Abstract
Background. Pegylated interferon, ribavirin, and telaprevir triple therapy is a new strategy expected to eradicate the hepatitis C virus (HCV) even in patients infected with difficult-to-treat genotype 1 strains, although adverse effects, such as anemia and rash, are frequent.
Methods. We assessed efficacy and predictive factors for sustained virological response (SVR) for triple therapy in 94 Japanese patients with HCV genotype 1. We included recently identified predictive factors, such as IL28B and ITPA polymorphism, and substitutions in the HCV core and NS5A proteins.
Results. Patients treated with triple therapy achieved comparatively high SVR rates (73%), especially among treatment-naive patients (80%). Of note, however, patients who experienced relapse during prior pegylated interferon plus ribavirin combination therapy were highly likely to achieve SVR while receiving triple therapy (93%); conversely, prior nonresponders were much less likely to respond to triple therapy (32%). In addition to prior treatment response, IL28B SNP genotype and rapid viral response were significant independent predictors for SVR. Patients with the anemia-susceptible ITPA SNP rs1127354 genotype typically required ribavirin dose reduction earlier than did patients with other genotypes.
Conclusions. Analysis of predictive factors identified IL28B SNP, rapid viral response, and transient response to previous therapy as significant independent predictors of SVR after triple therapy.
Hepatitis C virus (HCV) establishes a chronic infection in 80% of infected individuals, and currently, >100 million persons are chronically infected and at increased risk of cirrhosis, hepatocellular carcinoma, and end-stage liver disease [1–3]. The current standard of care is combination treatment with pegylated interferon (PEG-IFN) and ribavirin, but this costly and poorly tolerated treatment achieves sustained virological response in only 50% of patients [4]. Options are limited in the event of treatment failure, and alternative therapies are needed.
Of the many drugs under investigation, the most promising are the direct-acting antiviral agents, which directly target essential aspects of viral replication, including internal ribosome entry site inhibitors, protease and polymerase inhibitors, and assembly inhibitors [5]. Several protease inhibitors, including telaprevir and boceprevir, are in phase III clinical trials and will likely become the first direct-acting antiviral agents approved for clinical use [6].
The HCV genome is initially translated as a large polyprotein and must be processed to produce functional viral proteins. Host proteases cleave the N-terminal structural proteins, but the viral NS3-4A serine protease is essential for cleaving the nonstructural proteins. NS3-4A also interferes with the immune response by degrading immune-signaling molecules [7]. Consequently, targeting this protease using the peptidomimetic inhibitor telaprevir both interferes with viral replication and may help rescue immune signaling, leading to a rapid decrease in HCV RNA level [8, 9]. In most patients, however, viral decline after telaprevir monotherapy is short-lived, followed by viral breakthrough because of strong selection for escape mutants within several weeks. Combination therapy with IFN alone yields unsatisfactory results, and ribavirin appears to be required to avoid relapse [10]. Because telaprevir triple therapy is an extension of the current standard of care instead of an IFN-free alternative, it does not address problems associated with the cost or adverse effects of combination therapy and may limit options for retreatment; however, it is particularly promising for patients who showed at least a transient response after prior combination therapy [11]. Nonetheless, telaprevir monotherapy may provide an alternative treatment for patients unable to tolerate IFN and/or ribavirin—at least in patients with low viral loads [12]. Additional research is needed to identify factors predicting outcome of treatment and incidence of adverse effects in different populations.
A number of host factors are known to affect outcome of PEG-IFN plus ribavirin combination therapy, including age, fibrosis, obesity, hepatic steatosis, [13] low-density lipoprotein cholesterol, γ-gamma-glutamyl transpeptidase (GTP) [14], and insulin resistance [15]. A number of recent studies have also shown that common genetic variation in the IL28B locus on chromosome 19 is strongly associated with spontaneous clearance and outcome after combination therapy [16–19]. Viral factors have also been shown to predict response to combination therapy, including HCV genotype [20], baseline viral titer [13, 20], amino acid substitutions at positions 70 and 91 of the HCV core protein, and the NS5A IFN Sensitivity Determining Region (ISDR) [21, 22]. Because telaprevir directly targets the virus and often results in selection for escape mutants, it is likely that additional predictive factors affecting response to treatment will be uncovered.
Combination therapy is poorly tolerated among some patients, and ribavirin-induced anemia is a serious adverse effect of the therapy that may result in dose reduction or discontinuation. Recent studies have shown an association between genetic variation in the ITPA locus and change in hemoglobin levels during treatment [23–25]. Although it does not appear to affect outcome of therapy [23, 24] (but see [25]), patients with an anemia-susceptible genotype may require greater reductions in ribavirin dose, which is associated with poorer response to therapy [26]. Telaprevir also moderately affects hemoglobin levels, but rash is the most common side effect of telaprevir therapy [10].
In the current study, we examined 94 patients with genotype 1 who received triple therapy to identify predictors for response to treatment and to assess effects of triple therapy on hemoglobin levels.
METHODS
Patients
Ninety-four Japanese patients who participated in a phase 3 clinical trial of the triple therapy in 2010 at Hiroshima University Hospital, Sapporo Kosei Hospital, and Toranomon Hospital (16, 17, and 61 patients, respectively) were investigated. Inclusion criteria for the study included remaining positive for genotype 1 HCV RNA for >6 months; having an HCV RNA level ≥5.0 log IU/mL, as determined by the COBAS TaqMan HCV test (Roche Diagnostics KK); and being aged 20–65 years, with a body weight >40 kg and <120 kg at the time of entry into the study. Exclusion criteria included cirrhosis; results positive for hepatitis B surface antigen or antibody against HIV; previous or current hepatocellular carcinoma; possible overlapping liver diseases, such as autoimmune hepatitis, hemochromatosis, Wilson disease, alcoholic liver disease, or renal disease; or creatinine clearance ≤50 mL/min at baseline, hemoglobin level <12 g/dL, neutrophil count <1500 neutrophils/mm3, or platelet count <100,000 platelets/mm3 at baseline. Patient profiles are shown in Tables 1 and 2.
