Abstract
Background & objectives:
Plasmid mediated AmpC β-lactamase (PMABL) resistance in Escherichia coli and Klebsiella spp. is an emerging problem worldwide. Phenotypic methods are commonly used for detection of PMABL production in Gram-negative isolates, but molecular data about the prevalence of plasmid-mediated AmpC-type resistance at the national level are needed. Hence, a prospective study was undertaken to determine the occurrence of PMABL gene and its types among clinical isolates of E. coli and K. pneumoniae obtained from six different hospitals in India.
Methods:
A total of 241 nosocomial isolates of K. pneumoniae (n=109) and E.coli (n=132) from six geographically distant hospitals in India were included. These were screened for cefoxitin resistance. AmpC disk test and modified three dimensional extraction test were used for phenotypic detection of PMABL production. Molecular types were determined by a multiplex PCR.
Results:
Among the 241 isolates, 187 (77.5%) were found to be cefoxitin resistant (K. pneumoniae n=83, E. coli n=104). AmpC activity was detectable in 153 (63.4%) isolates, (K. pneumoniae n=69, E. coli n=84). By PCR, the plasmid encoded AmpC genes were found in 92 (38.1%) isolates and the molecular types of the genes detected predominantly were DHA, CIT followed by MOX and ACC types.
Interpretation & conclusions:
A high percentage of plasmid-encoded AmpC enzymes was noted in E. coli and K. pneumonia isolates obtained from different parts of the country. Phenotypic methods alone may not reflect the true number of PMABL producers. Genotypic methods need to be employed in national surveillance studies.
Keywords: Escherichia coli, Indian isolates, Klebsiella pneumoniae multidrug resistance, multiplex PCR, plasmid-mediated AmpC β- lactamases
AmpC β-lactamase production is one of the mechanisms of resistance to β-lactam antibiotics in Gram negative bacteria conferring resistance to a wide variety of β-lactam antibiotics including 7-α-methoxy cephalosporins (cefoxitin or cefotetan), oxyimino cephalosporins (cefotaxime, ceftazidime, ceftriaxone), monobactam (aztreonam) and are not inhibited by clavulanic acid1. These are of two types, chromosomal inducible and plasmid mediated non-inducible. Plasmid mediated AmpC β-lactamases (PMABLs) have evolved by the movement of chromosomal genes on to plasmids and are found in Escherichia coli, Klebsiella pneumoniae, Salmonella spp, Proteus mirabilis, Citrobacter freundii, Enterobacter aerogenes which confer resistance similar to their chromosomal counterparts. Currently, there are over 30 known types derived from Enterobacter cloacae, Morganella morganii, Citrobacter freundii, Hafnia alvei and other of unknown origin2.
Organisms producing PMABLs such as E.coli and Klebsiella spp, are often associated with multidrug resistance, leaving a few therapeutic options. PMABLs can be detected by various phenotypic methods such as Amp C disk test3, three dimensional test4, cefoxitin agar method5, modified double disk test6, using inhibitors like boronic acids7, syn2160 compunds8 and broth micro dilution method7. Testing PMABL is not widely attempted by many laboratories; phenotypic tests may be ambiguous and unreliable resulting in misreporting and treatment failures. In addition, the co-existence of extended spectrum β-lactamases (ESBLs) may mask its detection phenotypically. There are no Clinical Laboratory Standards Institute (CLSI) guidelines available for its optimal detection and confirmation9. Phenotypic tests do not differentiate between chromosomal AmpC genes and AmpC genes that are carried on plasmids. Hence, genotypic characterization is considered as the gold standard10.
In view of the increasing reports of PMABL producing strains of Klebsiella spp. and E. coli and its types from around the world, and paucity of molecular studies in our country, the present work was conducted to examine the occurrence of PMABL gene types among the nosocomial isolates of K. pneumoniae and E.coli from six different hospitals.
