Abstract
Background & objectives:
Chronic myelogenous leukaemia (CML) is the commonest leukaemia in Asia. There is a paucity data on cytogenetic and molecular analyses of Indian CML patients. This apparently reflects the low availability of cytogenetic and molecular techniques in our country. This study aimed to document various types of BCR-ABL fusion transcripts in different phases of CML and to compare the Ph chromosome positivity/negativity vis-a-vis BCR-ABL fusion transcripts in adult CML patients.
Methods:
Between June 2004 and February 2009, 208 patients were diagnosed as CML in chronic phase (CP), accelerated phase (AP) and blast crisis (BC), according to standard criteria. Cytogenetic and molecular genetic analyses were performed in all patients. Various types of BCR-ABL hybrid transcripts were compared with phases of CML and cytogenetic abnormalities.
Results:
Among 208 CML patients, b3a2 BCR-ABL transcripts were most commonly detected (66.82%) followed by b2a2 (28.84%), b3a2 + b2a2 (3.36%), b3a2 + e19a2 (0.48%) and b2a2 + e19a2 (0.48%). b3a2 transcripts were more frequently detected than b2a2 transcripts, in the whole group of 208 as well as in 183 CML-CP patients (P<0.0001). Ph chromosome was positive in 135 of 139 patients with b3a2 transcripts and 56 of 60 patients with b2a2 transcripts, difference not being significant. Additional cytogenetic abnormalities detected in 3.8 per cent patients in CML-CP and 44 per cent patients in CML-AP/BC, did not show predilection for any BCR-ABL transcript type.
Interpretation & conclusions:
This study documents higher Ph positivity (96.15%) by cytogenetic analysis among CML patients, as confirmed by qualitative reverse transcriptase-polymerase chain reaction (RT-PCR) analysis in a large patient group from north India. Both the techniques contribute towards understanding the disease biology, and have important implications for diagnosis and management of CML patients.
Keywords: BCR-ABL fusion transcripts, chronic myelogenous leukaemia, cytogenetic analysis, polymerase chain reaction, reverse transcriptase
Chronic myelogenous leukaemia (CML), the prototype chronic myeloproliferative neoplasm (CMPN) is one of the leukaemias which can be easily diagnosed in view of typical haematological and morphological findings, in an appropriate clinical setting. CML is diagnosed in the presence of features such as splenomegaly, leucocytosis with myelocyte and neutrophil predominance, low neutrophil alkaline phosphatase (NAP) score, hypercellular bone marrow (BM) with granulocytic or granulocytic/megakaryocytic hyperplasia. Patients lacking typical features of specific CMPN, need to be differentiated from other primary haematological and secondary disorders showing myeloproliferation1–3.
The sine qua non of CML, t(9;22)(q34;q11) and/or BCR-ABL positivity must be detected in all cases of CML in chronic phase (CP) and accelerated phase (AP)/blast crisis (BC)1. t(9;22) or Philadelphia (Ph) chromosome is detected in 90-95 per cent patients on cytogenetic analysis. A few CML patients may not demonstrate Ph chromosome (Ph chromosome negative and BCR-ABL positive), yet have clinical course and morphological features just like typical CML1–3. Additional cytogenetic abnormalities are associated with disease progression from CML-CP towards CML-AP/BC in nearly 70-80 per cent patients2,3.
Ph chromosome/t(9;22) is a diagnostic hallmark of CML, however it is detected in other haematological malignant disorders as well [10-20% adult acute lymphoblastic leukaemia (ALL), 2-5 per cent paediatric ALL, <5 per cent cases of acute myeloblastic leukaemia (AML), and rarely in multiple myeloma, lymphoma and chronic neutrophilic leukaemia (CNL) patients]2.
Presence of molecular counterpart of Ph chromosome/t(9;22) in CML patients (Ph positive and Ph negative) is confirmed by reverse transcriptase-polymerase chain reaction (RT-PCR) or fluorescence in situ hybridization (FISH). In addition, molecular analysis contributes to identify various molecular subtypes of CML-like disorders and makes differential diagnoses on the basis of 3 breakpoint cluster (bcr) regions involved in the translocation between BCR and ABL genes. Disease phenotype of patients varies with different breakpoints involved, resulting in varying sizes of fusion mRNA transcripts and chimeric proteins1.