Table 1.
Patient Characteristics
| Total (n = 94) | SVR (n = 69) | Non-SVR (n = 25) | |
| Response to previous therapy (naive/relapser/NR) | 25/44/25 | 20/41/8 | 5/3/17 |
| Age | 57 (23–65) | 57 (23–65) | 56 (40–65) |
| Sex (M/F) | 52/42 | 42/27 | 10/15 |
| Height (cm) | 163.6 (141.8–189.2) | 164.7 (141.8–189.2) | 157.7 (148.5–181.5) |
| Weight (kg) | 61 (41–92.5) | 61.7 (41–92.5) | 58.8 (44.9–80.3) |
| rs8099917 (TT/TG/GG) | 50/41/3 | 47/21/1 | 3/20/2 |
| rs1127354 (CC/CA/AA) | 75/18/1 | 55/13/1 | 20/5/0 |
| Viral genotype (1b/others) | 93/1 | 69/0 | 24/1 |
| Core 70 (W/M/ND) | 50/43/1 | 43/26/0 | 7/17/1 |
| Core 91 (W/M/ND) | 48/45/1 | 39/30/0 | 9/15/1 |
| ISDR (0–1/≥2/ND) | 82/8/4 | 61/5/3 | 3/21/1 |
| WBC (/mm3) | 4800 (2800–8100) | 4900 (2800–8100) | 4660 (3000–7900) |
| Plt (×104/mm3) | 17.7 (9.1–33.8) | 18 (9.9–33.8) | 16 (9.1–23.9) |
| Hb (g/dL) | 14.3 (12.3–16.6) | 14.5 (12.5–16.5) | 14.1 (12.3–16.6) |
| ALT (IU/L) | 39 (12–302) | 38 (12–302) | 46 (17–135) |
| γGTP (IU/L) | 36 (11–233) | 33 (11–233) | 53 (19–226) |
| Virus titer (log IU/mL) | 6.7 (5.1–7.7) | 6.8 (5.1–7.7) | 6.7 (5.4–7.6) |
| Days to first ribavirin reduction | 17 (2–168) | 18 (2–168) | 14 (7–73) |
| Duration of telaprevir administration (days) | 85 (29–85) | 85 (29–85) | 84 (35–85) |
| Duration of peg-interferon injection (days) | 162 (22–165) | 162 (22–165) | 162 (30–165) |
| Duration of ribavirin administration (days) | 169 (29–169) | 169 (29–169) | 168 (36–169) |
| Effect of therapy (SVR/BT/TR/NR) | 69/4/19/2 | – | – |
NOTE. All patients were infected with genotype 1. Counts are listed for categorical values and the median and range are reported for continuous variables. ND, not determined, data unavailable.
Table 2.
Patient Characteristics Grouped by Treatment History
| Total (n = 94) | Naive (n = 25) | Relapser (n = 44) | NR (n = 25) | |
| Age | 56.5 (23–65) | 54 (23–64) | 57.5 (44–65) | 57 (40–65) |
| Sex (M/F) | 52/42 | 13/12 | 27/17 | 12/13 |
| Height (cm) | 163.5 (142–189) | 163 (147–189) | 167.5 (142–177) | 160 (149–174) |
| Weight (kg) | 61 (41–93) | 57 (42–80) | 63.5 (41–93) | 59 (45–77) |
| rs8099917 (TT/GT/GG) | 50/41/3 | 15/9/1 | 33/11/0 | 2/21/2 |
| rs1127354 (CC/CA/AA) | 75/18/1 | 18/6/1 | 34/10/0 | 23/2/0 |
| Viral genotype (1b/others) | 93/1 | 25/0 | 44/0 | 24/1 |
| Core 70 (W/M/ND) | 50/43/1 | 13/12/0 | 28/16/0 | 9/15/1 |
| Core 91 (W/M/ND) | 48/45/1 | 14/11/0 | 23/21/0 | 11/13/1 |
| ISDR (0–1/≥2/ND) | 82/8/4 | 25/0/0 | 38/4/2 | 19/4/2 |
| WBC (/mm3) | 4800 (2800–8100) | 5390 (3000–7500) | 4750 (2800–8100) | 4700 (3040–8000) |
| Plt (×104/mm3) | 18 (9–34) | 20 (15–30) | 16.5 (10–34) | 16 (9–24) |
| Hb (g/dL) | 14.3 (12.3–17) | 14.1 (12.5–16.1) | 14.45 (12.3–17) | 14.4 (12.3–16.6) |
| ALT (IU/L) | 38.5 (12–302) | 35 (12–113) | 39.5 (16–302) | 45 (17–135) |
| γGTP (IU/L) | 36 (11–233) | 31 (11–141) | 34 (14–233) | 49 (21–226) |
| Virus titer (log IU/mL) | 6.7 (5.1–7.7) | 6.7 (5.1–7.4) | 6.7 (5.4–7.6) | 6.7 (5.8–7.7) |
| Days to first ribavirin reduction | 18 (3–168) | 18 (3–52) | 18 (3–168) | 15 (8–52) |
| Duration of telaprevir administration (days) | 85 (29–85) | 85 (29–85) | 85 (32–85) | 85 (35–85) |
| Duration of peg-interferon injection (days) | 162 (22–165) | 163 (22–165) | 162.5 (30–165) | 162 (30–165) |
| Duration of ribavirin administration (days) | 169 (29–169) | 168 (29–169) | 169 (32–169) | 169 (36–169) |
| Effect of therapy (SVR/BT/TR/NR) | 69/4/19/2 | 20/0/5/0 | 41/1/2/0 | 8/3/12/2 |
NOTE. Counts are listed for categorical values and the median and range are reported for continuous variables.