Material & Methods
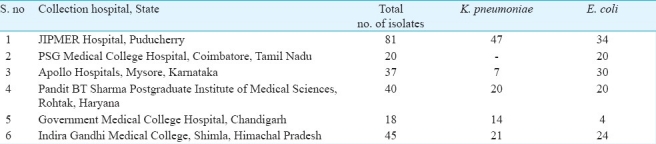
Bacterial isolates: In this study, during a period of five months (September 2009-January 2010), a total of 241 non-duplicate clinical isolates of K. pneumoniae (n=109) and E.coli (n=132) from hospitalized patients obtained from six geographically distant hospitals in India were investigated (Table I). All the isolates were identified as per the standard bacteriological procedures11, stocked in 0.2 per cent semi-solid agar tubes and transferred to the m0 icrobiology department of the study centre Jawaharlal Institute of Postgraduate Medical Education and Research (JIPMER), Puducherry for further characterization. This study was restricted to E. coli and K. pneumoniae isolates only as the CLSI guidelines for ESBL screening and confirmation are available only for these two9.
Table I.
Distribution and source of the clinical isolates from various parts of the country
Antibiotic susceptibility testing: Antibiotic susceptibilities were determined by the standard disk diffusion test for the following antibiotics (concentration/disk in μg); ceftazidime (30), ceftriaxone (30), cefotaxime (30), cefoxitin (30), aztreonam (30), amikacin (30), tetracycline (30), ampicillin (10), co-trimoxazole (10), cefepime (30), gentamicin (10), meropenem (10), imipenem (10), ciprofloxacin (5), amoxycillin-clavulanic acid (30/10), piperacillin-tazobactam (100/10) (Hi-Media, Mumbai, India). E. coli ATCC 25922 was used as the control and the results were interpreted as per CLSI criteria9.
Isolates showing resistance to cefoxitin (inhibition zone <18 mm), a 3rd generation cephalosporins (3GC), intermediate or resistant to amoxycillin-clavulanic acid and showing no cephalosporin/clavulanate synergism were considered as putative AmpC producers.
Amp C disk test: AmpC disk test was done for the phenotypic detection of AmpC β-lactamases production on a Muller-Hinton agar (MHA) plate as described previously3. A flattening or indentation of the cefoxitin inhibition zone in the vicinity of the disk with test strain was interpreted as positive for the production of AmpC β-lactamase. An undistorted zone was considered as negative.
Modified three-dimensional test: Plasmid mediated AmpC beta lactamases production was further confirmed by the modified three-dimensional test12. Briefly, crude enzyme extract of the test organism was prepared by repeated freeze thawing in -80°C for seven times. A lawn culture of E. coli ATCC 25922 was made on MHA and a cefoxitin (30 μg) disk was placed at the centre. Linear slit was cut, 3 mm away from the disk and 30 μl of the enzyme extract was added to a well made at the outer edge of the slit. The plate was incubated overnight at 37°C. Clear distortion of zone of inhibition of cefoxitin is considered as positive test. Quality control was achieved by using known in-house AmpC positive isolate of K. pneumoniae.
Detection of Plasmid encoded AmpC genes: All the isolates that were phenotypically positive for AmpC activity were tested by a multiplex PCR assay which identifies six family-specific AmpC genes carried on the plasmids such as MOX, FOX, EBC, ACC, DHA and CIT13. Template DNA was obtained by boiling lysis method. Amplification was performed on a Corbett Research thermal cycler (HP, USA) as per primers and condition described13. ACC, DHA and CIT positive E.coli controls were included (Kindly provided by Dr. John Hays, Erasmus Medical College, The Netherlands) and the PCR products were analyzed by electrophoresis in 2 per cent agarose gels stained with ethidium bromide (Fig. a & b).
Fig.
