Abstract
Cerebral amyloid β (Aβ) accumulation is pathogenically associated with sporadic Alzheimer’s disease (SAD). BACE-1 is involved in Aβ generation while insulin-degrading enzyme (IDE) partakes in Aβ proteolytic clearance. Vulnerable regions in AD brains show increased BACE-1 protein levels and enzymatic activity while the opposite occurs with IDE. Another common feature in SAD brains is Notch1 overexpression. Here we demonstrate an increase in mRNA levels of Hey-1, a Notch target gene, and a decrease of IDE transcripts in the hippocampus of SAD brains as compared to controls. Transient transfection of Notch intracellular domain (NICD) in N2aSW cells, mouse neuroblastoma cells (N2a) stably expressing human amyloid precursor protein (APP) Swedish mutation, reduce IDE mRNA levels, promoting extracellular Aβ accumulation. Also, NICD, HES-1 and Hey-1 overexpression result in decreased IDE proximal promoter activity. This effect was mediated by 2 functional sites located at −379/−372 and −310 −303 from the first translation start site in the −575/−19 (556 bp) fragment of IDE proximal promoter. By site-directed mutagenesis of the IDE promoter region we reverted the inhibitory effect mediated by NICD transfection suggesting that these sites are indeed responsible for the Notch-mediated inhibition of the IDE gene expression. Intracranial injection of the Notch ligand JAG-1 in Tg2576 mice, expressing the Swedish mutation in human APP, induced overexpression of HES-1 and Hey-1 and reduction of IDE mRNA levels, respectively. Our results support our theory that a Notch-dependent IDE transcriptional modulation may impact on Aβ metabolism providing a functional link between Notch signaling and the amyloidogenic pathway in SAD.
Keywords: Amyloid β metabolism, Notch signaling, HES, Hey, Transcriptional repressors, IDE gene promoter
1. Introduction
In the rare familial Alzheimer’s disease (FAD) cases, mutations in the amyloid precursor protein (APP) or presenilin genes result in increased amyloid β (Aβ) concentration in the brain [1]. Yet, the mechanisms of Aβ accumulation in sporadic AD (SAD) remain largely unknown. Notably, Down Syndrome (DS) patients, who harbor a chromosome 21 trisomy, almost invariably exhibit Aβ accumulation in senile plaques at adulthood. The fact that these patients carry an extra APP gene copy may be a plausible explanation for the observed AD-like brain pathology [2]. However, recent work has shown that APP was not over-expressed in a cohort of adult DS brains as assessed by microarray QPCR [3] whereas, as expected, a subset of chromosome 21 genes was found to be up-regulated. The lack of APP over-expression suggests that post-translational disturbances in APP processing, trafficking or Aβ metabolism may be more relevant than the levels of APP to amyloid deposition in DS brain. In addition, the brain of adult DS patients showed up-regulation of several genes involved in developmental processes including components of the Notch signaling pathway. This observation was in agreement with previous works indicating an increased Notch1 immunoreactivity in the cerebellum and in the hippocampal formation of SAD brain as compared to age-matched controls with a strong signal in neurons of CA4, CA3 and CA2 fields and a weaker staining in the dentate gyrus. In that report, neither neurofibrillary tangles, senile plaques, astrocytes nor microglial cells were positive for Notch1 labeling [4]. Taken together, these evidences raise the possibility that Notch activation is a common feature of AD and DS with pathogenic implications.
Notch1 is a single-pass type I transmembrane receptor that is critical during development through the spatial and temporal regulation of cell proliferation, fate specification and differentiation in multiple tissues and organs [5]. In adult brain, Notch signaling pathway has been involved in neurogenesis, regulation of neurite growth, neuronal plasticity and long-term memory [5–7]. Activation of the mammalian Notch pathway occurs when a specific ligand Delta/Jagged binds to Notch extracellular domain. Sequential proteolytic events result in a γ-secretase-mediated release of a Notch intracellular domain (NICD). Then, NICD translocates to the cell nucleus and elicits expression of two independent primary target genes, HES and Hey, which are members of the bHLH family of transcriptional repressors [8]. Each works either individually or cooperatively to repress target gene expression through its specific DNA-binding sites [9].
Aβ peptides are generated and released after a sequential proteolytic processing of APP by β- and γ-secretases [10]. The first cleavage is mediated by β-secretase (BACE-1), the rate-limiting step in Aβ generation. Interestingly, BACE-1 protein levels and enzymatic activity are increased in AD brains as compared to age-matched controls [11], suggesting that BACE-1 may participate in AD pathogenesis by accelerating the rate of Aβ production. In addition, Aβ concentration in the brain is dependent upon its bi-directional transport across the blood–brain barrier and its proteolytic degradation in situ.