All patients were treated with PEG-IFN-α-2b, ribavirin, and telaprevir triple therapy. Telaprevir (750 mg; MP-424; Mitsubishi Tanabe Pharma) was administered every 8 h after meals. PEG-IFN-α-2b (Schering Plough) was injected subcutaneously at a median dose of 1.5 μg/kg per week. Ribavirin (Schering Plough) dose was adjusted by body weight (600 mg for ≤60 kg; 800 mg for >60 to ≤80 kg; and 1000 mg for >80 kg), based on guidelines by the Ministry of Health, Labor and Welfare of Japan [27], and the drug was administered orally after breakfast and dinner. Triple therapy with telaprevir was given for 12 weeks, followed by an additional 12 weeks of PEG-IFN-α-2b and ribavirin combination therapy. Triple therapy was withdrawn if hemoglobin levels were <8.5 g/dL. Ribavirin dose was reduced by 200 mg/day in patients who were receiving 600 or 800 mg/day (or by 400 mg in those receiving 1000 mg/day) when hemoglobin levels decreased to <12 g/dL and by an additional 200 mg if levels decreased to <10 g/dL. In addition, ribavirin dose was also reduced by 200 mg in patients with a hemoglobin level <13 g/dL at baseline and in those in whom the level decreased by 1 g/dL to <13 g/dL within 1 week. PEG-IFN dose was decreased to one-half when leukocyte count decreased to <1500 leukocytes/mm3, neutrophil count decreased to <750 neutrophils/mm3, or platelet count decreased to <80 × 103platelets/mm3; PEG-IFN was withdrawn if these factors decreased to <1000 leukocytes/mm3, 500 neutrophils/mm3, or 50 × 103platelets/mm3, respectively. Triple therapy was suspended temporarily when hemoglobin levels decreased to <8.5 g/dL. Treatment was resumed with PEG-IFN and 200 mg ribavirin if hemoglobin levels increased to ≥8.5 g/dL within 2 weeks after withdrawal. Reduction of telaprevir dose was not permitted. It was discontinued if severe adverse effects appeared, and therapy was continued with PEG-IFN and ribavirin alone. Erythropoietin was not used to elevate hemoglobin levels.
Virologic response was analyzed on an intent-to-treat basis. The successful end point of treatment was sustained virological response (SVR) for patients who showed undetectable HCV RNA for 24 weeks after cessation of treatment. In transient responders (or persons who experienced relapse), HCV RNA levels became undetectable by the end of treatment but became positive again during the follow-up period. In patients with viral breakthrough, HCV RNA became undetectable during the treatment period but then became positive again before the end of the treatment period. The remaining patients whose HCV RNA never became undetectable were nonresponders. We also defined rapid virological response (RVR) as undetectable HCV RNA at week 4 of treatment and early virological response as a >2 log10 decrease in HCV RNA levels by week 12 of treatment. All participants gave written informed consent to participate in the study according to the process approved by the ethical committee of each hospital and conforming to the ethical guidelines of the 1975 Declaration of Helsinki.
HCV RNA Levels
HCV RNA levels were measured using the TaqMan reverse-transcription polymerase chain reaction (PCR) test. The measurement range of this assay was 1.2–7.8 log IU/mL. Samples that exceeded the measurement range were diluted with phosphate-buffered saline and reanalyzed.
ISDR and Core Amino Acid Substitutions
Amino acid substitutions in the HCV core and ISDR regions were determined using direct sequencing of PCR products after extraction and reverse transcription of HCV RNA with use of serum samples kept frozen at −80°C. Core amino acid substitutions at positions 70 and 91 (core70 and core91, respectively) were determined according to Akuta et al [14, 28], and the number of ISDR substitutions was determined using the methods of Enomoto et al [21, 29, 30].
Single-Nucleotide Polymorphism (SNP) Genotyping
We genotyped each patient for 2 SNPs: rs8099917, an IL28B SNP previously reported to be associated with therapy outcome, and rs1127354 [31], an ITPA SNP reported to be associated with ribavirin-induced anemia [23]. Samples were genotyped using the Illumina HumanHap610-Quad Genotyping BeadChip or with the Invader or TaqMan assay, as described elsewhere [32, 33].
Statistical Analysis
Statistical analysis was performed using PASW Statistics, version 18 (SPSS) and R, version 2.11. Categorical data were analyzed using χ2 and Fisher's exact tests, and continuous data were analyzed using the nonparametric Mann-Whitney U test. To identify independent predictive factors, variables that were significant at the .05 level in univariate tests were considered as candidate factors for multiple logistic regression analysis. The model was reduced using AIC-based forward and/or backward stepwise selection with bootstrap validation. Odds ratios (ORs) were corrected for over-optimism with use of penalized maximum likelihood.
RESULTS
Effect of the Triple Therapy by Previous Response to PEG-IFN Plus Ribavirin Therapy
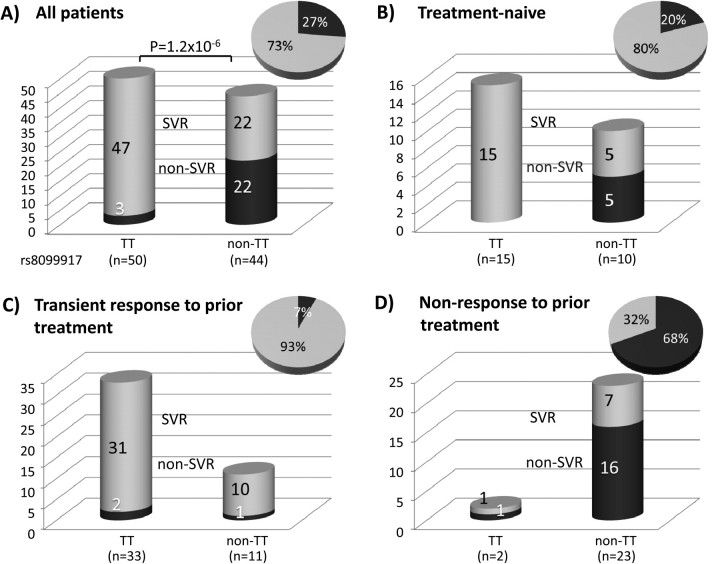
Patient profiles are shown in Tables 1 and 2. After triple therapy, 69 (73%) of 94 patients achieved SVR. Of the 25 treatment-naive patients, 20 (80%) eradicated the virus, and the remaining 5 achieved transient response. Similarly, 49 (71%) of the 69 patients who had received prior treatment achieved SVR with triple therapy. Of note, however, 41 (93%) of 44 patients who had responded transiently to previous treatment were able to eradicate the virus with use of triple therapy. Conversely, only 8 (32%) of 25 patients who had failed to respond to prior treatment were able to achieve SVR with use of triple therapy, and 2 of these patients also failed to respond to triple therapy. None of the 4 patients in whom viral breakthrough occurred were treatment naive, and 3 of the 4 were nonresponders to prior treatment.