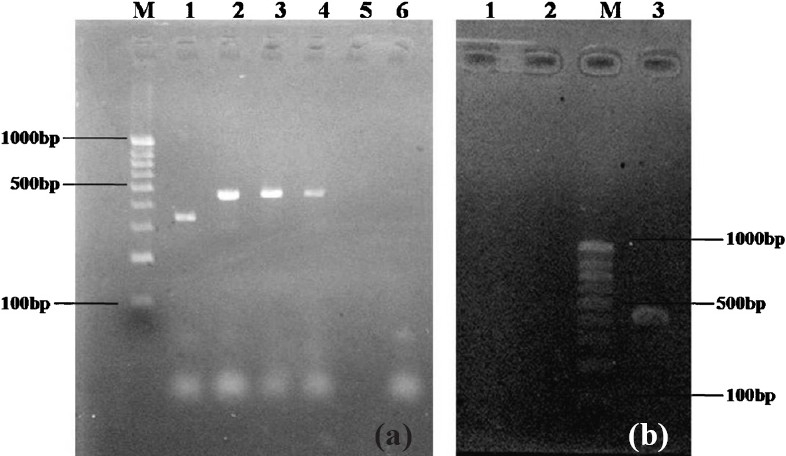
PCR for Plasmid-encoded AmpC β-lactamase genes: 2 per cent Agarose gel showing products of PCR amplification. (a) M-100bp molecular size standard DNA ladder. Lane 1-E.coli with 342bp ACC gene, Lane 2-K. pneumoniae with 462bp CIT, Lane 3 & 4 -E. colishowing 462bp CIT gene, Lane 5- negative K. pneumoniae & Lane 6- negative water control. (b) Lane 1-negative K. pneumoniae, Lane 2- negative water control, M-100bp molecular size standard DNA ladder, Lane 4- E.coli with 405 DHA positive, the 520bp MOX gene was not shown in the figure.
Results
Of the 241 isolates tested, 187 (77.5%) were cefoxitin resistant (K. pneumoniae n=83, E. coli n=104) and were thus considered as putative AmpC producers. Phenotypically, AmpC production was present in 63.4 per cent (153/241) isolates (K. pneumoniae n=69, E. coli n=84). Using AmpC disk test and modified three-dimensional tests, PMABL production was detected in 137 (73.2%) and 149 (79.6%) of cefoxitin resistant isolates, respectively.
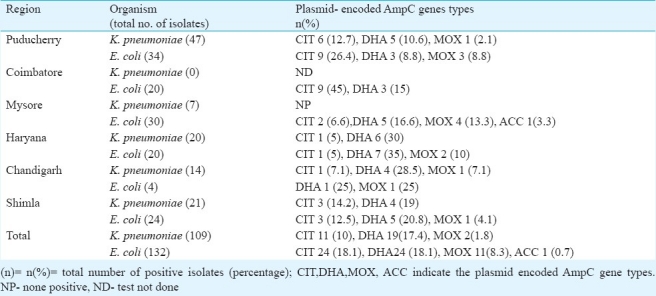
Among the 241 total isolates tested, plasmid-encoded AmpC genes were detected by PCR in 92 (38.1%), which included K. pneumoniae (n=32) and E. coli (n=60). Of these, plasmid-encoded AmpC genes belonging to the DHA family were detected in 43 (46.7%) isolates. Of the 43 DHA positive isolates, 19 (44.1%) were K. pneumoniae and 24 (55.8%) were E. coli. Plasmid-encoded AmpC genes belonging to the CIT family were detected in 38 per cetn (35/92) isolates24. E. coli isolates (68.5%) and 11(31.4%) K. pneumoniae.
AmpC genes belonging to the MOX family were detected in 14.1 per cent (13/92) isolates, [E. coli (n=11, 84.6%) and K. pneumoniae (n=2, 15.3%)]. Gene of the family ACC type was present in only one isolate. No genes belonging to the FOX or EBC family were detected.
Of the 92 isolates identified as possessing plasmid-mediated AmpC, the distribution of the sources were from blood cultures (n=17, 18.4%), urine (n=44, 47.8%), and other body fluids (n=31, 33.6%).
Among the 84 E. coli that showed AmpC production phenotypically, non-plasmid-derived AmpC activity was present in 28.5 per cent (n=24). Similarly, among the 69 K. pneumoniae isolates non-plasmid-derived AmpC activity was present in 53.6 per cent (n=37).
By antibiotic susceptibility testing, all the PMABL producers were resistant to piperacillin/tazobactam, amoxycillin/clavulanate combination and 84 (91%) were resistant to co-trimoxazole, gentamicin, tetracycline and amikacin thus showing multidrug resistance. Among the PMABL producers, (67%) had shown cefepime resistant. A total of 26 (10.7%) and 13(5.3%) isolates were resistant to meropenem and imipenem, respectively.