Major bcr (M-bcr) is almost always involved in CML patients, resulting in b2a2 and b3a2 mRNA transcripts, p210 chimeric protein, and classical CML phenotype. Minor bcr (m-bcr), e1a2 mRNA transcripts and p190 chimeric protein, are most frequently associated with Ph positive ALL. However small amount of e1a2 mRNA transcripts, due to alternate splicing, can also be detected in many patients with classical CML phenotype. m-bcr involvement may also be seen in rare CML patients with monocytosis, thus resembling chronic myelomonocytic leukaemia (CMML). CML patients with breakpoint in micro bcr (μ-bcr), e19a2 and p230 chimeric protein, may demonstrate prominent neutrophilic and/or thrombocytosis; these should not be diagnosed as chronic neutrophilic leukaemia (CNL) or essential thrombocythaemia (ET)1. Therefore, in the current scenario cytogenetic and molecular analyses of CML patients have become mandatory in order to undertake diagnostic evaluation and monitor/predict response to newer molecular targeted treatment modalities like imatinib mesylate (IM)1–5. Variable frequencies of BCR-ABL fusion transcripts have been reported from different parts of the world2,6–11; a few documented b2a2 to be more common7,8, whereas other studies found b3a2 to be more common9–11. Co-expression of BCR-ABL fusion transcripts, though rare, presents an interesting scenario for future exploration.
Despite being the commonest leukaemia in Asia12, there are very few studies published from India, documenting the frequency of BCR-ABL fusion transcripts13–15. A very wide range of Ph positivity (67-95%) has been reported in various studies on CML patients from India, without having the benefit of gold standard molecular analysis16–22. However, Ph chromosome positivity/negativity (and other cytogenetic abnormalities) in relation to BCR-ABL fusion transcripts has not been analyzed earlier among Indian patients.
Very few patients can afford to purchase IM on their own in India. Approximately 95 per cent CML patients are provided IM as a first-line treatment, through the Glivec International Patient Assistance Program (GIPAP)23. Several CML patients are prescribed some form of treatment, without optimal diagnostic work up. This may alter their haematological and/or cytogenetic profiles though molecular analysis always remains informative23.
We present here our experience of analysis of a large number of CML patients being diagnosed and treated at Postgraduate Institute of Medical Education and Research (PGIMER), Chandigarh. This study was planned to document in a large group of CML patients various types of BCR-ABL fusion transcripts in different phases of CML (single or co-expression of more than one type of fusion transcript) and to compare the Ph chromosome positivity/negativity vis-a-vis BCR-ABL fusion transcripts.
Material & Methods
Consecutively diagnosed CML patients in chronic phase, accelerated phase and blast crisis were recruited in this study, during June 2004 to February 2009. These patients were managed in the OPD and wards, under the adult haematology services of Internal Medicine department of PGIMER, Chandigarh. Diagnosis of different phases of CML was made according to the WHO criteria1. This study protocol was approved by the institutional ethics committee. Informed consent was obtained from all the subjects.
Blood counts, morphology and lineage assignment: Following parameters along with careful morphological appraisal, were routinely evaluated in all the patients: complete blood counts (CBC), differential leucocyte count (DLC), NAP score, BM aspirate smears, BM biopsy imprint smears, BM biopsy sections [hematoxylin & eosin (H & E) stain, reticulin stain]. Special attention was paid to blasts, eosinophils, and basophils. Normal range of NAP score in our laboratory is 36-140. Cytochemistry [myeloperoxidase (MPO), Periodic acid Schiff (PAS) and sudan black B stains], immunocytochemistry and/or flowcytometry techniques were employed for characterization of blast cells in CML-BC patients, wherever indicated.
Cytogenetic analysis: Bone marrow samples, with and without peripheral blood leucocyte cultures (PBLC) were processed and analyzed to identify acquired cytogenetic abnormalities (Ph chromosome and others) in all CML patients, as baseline diagnostic work up. Ph chromosome positivity was expressed as percentage of Ph positive metaphases out of all metaphases analyzed. A minimum of 20 metaphases were analyzed routinely according to ISCN guidelines24.