IL28B SNP Genotypes
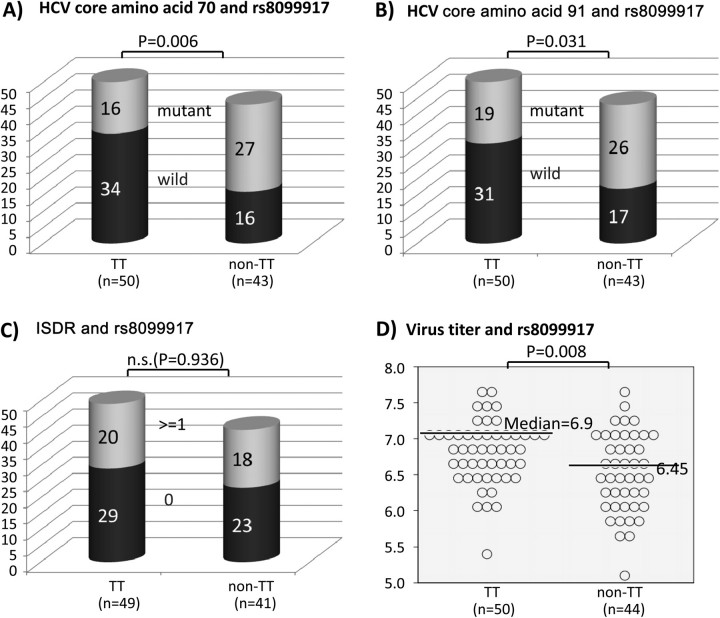
The genotype of IL28B SNP rs8099917 was determined for each patient. The frequency of the rs8099917 risk allele (G) was 0.25 among all patients, 0.17 among patients who achieved SVR, 0.38 among patients with viral breakthrough, and 0.5 among both transient responders and nonresponders. Patients with the rs8099917 TT genotype were significantly more likely to achieve SVR (94% vs 50%; P = 4.6E-6; Figure 1) and had significantly higher baseline viral loads (6.9 vs 6.45 log IU/mL; P = .0056; Figure 2D), compared with patients with GT or GG genotypes.
Figure 1.
SVR frequency after triple therapy grouped by IL128B SNP rs8099917 genotype and by response to previous interferon (IFN) treatment. A, All patients. B, Treatment-naive patients. C, Previously treated patients who responded transiently to therapy. D, Previously treated patients who failed to respond to therapy. Inset pie charts indicate percentage of SVR (light gray) and non-SVR (dark gray) patients.
Figure 2.
Viral factors and IL128B SNP rs8099917 genotype. A, Substitutions at HCV core amino acid 70. B, Substitutions at core amino acid 91. C, Frequency of patients with ≥2 substitutions in the NS5A interferon sensitivity determining region. D, Baseline viral load.
Loss of Hemoglobin During and After Triple Therapy
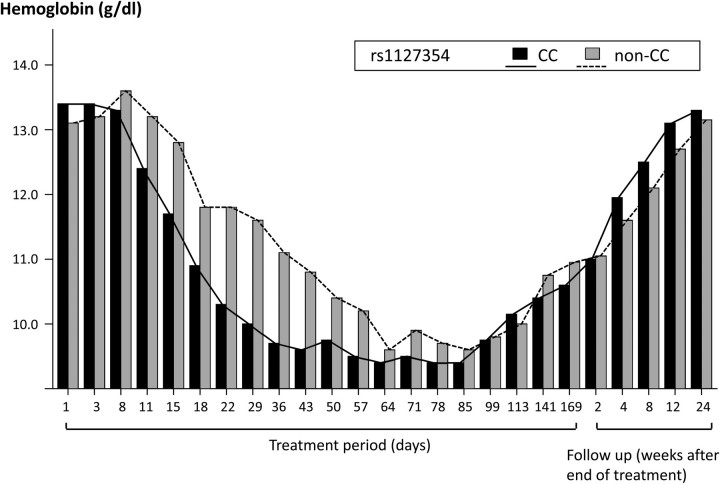
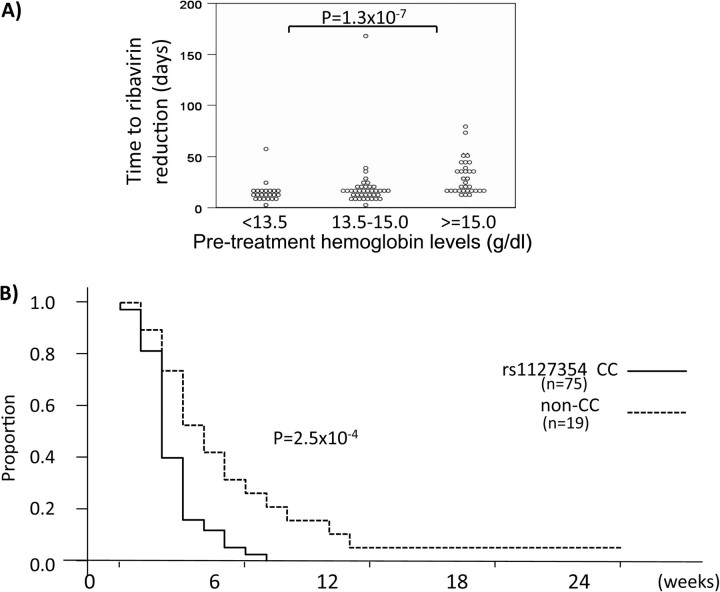
The triple therapy resulted in hemoglobin loss in all patients, but the pattern differed by ITPA SNP rs1127354 genotype (Figure 3). The frequency of the rs1127354 minor allele (A) was 0.11 among all patients, 0.11 among patients who achieved SVR, .13 among transient responders, and 0 in both patients with viral breakthrough and nonresponders. There was no effect of rs1127354 genotype on SVR (73% for both CC and non-CC genotypes), but ribavirin dosage reduction was required significantly earlier in patients with genotype CC than in those with non-CC genotypes (18 days vs 29 days, respectively; P = 3.2E-5; Figure 4). Although hemoglobin loss resulted in dose reduction according to the treatment protocol, no significant effects on SVR rate resulting from dose reduction were observed.
