J. 2006;3:3. doi: 10.1186/1743-422X-3-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goldmann C, Petry H, Frye S, et al. Molecular cloning and expression of major structural protein VP1 of the human polyomavirus JC virus: formation of virus-like particles useful for immunological and therapeutic studies. J Virol. 1999;73:4465–9. doi: 10.1128/jvi.73.5.4465-4469.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Komagome R, Sawa H, Suzuki T, et al. Oligosaccharides as receptors for JC virus. J Virol. 2002;76:12992–3000. doi: 10.1128/JVI.76.24.12992-13000.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weber F, Goldmann C, Kramer M, et al. Cellular and humoral immune response in progressive multifocal leukoencephalopathy. Ann Neurol. 2001;49:636–42. [PubMed] [Google Scholar]
- 18.Klenk E. On the discovery and chemistry of neuraminic acid and gangliosides. Chem Phys Lipids. 1970;5:193–7. doi: 10.1016/0009-3084(70)90017-4. [DOI] [PubMed] [Google Scholar]
- 19.Posse de Chaves E, Sipione S. Sphingolipids and gangliosides of the nervous system in membrane function and dysfunction. FEBS Lett. 2010;584:1748–59. doi: 10.1016/j.febslet.2009.12.010. [DOI] [PubMed] [Google Scholar]
- 20.Dorries K, ter Meulen V. Progressive multifocal leucoencephalopathy: detection of papovavirus JC in kidney tissue. J Med Virol. 1983;11:307–17. doi: 10.1002/jmv.1890110406. [DOI] [PubMed] [Google Scholar]
- 21.Randhawa P, Baksh F, Aoki N, Tschirhart D, Finkelstein S. JC virus infection in allograft kidneys: analysis by polymerase chain reaction and immunohistochemistry. Transplantation. 2001;71:1300–3. doi: 10.1097/00007890-200105150-00020. [DOI] [PubMed] [Google Scholar]
- 22.Major EO, Amemiya K, Tornatore CS, Houff SA, Berger JR. Pathogenesis and molecular biology of progressive multifocal leukoencephalopathy, the JC virus–induced demyelinating disease of the human brain. Clin Microbiol Rev. 1992;5:49–73. doi: 10.1128/cmr.5.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Atwood WJ, Amemiya K, Traub R, Harms J, Major EO. Interaction of the human polyomavirus, JCV, with human B-lymphocytes. Virology. 1992;190:716–23. doi: 10.1016/0042-6822(92)90909-9. [DOI] [PubMed] [Google Scholar]
- 24.Aksamit AJ, Mourrain P, Sever JL, Major EO. Progressive multifocal leukoencephalopathy: investigation of three cases using in situ hybridization with JC virus biotinylated DNA probe. Ann Neurol. 1985;18:490–6. doi: 10.1002/ana.410180412. [DOI] [PubMed] [Google Scholar]
- 25.Major EO, Miller AE, Mourrain P, Traub RG, de Widt E, Sever J. Establishment of a line of human fetal glial cells that supports JC virus multiplication. Proc Natl Acad Sci USA. 1985;82:1257–61. doi: 10.1073/pnas.82.4.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Watanabe I, Preskorn SH. Virus–cell interaction in oligodendroglia, astroglia and phagocyte in progressive multifocal leukoencephalopathy. An electron microscopic study. Acta Neuropathol. 1976;36:101–15. doi: 10.1007/BF00685273. [DOI] [PubMed] [Google Scholar]
- 27.Neu U, Maginnis MS, Palma AS, et al. Structure–function analysis of the human JC polyomavirus establishes the LSTc pentasaccharide as a functional receptor motif. Cell Host Microbe. 2010;8:309–19. doi: 10.1016/j.chom.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roth J. Cellular sialoglycoconjugates: a histochemical perspective. Histochem J. 1993;25:687–710. doi: 10.1007/BF00211765. [DOI] [PubMed] [Google Scholar]
- 29.Varki A. Diversity in the sialic acids. Glycobiology. 1992;2:25–40. doi: 10.1093/glycob/2.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bauer PH, Bronson RT, Fung SC, et al. Genetic and structural analysis of a virulence determinant in polyomavirus VP1. J Virol. 1995;69:7925–31. doi: 10.1128/jvi.69.12.7925-7931.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Persidsky Y, Ramirez SH, Haorah J, Kanmogne GD. Blood-brain barrier: structural components and function under physiologic and pathologic conditions. J Neuroimmune Pharmacol. 2006;1:223–36. doi: 10.1007/s11481-006-9025-3. [DOI] [PubMed] [Google Scholar]
- 32.Petito CK, Cash KS. Blood-brain barrier abnormalities in the acquired immunodeficiency syndrome: immunohistochemical localization of serum proteins in postmortem brain. Ann Neurol. 1992;32:658–66. doi: 10.1002/ana.410320509. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.