Qualitative multiplex RT-PCR: Multiplex RT-PCR for BCR/ABL hybrid transcripts was performed according to the previously described methods25,26. Total cellular RNA from peripheral blood mononuclear cells (PBMC) was extracted by Qiagen kits (QIAamp blood mini kit, Germany). RNA was reverse transcribed to make complimentary DNA (cDNA) copies using a commercially available kit (Revert Aid First strand cDNA synthesis kit, MBI Fermentas, European Union). Integrity of cDNA was checked by amplifying β-actin gene. PCR primers specific to the DNA sequences of portions of BCR/ABL were used to amplify the cDNA. The PCR products were separated by agarose gel electrophoresis and visualized by UV transillumination.
The sequences of oligonucleotide primers (Sigma-Aldrich Chemicals Pvt. Ltd., Bangalore, India) used in multiplex RT-PCR for BCR-ABL fusion transcripts as the target gene are as follows:

Co-expression of b3a2 and e19a2, and b2a2 and e19a2 has not been validated by sequencing. Non-parametric Kruskal-Wallis and Mann-Whitney tests were used for statistical analysis.
Results
All 208 CML patients included in this study were positive for BCR-ABL transcripts. CML-CP was diagnosed in 183, CML-AP in 6 and CML-BC in 19 patients (Table I). Among 208 CML patients, b3a2 transcripts were most commonly detected (139, 66.82%) followed by b2a2 (60, 28.84%), b3a2 + b2a2 (7, 3.36%), b3a2 + e19a2 and b2a2 + e19a2 (1 each, 0.48%).
Table I.
Results of cytogenetic & molecular analysis in CML patients (n=208)
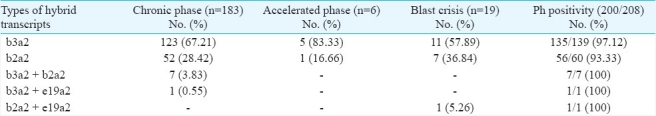
Among 183 CML-CP patients, b3a2 transcripts were most commonly detected (123, 67.21%) followed by b2a2 (52, 28.42%), b3a2 + b2a2 (7, 3.83%) and b3a2 + e19a2 (1, 0.55%). The frequency of b3a2 and b2a2 types of hybrid transcripts was significantly different, among the whole group of 208 patients as well as in CML-CP patients (P<0.0001). The frequency of b3a2 and b2a2 types of hybrid transcripts was not significantly different, among the 6 CML-AP patients as well as in 19 CML-BC patients. Co-expressed hybrid transcripts were detected in 8 of 183 (4.38%) CML-CP patients (b3a2 + b2a2 in 7 and b3a2 + e19a2 in 1) and in 1 of 19 (5.26%) CML-BC patient (b2a2 + e19a2) (Table I).
At the cytogenetic level, 200 (96.15%) patients were positive (Ph positive & BCR-ABL positive CML) and 8 negative for Ph chromosome (Ph negative & BCR-ABL positive CML). All of these Ph negative BCR-ABL positive patients were in CML-CP. Four of the 123 (3.25%) had b3a2 transcripts (UPN 23, 139, 151 and 188) and 4 of 52 (7.69%) had b2a2 transcripts (UPN 23, 64, 162 and 199); the difference in the frequencies was not significant.
Additional cytogenetic abnormalities were detected in 3.8 per cent CP patients and 44 per cent in AP/BC patients. Double Ph chromosome was observed in 4 CP, 2 AP and 3 BC patients; +8 in 4 CP, 1 AP and 2 BC patients; +19 in 2 BC patients. b3a2 and b2a2 types of hybrid transcripts did not show significantly different predilection for additional cytogenetic findings. Rarely encountered co-expressed transcripts b3a2 + b2a2, b3a2 + e19a2 and b2a2 + e19a2 were not associated with additional cytogenetic abnormalities (Table II).
Table II.