Figure 3.
Change in hemoglobin level by ITPA SNP during triple therapy. Hemoglobin levels in patients grouped by ITPA SNP rs1127354 genotype (solid line represents CC; dashed line represents non-CC).
Figure 4.
Ribavirin dose reduction during triple therapy. A, Number of days of treatment until first ribavirin dose reduction, by pretreatment hemoglobin levels. B, Kaplan-Meier curve for dose reduction grouped by ITPA SNP rs1127354 genotype (solid line represents CC; dashed line represents non-CC).
Viral Substitutions
The 43 patients (46%) with a substitution at position 70 of the HCV core protein (core70) were significantly less likely to achieve SVR than were patients with wild-type core70 (60% vs 86%; P = .01). There was no difference in SVR rate due to substitution at position 91 (core91; 81% vs 67%; P = .17) (Figure 2). There was also no difference in SVR rate due to substitutions in the NS5A ISDR region (P = .43). Patients with rs8099917 genotype TT were significantly more likely to be associated with wild-type core70 or core91 (P = .006 and P = .031, respectively). There was no association between rs809917 genotype and ISDR substitutions (P = .94).
Predictive Factors for RVR
RVR, defined as undetectable HCV RNA levels at week 4 of treatment, is a strong on-treatment predictor of SVR [34]. Previous IFN treatment, time to first ribavirin dose reduction, and baseline hemoglobin levels were each significant univariate predictors, but only hemoglobin level was a significant independent predictor of RVR under multiple logistic regression (P = .028; OR, 3.11).
Predictive Factors for SVR
Significant univariate predictors for SVR included clinical factors (γGTP level; rs8099917 genotype), viral factors (core70 substitutions), response to prior treatment (relapse or nonresponse), and on-treatment factors (RVR) (Table 3). Of these, nonresponse to prior treatment, rs8099917 genotype, RVR, and core70 substitutions were retained in the multivariate model, and nonresponse to prior treatment (OR, .17; P = .01), rs8099917 genotype (OR, .12; P = .014), and RVR (OR, 14.0; P = .0064) were identified as significant independent predictors for SVR. When only pretreatment factors were considered, nonresponse to prior treatment (OR, .14; P = .0028) and rs8099917 genotype (OR, .19; P = .027) were the only independent predictors.
Table 3.
Predictive Factors Associated With SVR in Chronic Hepatitis C Virus Genotype 1 Patients Who Received Pegylated Interferon/Ribavirin/Telaprevir Triple Therapy
| Simple |
Multiple |
|||||||
| Variable | n | OR | P | OR | (95% CI) | P | ||
| Treatment-naive | 94 | 1.6 | .389 | |||||
| Previous non-responder | 94 | 0.1 | 5.5E-08 | *** | 0.17 | (.04–.66) | .010 | * |
| Previous relapser | 94 | 10.7 | 5.2E-05 | *** | ||||
| Age | 94 | 0.8 | .939 | |||||
| Sex (male vs female) | 94 | 1.5 | .100 | |||||
| BMI (kg/m2) | 94 | 0.9 | .558 | |||||
| rs8099917 (TT vs GT/GG) | 94 | 0.1 | 1.7E-06 | *** | 0.12 | (.02–.65) | .014 | * |
| rs1127354 (CC vs AC/AA) | 94 | 1.0 | .980 | |||||
| Core aa70 (wt vs mutant) | 93 | 0.2 | .0053 | ** | 0.35 | (.09–1.31) | .119 | |
| Core aa91 (wt vs mutant) | 93 | 0.5 | .111 | |||||
| ISDR (0–1 vs ≥2) | 90 | 1.7 | .308 | |||||
| Viral load | 94 | 1.1 | .560 | |||||
| ALT (IU/L) | 94 | 0.9 | .142 | |||||
| gammaGTP | 94 | 0.7 | .0009 | *** | ||||
| Hemoglobin (g/dL) | 94 | 1.4 | .292 | |||||
| WBC (/mm3) | 94 | 1.3 | .271 | |||||
| Platelets (×104/mm3) | 94 | 1.7 | .165 | |||||
| Total cholesterol (mg/dL) | 94 | 1.7 | .160 | |||||
| LDL cholesterol (mg/dL) | 94 | 2.6 | .018 | * | ||||
| Days to first ribavirin dose reduction | 94 | 1.2 | .129 | |||||
| RVR | 94 | 10.8 | 4.4E-05 | *** | 14.00 | (2.10–93.2) | .006 | ** |
| EVR | 94 | 7992.0 | .004 | ** | ||||
NOTE. Results of simple and multiple logistic regression are shown. The multivariate model was constructed using stepwise selection of univariate terms significant at the .05 level. Symbols: * (P < .05), ** (P < .01), *** (P < .001).