Discussion
Plasmid mediated AmpC β-lactamases represent a new threat since these confer resistance to cephamycins and are not affected by β -lactamase inhibitors, and can, in strains with loss of outer membrane porins, provide resistance to carbapenems. This resistance mechanism in E.coli and K. pneumoniae has been found around the world causing nosocomial outbreaks14,15.
Distinguishing between cefoxitin-resistant AmpC producers from cefoxitin-resistant non-AmpC producers could guide treatment options, i.e. extended spectrum cephalosporins for cefoxitin-resistant non-AmpC, non-ESBL producers and carbapenems for the cefoxitin-resistant AmpC producers. Differentiation between these types of organisms would prevent the unnecessary usage of cephalosporins and carbapenems resulting in the selective pressure driving the AmpC or plasmid mediated class A carbapenem resistance gene propagation16. There are also concerns that treatment failures will occur with certain cephalosporins due to incorrect susceptibility tests when organisms producing PMABL appears falsely susceptible. Hence, detection of PMABL producing organisms is important to ensure effective therapeutic intervention and optimal clinical outcome8,17.
In this study, occurrence of a large percentage of multidrug resistance has been observed among the PMABL producing strains. Though AmpC producers are susceptible to tazobactam compared to other β-lactamases inhibitors, a high resistant to piperacillin/tazobactam combination was noticed in our isolates, which may be due to hyperproduction of β-lactamases and inhibitor-resistant TEM β-lactamases, which are responsible for resistance to inhibitor combinations among E. coli and K. pneumoniae1. Further, the co-presence of ESBLs adds to the mechanism of resistance to piperacillin/tazobactam and cefepime (Data not shown). The resistance to piperacillin/tazobactam among the AmpC producers in our study was similar to the findings of a study from south India18 and contrary to that of Taneja et al19. With the continuing use of cefoxitin and cefotetan and the clinical introduction of β-lactamase combinations such as clavulanate with amoxicillin or ticarcillin, sulbactam with ampicillin, and tazobactam with piperacillin, plasmids encoding class C β-lactamases appeared. Such enzymes provide a broader spectrum of resistance than ESBLs1.
In the last one decade, AmpC production has been reported from various parts of the country. From north India, 6.97 per cent E.coli and 6.18 per cent K. pneumoniae (New Delhi)20, 9.9 per cent E. coli and 31.1 per cent K. pneumoniae21 were reported as PMABL producers. From eastern part of the country (Kolkata) 47.8 per cent E. coli, 13 per cent K. pneumoniae were reported as AmpC β-lactamase producers22. From southern States, 24.1 per cent of Klebsiella spp. and 37.5 per cent of E. coli (Chennai), 9.2 per cent was reported in an another study from Chennai; 3.3 per cent of E. coli, 2.2 per cent of K. pneumoniae (Karnataka) and 3.4 per cent of E. coli, 4.8 per cent of K. pneumoniae (Andhra Pradesh) were found to harbour AmpC enzymes19,23–25. However, these studies were based on phenotypic tests which do not differentiate between the plasmid-mediated enzymes producers and the chromosomal hyper producers or porin loss mutants. Also these studies did not differentiate the types or families of plasmid-mediated AmpC β-lactamase. Thus, molecular studies will help us to know the actual prevalence of these enzymes13,27.
It is pertinent to note that in this study 10.4 per cent of the cefoxitin resistant isolates were not detected by AmpC disk test and 2.6 per cent by modified three dimensional test. Phenotypic tests alone may not reflect the true number of PMABL producers, hence, molecular studies, although not possible routinely in clinical laboratories, need to be employed in surveillance studies.
The knowledge about the molecular types and the prevalence of plasmid-mediated AmpC-type resistance at the national level is important to provide useful information needed for targeted antimicrobial therapy and better infection control16. In this study, of the 241 isolates tested, 92 carried plasmid-encoded AmpC genes (38.1%), with an occurrence of 29.3 per cent in K. pneumoniae and 45.5 per cent in E.coli thus showing their presence among Indian isolates.