Additional cytogenetic findings and BCR-ABL transcripts in CML patients

Median age of the total study group was 38 years (range 13-74 yr). Median age of CML patients with b2a2 transcripts was 42 in CP, 59 in AP and 35 years in BC; and for those with b3a2 transcripts was 37 in CP, 40 in AP and 31 years in BC. Although median age of CML patients with b2a2 transcripts was higher than those with b3a2 transcripts, the difference was not statistically significant.
Male:female ratio was 1.85:1 for CML-CP, 6:1 for CML-AP and 3.75:1 for CML-BC group, with relative over-representation of males in CML-AP and CML-BC groups. There was no significant difference in the sex distribution with respect to b3a2 or b2a2 transcripts in CML-CP and CML-BC patients.
Discussion
Guidelines have been formulated by the WHO to facilitate the diagnosis of CML and various CMPNs/related disorders1. However, a few CMPN cases possess overlapping features. Therefore, the entities which must be differentiated from CML are other CMPNs [(essential thrombocythemia - ET, chronic idiopathic myelofibrosis - CIMF and polycythemia vera - PV); morphological variants (depending upon the predominant granulocyte cell type) e.g. chronic neutrophilic leukaemia (CNL) and chronic eosinophilic leukaemia (CEL)] and myelodysplastic/myeloproliferative neoplasms (chronic myelomonocytic leukaemia - CMML, juvenile myelomonocytic leukaemia - JMML, atypical CML - aCML and unclassifiable myelodysplastic/myeloproliferative neoplasm). Leukemoid reaction always needs to be excluded1–3.
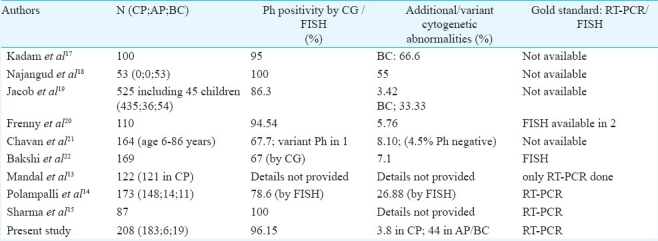
Detection of various types of the transcripts in CML patients has been addressed in a few studies from different geographic locations6–11 and from India13–15. Very few studies have detected co-expressed transcripts and included large number of CML patients (Table III). CML is the commonest leukaemia in adult patients in Asia12, however there are only few studies published encompassing the molecular analyses of adult CML patients in India13–15. Possibly due to cytogenetic analysis being a very time consuming and labour intensive technique, these studies have focused either on RT-PCR alone13 or RT-PCR and FISH techniques only14. RT-PCR along with brief mention of cytogenetic analysis (only Ph positivity) has been reported in one study15.
Table III.
Comparison of Indian studies on frequency of Ph positivity with and without molecular studies
The frequency of b3a2 was reported as 55 per cent and b2a2 as 40 per cent in Caucasian population6. Two other studies found higher frequency of b2a2 transcripts7,8, whereas three reports document the comparison of frequencies of b3a2 and b2a2 transcripts among adult CML patients from western hemisphere9–11. Only one study has included more than 100 CML patients9. Paz-y-Miño et al7 found frequencies of 5.4 per cent for the b3a2 transcripts and 94.6 per cent for the b2a2 transcripts in Ecuadorian Mestizos CML patients.
Rosas-Cabral et al8 detected b3a2 BCR-ABL transcripts in 28 per cent cases, b2a2 in 59 per cent cases, and 13 per cent with both b3a2/b2a2 transcripts among 97 Philadelphia-positive CML Mexican cases. Ruiz-Argüelles et al9 studied a group of 238 Mexican Mestizos patients with Ph positive CML; 54.2 per cent showed b3a2 subtype, 43.2 per cent b2a2 and 2.5 per cent b3a2/b2a2.
de Lemos et al10 from Brazil performed RT-PCR for BCR-ABL in 22 CML patients. Of these patients, 15 (68%) were in chronic phase, five (23%) in accelerated phase and two (9%) in blastic phase; 59 per cent patients had b3a2 and 41 per cent had b2a2 transcripts.
Yaghmaie et al11 studied 75 adult Iranian CML patients; 83 per cent patients expressed one of the p210 BCR-ABL transcripts (b3a2, 63% and b2a2, 20%), while the remaining showed one of the transcripts of b3a3, b2a3, e1a2 or co-expression of b3a2 and b2a2. b3a2 and b2a2 were co-expressed in 5 per cent patients.