DISCUSSION
This study showed that patients undergoing PEG-IFN, ribavirin, and telaprevir triple therapy for chronic hepatitis C genotype 1 infection achieve a higher SVR rate than typically expected under combination therapy alone in Japanese patients. Moreover, patients who showed transient response in previous treatment were more likely to achieve SVR after triple therapy, whereas nonresponders to prior treatment remained unlikely to eradicate the virus. Considering that telaprevir has a mode of action different from that of IFN and ribavirin, [5] it is surprising that triple therapy does not better improve SVR rates among prior nonresponders, suggesting that additional unknown factors contribute to nonresponse. However, the duration of triple therapy, followed by standard of care, was limited to 24 weeks in this study; therefore, it is possible that prior nonresponders and patients who experienced relapse may benefit from a longer duration of therapy.
The most interesting result from this study is the high SVR rate among patients who previously experienced relapse, even compared with that of naive patients. This is partly because of the higher frequency of the favorable rs8099917 TT genotype among patients who previously experienced relapse (33 [75%] of 44) than among naive patients (15 [60%] of 25), which perhaps reflects the fact that all patients who previously experienced relapse demonstrated at least a transient response to combination therapy and that this group is less likely to include as many patients with non-TT genotypes. All of the treatment-naive patients with the favorable genotype (15 [100%] of 15) achieved SVR, compared with 31 (94%) of 33 patients who previously experienced relapse; conversely, only one-half of the treatment-naive patients with unfavorable rs8099917 genotypes (5 [50%] of 10) achieved SVR, compared with only 1 (9%) of 11 of the patients who previously experienced relapse. This suggests that, although patients who previously experienced relapse have a demonstrated potential to respond to the therapy, there should be more variability among naive patients. Another consideration is that the frequency of the favorable wild-type core70 amino acid was slightly higher among patients who previously experienced relapse (28 [64%] of 44) than among naive patients (13 [52%] of 25). It should be noted, however, that the small number of patients in this study limits the conclusions that can be drawn, and results should be verified in a larger study, perhaps using stratified sampling based on patient background with regard to treatment history to establish more homogeneous patient populations.
In this and a number of other studies, variation in the IL28B locus remains the strongest predictor of SVR reported to date [16–18, 35]. It is unclear which, if any, of the reported SNPs is the primary or functional SNP, but most studies report results for rs8099917 and/or rs12979860, which are under strong linkage disequilibrium in Japanese patients and fall within the intergenic region upstream of IL28B. Although the mechanism is unknown, IL28B and the other 2 members of the IFN-λ family, IL28A and IL29, code for type III IFNs, which are similar to type I IFNs but use a highly tissue-specific receptor [36, 37]. IFN-stimulated genes appear to be initially down-regulated in patients with the favorable rs8099917 TT genotype [38], which may help to prevent desensitization and promote maximal induction of IFN-stimulated genes, although mechanistic studies are needed to understand the connection between IL28B and SVR.
In addition to IL28B polymorphisms, a number of studies have reported that amino acid substitutions in the HCV core protein and the ISDR region of NS5A independently predict treatment outcome after combination therapy [14, 22, 28, 30], and these findings have recently been extended to triple therapy[39, 40]. In this study, substitution at core70 was significant in univariate tests and was selected for inclusion in the multivariate model, but it was not significant in multiple logistic regression. One reason for this may be that core substitutions were initially reported to be associated with nonresponse [22], whereas this study focused on SVR because of the very small number of nonresponders. Terms that are significant in univariate but not multivariate tests may be correlated with each other, and only the factor with the strongest effect remains significant. In this case, core70 is significantly correlated with the stronger rs8099917 genotype (r = .31; P = .0027), although other studies have shown that these terms contribute independently, especially when a larger number of patients are included [39]. Without knowing the mechanism underlying either factor, it is not possible to determine whether the underlying factors that they represent are in fact independent or whether they represent different aspects of a common unknown factor.
Although novel therapies that are not based on IFN and ribavirin are urgently needed, the pending introduction of protease inhibitors represents a pivotal addition to the treatment arsenal, especially for patients who show at least partial response to combination therapy. Because telaprevir is effective as monotherapy, even if only briefly until resistant mutations emerge, alternate combination therapies based on telaprevir and another component designed to raise the barrier to resistance may provide an adequate alternative for older patients and patients unable to tolerate IFN or ribavirin. Furthermore, identification of additional SNPs associated with anemia and other adverse effects will help reduce complications and the need for dose reductions and may lead to treatment guidelines for at-risk patients, such as administration of erythropoietin to stimulate erythropoiesis [41]. Ribavirin dose reductions were required significantly earlier in patients with ITPA SNP genotype CC, compared with patients with non-CC genotypes, which may contribute to poorer response if cumulative ribavirin administration decreases to <80% of the planned dose [26], although ribavirin dose reduction did not affect SVR rate in this study.
In conclusion, triple therapy with PEG-IFN, ribavirin, and telaprevir resulted in higher rates of SVR, compared with PEG-IFN plus ribavirin combination therapy, especially among treatment-naive patients and patients who showed transient response to prior treatment. ITPA polymorphisms predict ribavirin-induced anemia but are not associated with SVR, whereas IL28B polymorphisms and early viral kinetics remain the strongest predictors of SVR with use of triple therapy. Considering both host and viral factors, we identified 2 subgroups of patients who responded well to triple therapy: patients with the favorable rs8099917 TT genotype (47 [94%] of 50) and patients with non-TT genotypes who had wild-type core70 and core91 amino acids (7 [78%] of 9). Patients matching these conditions would benefit most from this 24-week triple therapy, whereas a longer duration of therapy should perhaps be considered for the remaining difficult-to-treat patients.
Funding
This work was supported by Grants-in-Aid for scientific research and development from the Ministry of Health, Labor and Welfare, Government of Japan.
Acknowledgments
We thank Mika Tsuzuno, Sakura Akamatsu, Sanae Furuya, and other members of the clerical and medical staff at Hiroshima University Hospital for their help.