In a nationwide study from China26, the prevalence of plasmid-mediated AmpC β-lactamases was 10.1 per cent in K. pneumoniae and 2.0 per cent in E. coli strains. Similarly, from the United States27, 8.5 per cent of the Klebsiella spp. and 4 per cent of the E. coli strains contained plasmid-mediated AmpC-type enzymes. From Singapore28, 26 per cent of PMABL genes were reported and the lowest rates of AmpC genes were reported from Switzerland as 0.2 per cent in E. coli and 0.4 per cent in K. pneumoniae29. Thus, the percentage of PMABL genes production was found to be higher in our country compared to China, US and Singapore and Switzerland. On the contrary, compared to our results, the highest prevalence of AmpC genes were reported in a Korean surveillance30 showing 73 per cent of E. coli and 77 per cent of K. pneumoniae carrying PMABL genes.
Plasmid mediated AmpC β-lactamases from K. pneumoniae isolates was first reported in 1989 in Seoul, South Korea14. Subsequently, these have been reported worldwide and 29 different plasmid-mediated AmpC genes have been identified to date and deposited in Gene Bank15. In 1998, CMY-4 was reported from India from a strain of K. pneumoniae1 and CMY-6 type (CIT family) was reported in 2009 from Uttar Pradesh21. In the present study, DHA and CIT type genes were predominantly present in K. pneumoniae and E.coli, followed by MOX and ACC types in E.coli (Table II). The ACC genes were recovered only from the southern part of the country. Whereas, FOX or EBC family genes were not detected from any region. The current study was restricted mainly to detect plasmid mediated AmpC gene types and hence the co-existence of ESBLs was not shown.
Table II.
Distribution of the types of plasmid-encoded AmpC β-lactamase genes from various region of the country
This study had certain limitations. The presence of only plasmid mediated AmpC β-lactamase genes was targeted and the other possible mechanisms of cefoxitin resistance like chromosomal hyper producers or porin loss mutants were not detected. Further, we have come across strains with imipenem and cefoxitin resistance and the possible mechanism of this phenotype could be due to the presence of carbapenemases like K. pneumoniae carbapenemase (KPCs) along with AmpC31 and their occurrence were not looked for.
In conclusion 38.1 per cent isolates of K. pneumoniae and E. coli showed the occurrence of PMABL which is alarming. Dissemination of these organisms within the hospital or between the different regions of the country may become an important public health issue. Hence, identification of types of AmpC may aid in hospital infection control and help the physician to prescribe the most appropriate antibiotic, thus decreasing the selective pressure, which generates antibiotic resistance. Further studies such as sequencing and typing the strains may be required to better understand the genetic relatedness and the molecular epidemiology of this resistance mechanism.
Acknowledgments
The authors acknowledge the following contributors from various parts of the country who provided the clinical isolates for this study. Dr Jagdish Chander, Government Medical College Hospital, Chandigarh; Dr Puneeth Kumar Gupta, Indira Gandhi Medical College, Shimla; Dr Uma Chaudhary, Pandit BD Sharma PGIMS, Haryana; Dr Chaya, Apollo Hospitals, Mysore and Dr Appalaraju, PSG Medical College, Coimbatore. Part of the work was funded by University Grants Commission, New Delhi.