Mondal et al13 studied 122 CML patients and details of cytogenetic analysis are not provided. Out of 122, 112 (91.8%) patients were positive for one or more of the four junctional types tested.
Polampalli et al14 studied 202 CML patients; 138 (68%) had the b3a2 type BCR-ABL transcript and 64 (32%) had the b2a2 type. On comparing the RT-PCR with FISH in 186 patients, t(9;22) was detected in 78.6 per cent patients. FISH technique also found additional/variant cytogenetic abnormalities in 26.88 per cent patients. Sharma et al15 studied 87 CML-CP patients: b3a2 BCR-ABL transcripts were detected in 54 per cent, b2a2 transcripts in 38 per cent and b3a2 + b2a2 transcripts in 8 per cent patients. These authors mentioned 100 per cent Ph positive metaphases in the ‘pre-treated’ group of 57 patients (Table IV).
Table IV.
Comparison of frequency of BCR-ABL transcripts types in recent studies
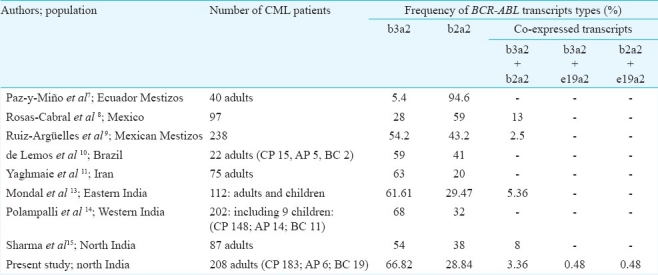
In our study b3a2 transcripts were most commonly detected (66.82%) among 208 CML patients, followed by b2a2 (28.84%). Similar was the case with 183 CML-CP patients; b3a2 transcripts were most commonly detected (67.21%) followed by b2a2 (28.42%). Co-expression of at least two types of hybrid transcripts was detected in 4.33 per cent patients: 7 CML-CP patients had b3a2 + b2a2 transcripts, 1 CML-CP patients had b3a2 + e19a2 transcripts, and 1 CML-BC patient had b2a2 + e19a2 transcripts. Latter two transcript types have not been detected in the earlier studies. Low expression of e1a2 (p190) transcripts may be found with classical b3a2/b2a2 (p210) transcripts in more than 90 per cent CML patients1, however, neither the aforementioned studies6–11,14,15, nor we detected e1a2 (p190) transcripts. These could have been detected by our PCR protocol. Co-expression of b3a2 and e19a2, and b2a2 and e19a2 was not validated by sequencing. All the studies6–11,14,15, except one13, have not performed sequencing for this purpose.
Association of b2a2 with younger age has been suggested by Mondal et al13 (without reaching statistical significance level). Median age of our CML patients with b2a2 transcripts was higher than those with b3a2 transcripts, though there was overlap in the range and the difference was not statistically significant. No sex predilection was found with regard to b3a2 versus b2a2 transcripts.
Cytogenetic study of CML patients was reported from our department in 197816. Table III shows studies published 1990s onwards, with Ph positivity being reported in 67-95 per cent Indian CML patients17–22. This leaves a lot of ‘CML’ patients whose diagnosis remains uncertain/questionable, according to the WHO criteria1. Therefore, it is important to have the gold standard molecular testing (FISH or RT-PCR) for comparison to ascertain the true frequency of Ph positivity or negativity in CML patients.
Ph positivity in our patients was higher compared to many Indian studies14,17,19–22. Additional cytogenetic abnormalities frequently associated with disease evolution include +Ph, +8, +19 and i(17q)1–3. We document here that the additional cytogenetic abnormalities are not associated with any particular BCR-ABL transcript type.
Cytogenetic and molecular analysis has become mandatory in order to make a correct diagnosis and predict/monitor response to newer molecular targeted treatment modalities like imatinib mesylate. Cytogenetic study and qualitative RT-PCR have prime importance at the time of diagnosis, whereas serial quantitative RQ-PCR is recommended for therapeutic monitoring4,5.