References
- 1.Alter MJ. Epidemiology of hepatitis C in the West. Semin Liver Dis. 1995;15:5–14. doi: 10.1055/s-2007-1007259. [DOI] [PubMed] [Google Scholar]
- 2.Shepard CW, Finelli L, Alter MJ. Global epidemiology of hepatitis C virus infection. Lancet Infect Dis. 2005;5:558–67. doi: 10.1016/S1473-3099(05)70216-4. [DOI] [PubMed] [Google Scholar]
- 3.Chevaliez S, Pawlotsky JM. Hepatitis C virus: virology, diagnosis and management of antiviral therapy. World J Gastroenterol. 2007;13:2461–6. doi: 10.3748/wjg.v13.i17.2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hadziyannis SJ, Sette H, Jr, Morgan TR, et al. Peginterferon-alpha2a and ribavirin combination therapy in chronic hepatitis C: a randomized study of treatment duration and ribavirin dose. Ann Intern Med. 2004;140:346–55. doi: 10.7326/0003-4819-140-5-200403020-00010. [DOI] [PubMed] [Google Scholar]
- 5.Jang JY, Chung RT. New treatments for chronic hepatitis C. Korean J Hepatol. 2010;16:263–77. doi: 10.3350/kjhep.2010.16.3.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fowell AJ, Nash KL. Telaprevir: a new hope in the treatment of chronic hepatitis C. Adv Ther. 2010;27:512–22. doi: 10.1007/s12325-010-0047-0. [DOI] [PubMed] [Google Scholar]
- 7.Foy E, Li K, Wang C, et al. Regulation of interferon regulatory factor-3 by the hepatitis C virus serine protease. Science. 2003;300:1145–8. doi: 10.1126/science.1082604. [DOI] [PubMed] [Google Scholar]
- 8.Reesink HW, Zeuzem S, Weegink CJ, et al. Rapid decline of viral RNA in hepatitis C patients treated with VX-950: a phase Ib, placebo-controlled, randomized study. Gastroenterology. 2006;131:997–1002. doi: 10.1053/j.gastro.2006.07.013. [DOI] [PubMed] [Google Scholar]
- 9.Sarrazin C, Kieffer TL, Bartels D, et al. Dynamic hepatitis C virus genotypic and phenotypic changes in patients treated with the protease inhibitor telaprevir. Gastroenterology. 2007;132:1767–7. doi: 10.1053/j.gastro.2007.02.037. [DOI] [PubMed] [Google Scholar]
- 10.McHutchison JG, Everson GT, Gordon SC, et al. Telaprevir with peginterferon and ribavirin for chronic HCV genotype 1 infection. N Engl J Med. 2009;360:1827–38. doi: 10.1056/NEJMoa0806104. [DOI] [PubMed] [Google Scholar]
- 11.McHutchison JG, Manns MP, Muir AJ, et al. Telaprevir for previously treated chronic HCV infection. N Engl J Med. 2010;362:1292–303. doi: 10.1056/NEJMoa0908014. [DOI] [PubMed] [Google Scholar]
- 12.Suzuki F, Suzuki Y, Akuta N, et al. Sustained virological response in a patient with chronic hepatitis C treated by monotherapy with the NS3-4A protease inhibitor telaprevir. J Clin Virol. 2010;47:76–8. doi: 10.1016/j.jcv.2009.09.029. [DOI] [PubMed] [Google Scholar]
- 13.Dienstag JL, McHutchison JG. American Gastroenterological Association technical review on the management of hepatitis C. Gastroenterology. 2006;130:231–64. doi: 10.1053/j.gastro.2005.11.010. quiz 214–7. [DOI] [PubMed] [Google Scholar]
- 14.Akuta N, Suzuki F, Kawamura Y, et al. Predictive factors of early and sustained responses to peginterferon plus ribavirin combination therapy in Japanese patients infected with hepatitis C virus genotype 1b: amino acid substitutions in the core region and low-density lipoprotein cholesterol levels. J Hepatol. 2007;46:403–10. doi: 10.1016/j.jhep.2006.09.019. [DOI] [PubMed] [Google Scholar]
- 15.Romero-Gómez M, Del Mar Viloria M, Andrade R, et al. Insulin resistance impairs sustained response rate to peginterferon plus ribavirin in chronic hepatitis C patients. Gastroenterology. 2005;128:636–41. doi: 10.1053/j.gastro.2004.12.049. [DOI] [PubMed] [Google Scholar]
- 16.Ge DL, Fellay J, Thompson AJ, et al. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461:399–401. doi: 10.1038/nature08309. [DOI] [PubMed] [Google Scholar]
- 17.Tanaka Y, Nishida N, Sugiyama M, et al. Genome-wide association of IL28B with response to pegylated interferon-alpha and ribavirin therapy for chronic hepatitis C. Nat Genet. 2009;41:1105–9. doi: 10.1038/ng.449. [DOI] [PubMed] [Google Scholar]
- 18.Suppiah V, Moldovan M, Ahlenstiel G, et al. IL28B is associated with response to chronic hepatitis C interferon-alpha and ribavirin therapy. Nat Genet. 2009;41:1100–U74. doi: 10.1038/ng.447. [DOI] [PubMed] [Google Scholar]
- 19.Thomas DL, Thio CL, Martin MP, et al. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature. 2009;461:798–801. doi: 10.1038/nature08463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zeuzem S, Franke A, Lee JH, Herrmann G, Ruster B, Roth WK. Phylogenetic analysis of hepatitis C virus isolates and their correlation to viremia, liver function tests, and histology. Hepatology. 1996;24:1003–9. doi: 10.1002/hep.510240505. [DOI] [PubMed] [Google Scholar]
- 21.Enomoto N, Sakuma I, Asahina Y, et al. Comparison of full-length sequences of interferon-sensitive and resistant hepatitis-C virus 1b—sensitivity to interferon is conferred by amino-acid substitutions in the NS5A region. J Clin Invest. 1995;96:224–30. doi: 10.1172/JCI118025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Akuta N, Suzuki F, Sezaki H, et al. Association of amino acid substitution pattern in core protein of hepatitis C virus genotype 1b high viral load and non-virological response to interferon-ribavirin combination therapy. Intervirology. 2005;48:372–80. doi: 10.1159/000086064. [DOI] [PubMed] [Google Scholar]