References
- 1.Philippon A, Arlet G, Jacoby GA. Plasmid-Determined AmpC-type β-lactamases. Antimicrob Agents Chemother. 2002;46:1–11. doi: 10.1128/AAC.46.1.1-11.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coudron PE, Moland ES, Thomson KS. Occurrence and detection of AmpC beta-lactamases among Escherichia coli, Klebsiella pneumoniae, and Proteus mirabilis isolates at a veterans medical center. J Clin Microbiol. 2000;38:1791–6. doi: 10.1128/jcm.38.5.1791-1796.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Black JA, Moland ES, Thomson KS. AmpC disk test for detection of plasmid-mediated AmpC β-lactamases in Enterobacteriaceae lacking chromosomal AmpC β -lactamases. J Clin Microbiol. 2005;43:3110–3. doi: 10.1128/JCM.43.7.3110-3113.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thomson KS, Sanders CC. Detection of extended spectrum beta-lactamases in members of the family Enterobacteriaceae: comparison of the double-disk and three-dimensional tests. Antimicrob Agents Chemother. 1992;36:1877–82. doi: 10.1128/aac.36.9.1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nasim K, Elsayed S, Pitout JD, Conly J, Church DL, Gregson DB. New method for laboratory detection of AmpC beta-lactamases in Escherichia coli and Klebsiella pneumoniae. J Clin Microbiol. 2004;42:4799–802. doi: 10.1128/JCM.42.10.4799-4802.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pitout JD, Reisbig MD, Venter EC, Church DL, Hanson ND. Modification of the double-disk test for detection of Enterobacteriaceae producing extended-spectrum and AmpC beta-lactamases. J Clin Microbiol. 2003;41:3933–5. doi: 10.1128/JCM.41.8.3933-3935.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coudron PE. Inhibitor-based methods for detection of plasmid-mediated AmpC beta-lactamases in Klebsiella spp., Escherichia coli and Proteus mirabilis. J Clin Microbiol. 2005;43:4163–7. doi: 10.1128/JCM.43.8.4163-4167.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Black JA, Thomson KS, Buynak JD, Pitout JDD. Evaluation of β-lactamase inhibitors in disk tests for detection of plasmid-mediated AmpC β-lactamases in well-characterized clinical strains of Klebsiella spp. J Clin Microbiol. 2005;43:4168–71. doi: 10.1128/JCM.43.8.4168-4171.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Performance standards for antimicrobial susceptibility testing. 17th ed. Wayne, Pa: Clinical and Laboratory Standards Institute; 2007. Clinical and Laboratory Standards Institute. CLSI document M100-S17. [Google Scholar]
- 10.Thomson KS. Controversies about extended-spectrum and AmpC β-lactamases. Emerg Infect Dis. 2001;7:333–6. doi: 10.3201/eid0702.010238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koneman EW, Allen SD, Janda WM, Schreckenberger PC, Winn WC., Jr . In: Color atlas and textbook of diagnostic microbiology. 4th ed. Philadelphia: J.B. Lippincott Co; 1992. The Enterobacteriaceae; pp. 105–84. editor. [Google Scholar]
- 12.Manchanda V, Singh NP. Occurrence and detection of AmpC β -lactamases among Gram negative clinical isolates using a modified three-dimensional test at Guru Tegh Bahadur Hospital, Delhi, India. J Antimicrob Chemother. 2003;51:415–8. doi: 10.1093/jac/dkg098. [DOI] [PubMed] [Google Scholar]
- 13.Perez-Perez FJ, Hanson ND. Detection of plasmid-mediated AmpC beta-lactamase genes in clinical isolates by using multiplex PCR. J Clin Microbiol. 2002;40:2153–62. doi: 10.1128/JCM.40.6.2153-2162.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bauernfeind A, Schneider I, Jungwirth R, Sahly H, Ullmann U. A novel type of AmpC β -lactamase, ACC-1, produced by a Klebsiella pneumoniae strain causing nosocomial pneumonia. Antimicrob Agents Chemother. 1999;43:1924–31. doi: 10.1128/aac.43.8.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dunne EF, Fey PD, Klutdt P, Reporter R, Mostashari F, Shillam P, et al. Emergence of domestically acquired ceftriaxone-resistant Salmonella 0infections associated with AmpC β-lactamase. JAMA. 2000;284:3151–6. doi: 10.1001/jama.284.24.3151. [DOI] [PubMed] [Google Scholar]