Diagnostic criteria of various disorders are evolving very rapidly and it is difficult to keep pace with the developments, especially in the developing countries. With the application of standard criteria, definite diagnosis of CML can be made most easily, as compared to other CMPDs1. Therefore, it is imperative to document the features of the diseases as completely as possible, for analysis of various features at the time of diagnosis.
References
- 1.Vardiman JW, Melo JV, Baccarani M, Thiele J. Myeloproliferative Neoplasms. In: Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, Thiele J, Vardiman JW, editors. WHO classification of tumours of haematopoietic and lymphoid tissues. Lyon: IARC Press; 2008. pp. 32–37. [Google Scholar]
- 2.Rabinowitz I, Larson RS. Chronic myeloid leukemia. In: Greer JP, Foerster J, Lukens JN, Rodgers GM, Paraskevas F, Glader B, editors. Wintrobe's clinical hematology. 11th ed. Philadelphia: Lippincott Williams & Wilkins; 2004. pp. 2235–58. [Google Scholar]
- 3.Lichtman MA, Lieveld JL. Chronic myelogenous leukemia and related disorders. In: Beutler E, Lichtman MA, Coller BS, Kipps TJ, editors. Williams hematology. 6th ed. New York: McGraw-Hill; 2001. pp. 1125–36. [Google Scholar]
- 4.Deininger MW. Milestones and monitoring in patients with CML treated with imatinib. Soc Hematol Educ Program. 2008:419–26. doi: 10.1182/asheducation-2008.1.419. [DOI] [PubMed] [Google Scholar]
- 5.Ou J, Vergilio JA, Bagg A. Molecular diagnosis and monitoring in the clinical management of patients with chronic myelogenous leukemia treated with tyrosine kinase inhibitors. Am J Hematol. 2008;83:296–302. doi: 10.1002/ajh.21096. [DOI] [PubMed] [Google Scholar]
- 6.Melo JV. The diversity of BCR-ABL fusion proteins and their relationship to leukemia phenotype. Blood. 1996;88:2375–84. [PubMed] [Google Scholar]
- 7.Paz-y-Miño C, Burgos R, Morillo SA, Santos JC, Fiallo BF, Leone PE. BCR-ABL rearrangement frequencies in chronic myeloid leukemia and acute lymphoblastic leukemia in Ecuador, South America. Cancer Genet Cytogenet. 2002;132:65–7. doi: 10.1016/s0165-4608(01)00515-5. [DOI] [PubMed] [Google Scholar]
- 8.Rosas-Cabral A, Martínez-Mancilla M, Ayala-Sánchez M, Vela-Ojeda J, Bahena-Reséndiz P, Vadillo-Buenfil M, et al. Analysis of Bcr-abl type transcript and its relationship with platelet count in Mexican patients with chronic myeloid leukemia. Gac Med Mex. 2003;139:553–9. [PubMed] [Google Scholar]
- 9.Ruiz-Argüelles GJ, Garcés-Eisele J, Reyes-Núñez V, Ruiz-Delgado GJ. Frequencies of the breakpoint cluster region types of the BCR/ABL fusion gene in Mexican Mestizo patients with chronic myelogenous leukemia. Rev Invest Clin. 2004;56:605–8. [PubMed] [Google Scholar]
- 10.de Lemos JA, de Oliveira CM, Scerni AC, Bentes AQ, Beltrão AC, Bentes IR, et al. Differential molecular response of the transcripts B2A2 and B3A2 to imatinib mesylate in chronic myeloid leukemia. Genet Mol Res. 2005;4:803–11. [PubMed] [Google Scholar]
- 11.Yaghmaie M, Ghaffari SH, Ghavamzadeh A, Alimoghaddam K, Jahani M, Mousavi SA, et al. Frequency of BCR-ABL fusion transcripts in Iranian patients with chronic myeloid leukemia. Arch Iran Med. 2008;11:247–51. [PubMed] [Google Scholar]