- 23.Fellay J, Thompson A, Ge D, et al. ITPA gene variants protect against anaemia in patients treated for chronic hepatitis C. Nature. 2010;464:405–8. doi: 10.1038/nature08825. [DOI] [PubMed] [Google Scholar]
- 24.Thompson A, Fellay J, Patel K, et al. Variants in the ITPA gene protect against ribavirin-induced hemolytic anemia and decrease the need for ribavirin dose reduction. Gastroenterology. 2010;139:1181–9. doi: 10.1053/j.gastro.2010.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ochi H, Maekawa T, Abe H, et al. ITPA polymorphism affects ribavirin-induced anemia and outcomes of therapy—a genome-wide study of Japanese HCV virus patients. Gastroenterology. 2010;139:1190–7. doi: 10.1053/j.gastro.2010.06.071. e3. [DOI] [PubMed] [Google Scholar]
- 26.McHutchison J, Manns M, Patel K, et al. Adherence to combination therapy enhances sustained response in genotype-1-infected patients with chronic hepatitis C. Gastroenterology. 2002;123:1061–9. doi: 10.1053/gast.2002.35950. [DOI] [PubMed] [Google Scholar]
- 27.Kumada H, Okanoue T, Onji M, et al. Guidelines for the treatment of chronic hepatitis and cirrhosis due to hepatitis C virus infection for the fiscal year 2008 in Japan. Hepatol Res. 2010;40:8–13. doi: 10.1111/j.1872-034X.2009.00634.x. [DOI] [PubMed] [Google Scholar]
- 28.Akuta N, Suzuki F, Sezaki H, et al. Predictive factors of virological non-response to interferon-ribavirin combination therapy for patients infected with hepatitis C virus of genotype 1b and high viral load. J Med Virol. 2006;78:83–90. doi: 10.1002/jmv.20507. [DOI] [PubMed] [Google Scholar]
- 29.Desmet VJ, Gerber M, Hoofnagle JH, Manns M, Scheuer PJ. Classification of chronic hepatitis–diagnosis, grading and staging. Hepatology. 1994;19:1513–20. [PubMed] [Google Scholar]
- 30.Enomoto N, Sakuma I, Asahina Y, et al. Mutations in the nonstructural protein 5A gene and response to interferon in patients with chronic hepatitis C virus 1b infection. New Engl J Med. 1996;334:77–81. doi: 10.1056/NEJM199601113340203. [DOI] [PubMed] [Google Scholar]
- 31.Rauch A, Kutalik Z, Descombes P, et al. Genetic variation in IL28B is associated with chronic hepatitis C and treatment failure: a genome-wide association study. Gastroenterology. 2010;138:1338–45. doi: 10.1053/j.gastro.2009.12.056. 1345 e1–7. [DOI] [PubMed] [Google Scholar]
- 32.Ohnishi Y, Tanaka T, Ozaki K, Yamada R, Suzuki H, Nakamura Y. A high-throughput SNP typing system for genome-wide association studies. J Hum Genet. 2001;46:471–7. doi: 10.1007/s100380170047. [DOI] [PubMed] [Google Scholar]
- 33.Suzuki A, Yamada R, Chang X, et al. Functional haplotypes of PADI4, encoding citrullinating enzyme peptidylarginine deiminase 4, are associated with rheumatoid arthritis. Nat Genet. 2003;34:395–402. doi: 10.1038/ng1206. [DOI] [PubMed] [Google Scholar]
- 34.Poordad F, Reddy KR, Martin P. Rapid virologic response: a new milestone in the management of chronic hepatitis C. Clin Infect Dis. 2008;46:78–84. doi: 10.1086/523585. [DOI] [PubMed] [Google Scholar]
- 35.Thompson AJ, Muir AJ, Sulkowski MS, et al. IL28B polymorphism improves viral kinetics and is the strongest pre-treatment predictor of SVR in HCV-1 patients. Gastroenterology. 2010;139:120–9.e18. doi: 10.1053/j.gastro.2010.04.013. [DOI] [PubMed] [Google Scholar]
- 36.Marcello T, Grakoui A, Barba-Spaeth G, et al. Interferons alpha and lambda inhibit hepatitis C virus replication with distinct signal transduction and gene regulation kinetics. Gastroenterology. 2006;131:1887–98. doi: 10.1053/j.gastro.2006.09.052. [DOI] [PubMed] [Google Scholar]
- 37.Kotenko SV, Gallagher G, Baurin VV, et al. IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat Immunol. 2003;4:69–77. doi: 10.1038/ni875. [DOI] [PubMed] [Google Scholar]
- 38.Honda M, Sakai A, Yamashita T, et al. Hepatic ISG expression is associated with genetic variation in IL28B and the outcome of IFN therapy for chronic hepatitis C. Gastroenterology. 2010;139:499–509. doi: 10.1053/j.gastro.2010.04.049. [DOI] [PubMed] [Google Scholar]
- 39.Akuta N, Suzuki F, Hirakawa M, et al. Amino acid substitution in hepatitis C virus core region and genetic variation near the interleukin 28B gene predict viral response to telaprevir with peginterferon and ribavirin. Hepatology. 2010;52:421–9. doi: 10.1002/hep.23690. [DOI] [PubMed] [Google Scholar]
- 40.Akuta N, Suzuki F, Hirakawa M, et al. Amino acid substitutions in the hepatitis C virus core region of genotype 1b affect very early viral dynamics during treatment with telaprevir, peginterferon, and ribavirin. J Med Virol. 2010;82:575–82. doi: 10.1002/jmv.21741. [DOI] [PubMed] [Google Scholar]
- 41.Dieterich D, Wasserman R, Bräu N, et al. Once-weekly epoetin alfa improves anemia and facilitates maintenance of ribavirin dosing in hepatitis C virus-infected patients receiving ribavirin plus interferon alfa. Am J Gastroenterol. 2003;98:2491–9. doi: 10.1111/j.1572-0241.2003.08700.x. [DOI] [PubMed] [Google Scholar]