- 16.Hanson DN. AmpC β-lactamases: what do we need to know for the future? J Antimicrob Chemother. 2003;52:2–4. doi: 10.1093/jac/dkg284. [DOI] [PubMed] [Google Scholar]
- 17.Hernandez-Alles S, Benedi VJ, Martinez-Martiner L, Pascual A, Aguilar A, Tomas JM, et al. Development of resistance during antimicrobial therapy caused by insertion sequence interruption of Porin genes. Antimicrob Agents Chemother. 1999;43:937–9. doi: 10.1128/aac.43.4.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Navaneeth BV. Extended-spectrum and AmpC β-lactamases in Escherichia coli and Klebsiella pneumoniae from a rural South Indian tertiary care hospital. Int J Antimicrob Agents. 2007;29:602–3. doi: 10.1016/j.ijantimicag.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 19.Taneja N, Rao P, Arora J, Dogra A. Occurrence of ESBL & Amp-C β-lactamases & susceptibility to newer antimicrobial agents in complicated UTI. Indian J Med Res. 2008;127:85–8. [PubMed] [Google Scholar]
- 20.Singhal S, Mathur T, Khan S, Upadhyay DJ, Chugh S, Gaind R, et al. Evaluation of methods for AmpC b-lactamase in Gram negative clinical isolates from tertiary care hospitals. Indian J Med Microbiol. 2005;23:120–4. doi: 10.4103/0255-0857.16053. [DOI] [PubMed] [Google Scholar]
- 21.Shahid M, Ensor VM, Hawkey PM. Emergence and dissemination of Enterobacteriaceae with plasmid-mediated CMY-6 and CTX-M-15 beta-lactamases in community in North-India. World J Microbiol Biotechnol. 2009;25:1439–6. [Google Scholar]
- 22.Arora S, Bal M. AmpC β-lactamase producing bacterial isolates from Kolkata hospital. Indian J Med Res. 2005;122:224–33. [PubMed] [Google Scholar]
- 23.Subha A, Renuka Devi V, Ananthan S. AmpC β-lactamase producing multidrug resistant strains of Klebsiella spp.& Escherichia coli isolated from children under five in Chennai. Indian J Med Res. 2003;117:13–18. [PubMed] [Google Scholar]
- 24.Ratna AK, Menon I, Kapur I, Kulkarni R. Occurrence & detection of AmpC β-lactamases at a referral hospital in Karnataka. Indian J Med Res. 2003;118:29–32. [PubMed] [Google Scholar]
- 25.Hemalatha V, Padma M, Sekar U, Vinodh TM, Arunkumar AS. Detection of Amp C beta lactamases production in Escherichia coli & Klebsiella by an inhibitor based method. Indian J Med Res. 2007;126:220–3. [PubMed] [Google Scholar]
- 26.Ding H, Yang Y, Lu Q, Wang Y, Chen Y, Deng L, et al. The prevalence of plasmid-mediated AmpC β-lactamases among clinical isolates of Escherichia coli and Klebsiella pneumoniae from five children's hospitals in China. Eur J Clin Microbiol Infect Dis. 2008;27:915–21. doi: 10.1007/s10096-008-0532-4. [DOI] [PubMed] [Google Scholar]
- 27.Alvarez M, Tran JH, Chow N, Jacoby GA. Epidemiology of conjugative plasmid-mediated AmpC beta lactamases in the United States. Antimicrob Agents Chemother. 2004;48:533–7. doi: 10.1128/AAC.48.2.533-537.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tan TY, Ng SY, Teo L, Koh Y, Teok CH. Detection of plasmid-mediated AmpC in Escherichia coli, Klebsiella pneumoniae and Proteus mirabilis. J Clin Pathol. 2008;61:642–4. doi: 10.1136/jcp.2007.053470. [DOI] [PubMed] [Google Scholar]
- 29.Adler H, Fenner L, Walter P, Hohler D, Schultheiss E, Oezcan S, et al. Plasmid-mediated AmpC β-lactamases in Enterobacteriaceae lacking inducible chromosomal ampC genes: prevalence at a Swiss university hospital and occurrence of the different molecular types in Switzerland. J Antimicrob Chemother. 2008;61:457–8. doi: 10.1093/jac/dkm472. [DOI] [PubMed] [Google Scholar]
- 30.Yong D, Choi YS, Park DY, Kim S, Lee H, Yum JH, et al., editors. Copenhagen, Denmark: Oxford; 2005. Apr 2-5, Prevalence and characteristics of plasmid-mediated AmpC beta-lactamase in Escherichia coli and Klebsiella pneumoniae isolates in a Korean hospital. Proceedings of the 15th European Congress of Clinical Microbiology and Infectious Diseases Conference; 2005. [Google Scholar]
- 31.Queenan AM, Bush K. Carbapenemases: the versatile β-lactamases. Clin Microbiol Rev. 2007;20:440–58. doi: 10.1128/CMR.00001-07. [DOI] [PMC free article] [PubMed] [Google Scholar]