- 12.Ries LAG, Harkins D, Krapcho M. SEER cancer statistics review, 1975-2003. Bethesda: National Cancer Institute; 2006. [Google Scholar]
- 13.Mondal BC, Bandyopadhyay A, Majumdar S, Mukhopadhyay A, Chandra S, Chaudhuri U, et al. Molecular profiling of chronic myeloid leukemia in eastern India. Am J Hematol. 2006;81:845–9. doi: 10.1002/ajh.20682. [DOI] [PubMed] [Google Scholar]
- 14.Polampalli S, Choughule A, Negi N, Shinde S, Baisane C, Amre P, et al. Analysis and comparison of clinicohematological parameters and molecular and cytogenetic response of two Bcr/Abl fusion transcripts. Genet Mol Res. 2008;7:1138–49. doi: 10.4238/vol7-4gmr485. [DOI] [PubMed] [Google Scholar]
- 15.Sharma P, Kumar L, Mohanty S, Kochupillai V. Response to Imatinib mesylate in chronic myeloid leukemia patients with variant BCR-ABL fusion transcripts. Ann Hematol. 2010;89:241–7. doi: 10.1007/s00277-009-0822-7. [DOI] [PubMed] [Google Scholar]
- 16.Das KC, Nayak J, Mohanty D, Garewal G. Cytology and cytogenetics in chronic myelogenous leukaemias. Indian J Med Res. 1978;68:148–63. [PubMed] [Google Scholar]
- 17.Kadam PR, Nanjangud GJ, Advani SH, Nair C, Banavali S, Gopal R, et al. Chromosomal characteristics of chronic and blastic phase of chronic myeloid leukemia. A study of 100 patients in India. Cancer Genet Cytogenet. 1991;51:167–81. doi: 10.1016/0165-4608(91)90129-i. [DOI] [PubMed] [Google Scholar]
- 18.Nanjangud G, Kadam PR, Saikia T, Bhisey AN, Kumar A, Gopal R, et al. Karyotypic findings as an independent prognostic marker in chronic myeloid leukaemia blast crisis. Leuk Res. 1994;18:385–92. doi: 10.1016/0145-2126(94)90023-x. [DOI] [PubMed] [Google Scholar]
- 19.Jacob RT, Gayatri K, Surath A, Rao DR. Cytogenetic profile of chronic myeloid leukemias. Indian J Cancer. 2002;39:61–5. [PubMed] [Google Scholar]
- 20.Vinsheth FJ, Sheth JJ, Patel AI, Shah AD, Verhest A. Usefulness of cytogenetics in leukemias. Indian J Cancer. 2002;39:139–42. [PubMed] [Google Scholar]
- 21.Chavan D, Ahmad F, Iyer P, Dalvi R, Kulkarni A, Mandava S, et al. Cytogenetic investigation in chronic myeloid leukemia: study from an Indian population. Asian Pac J Cancer Prev. 2006;7:423–6. [PubMed] [Google Scholar]
- 22.Bakshi SR, Brahmbhatt MM, Trivedi PJ, Shukla SN, Shah PM. A typical D-FISH patterns of BCR/ABL gene rearrangements in 169 chronic myeloid leukemia patients. J Assoc Genet Technol. 2006;32:164–7. [PubMed] [Google Scholar]
- 23.Au WY, Caguioa PB, Chuah C, Hsu SC, Jootar S, Kim DW, et al. Chronic myeloid leukemia in Asia. Int J Hematol. 2009;89:14–23. doi: 10.1007/s12185-008-0230-0. [DOI] [PubMed] [Google Scholar]
- 24.Shaffer LG, Tommerup N, editors. Basel, Switzerland: S Karger; 2005. ISCN (2005): An International System for Human Cytogenetic Nomenclature. [Google Scholar]
- 25.Jones CD, Yeung C, Zehnder JL. Comprehensive validation of a real-time quantitative bcr-abl assay for clinical laboratory use. Am J Clin Pathol. 2003;120:42–8. doi: 10.1309/60A9-C8WG-EGHR-NXEE. [DOI] [PubMed] [Google Scholar]
- 26.Cioc AM, Nuovo GJ. Expression of μ-BCR-ADL transcripts in chronic neutrophilic leukemia. Am J Clin Pathol. 2002;118:842–7. doi: 10.1309/GKCH-K9CW-R6J7-JR4T. [DOI] [PubMed] [Google Scholar]