The major peptidases that participate in the degradation of Aβ include the M13 zinc metallopeptidases, neprilysin (NEP) and its homologue endothelin-converting enzyme (ECE-1) and the M16 family member, insulin-degrading enzyme (IDE). Observations in human postmortem tissue suggest that hippocampal NEP, ECE-1 and IDE expression and/or activity are inversely related to the extent of AD pathology [12–17]. In addition, studies that focused on IDE in the cortex rather than hippocampus, showed different results in aging and AD [18] [19]. However, the mechanisms underlying the transcriptional inhibition of Aβ-degrading proteases in SAD hippocampus remain mostly unknown. To address this issue we have assessed the role of Notch signaling on the endogenous transcript levels of IDE, the most relevant extracellular protease involved in Aβ removal, which harbor putative HES/Hey-1 binding sites in its promoter region.
In the present study, we demonstrate by several approaches that, indeed, Notch signaling participates in the transcriptional regulation of IDE in vitro and in vivo with impact on Aβ metabolism providing a novel functional link between Notch activation and the amyloidogenic pathway in SAD.
2. Materials and methods
2.1. In silico promoter analysis
Genomic sequence of the 4799 bp corresponding to the promoter of the human IDE gene [20] (−4799/−18) up stream of the first ATG) was obtained from the GenBank database (accession number: NG 013012). Three different programs were run to detect putative Notch target genes consensus binding sites as follows: TESS (Transcription Element Search System), TF search software (www.cbrc.jp/research/db/TFSEARCH.html) and Regulatory Sequence Analysis Tool (RSAT) software (http://embnet.ccg.unam-mx/rsa-tools/). See Table 1 for classification, consensus and position of each site in human IDE promoter.
Table 1.
Summary of biological properties of the putative consensus binding sites of Notch target genes in human IDE promoter.
| Classification | Position* | Orientation | Consensus | bHLH protein | DNA binding activity• |
|---|---|---|---|---|---|
| N-box | −3711/−3715 | Foward (+) | 5′-CACNAG-3′ | HES-1 | ++ |
| −310/−305 | Reverse (−) | Hey-1 | + | ||
| Class C | −379/−374 | Reverse (−) | 5′-CACGNG-3′ | Hey-1 | ++ |
| E-box | −467/−462 | Forward (+) | 5′-CANNTG-3′ | HES-2 | ++ |
| −431/−425 | Foward/reverse (+)/(−) | ||||
| Class B | −308/−303 | Foward (+) | 5′-CANGTG-3′ | HES-1 | + |
| Hey-1 | ++ |
, From the first translation start site (ATG). Class C and N-box are mutually overlapping. Class B is a subtype of E-box.
, The data are based on gel mobility shift assay as reported [9]. ++, strong binding; +, weak binding.
2.2. Reagents
The γ-secretase inhibitor (GSI) IX (L-685,458) was purchased from Calbiochem (Darmstadt, Germany). The 17-mer JAG-1 peptide (CDDYYYGFGCNKFCRPR) was synthesized by Anaspec (Fremont, USA). Peptide was dissolved in DMSO (for cell culture experiments) or PBS (for in vivo experiments) at 50 mM, aliquoted, and stored at −20 °C.
2.3. Antibodies
BC2 rabbit polyclonal and 1C1 and 3A2 monoclonal antibodies anti-IDE were generated in our laboratory [21]. Rabbit anti-NICD was purchased from Cell Signaling (Danvers, USA). This antibody targets the cleaved Val1754 of the Notch molecule. Polyclonal antibodies anti-heterochromatin protein-1 (HP-1) and tubulin were from Sigma (St. Louis, USA). Anti-HA antibody was purchased from Invitrogen (Carlsbad, USA).
2.4. Human tissues
Frozen samples of hippocampus from late-onset SAD Braak stage V (n=5) and age-matched controls (NDC, n=5) were kindly provided by the Harvard Brain Tissue Resource Center (Boston, USA). See Table 2 for clinico-pathological features of each group. Frozen tissues were homogenized as previously described [22] and nuclear fractions confirmed by immunoblotting for HP1.
Table 2.
Summary of clinico-pathological features of the autopsied cases of demented and control groups.
| Group | Number of cases | Sex (M:F) | PMD (hours) | AAD• (years) |
|---|---|---|---|---|
| SAD (Braak V) | 5 | 2:3 | 22.80±1.44 | 78.2±6.13 |
| NDC | 5 | 5:0 | 25.29±1.15 | 67.0±3.0 |
AAD, Age at death;
, no statistical differences were detected between SAD and NDC; F, female; M, male; PMD, post mortem delay; SAD, sporadic Alzheimer’s disease; NDC, non-demented control
2.5. cDNA constructs
HA-tagged Notch Intracellular Domain (NICD) cDNA was kindly provided by Dr. Bettina Kempkes (German Research Center for Environmental Health, Munich, Germany). A myc-tagged mouse Notch construct lacking the extracellular domain (ΔE-Notch) was kindly provided by Dr. Raphael Kopan (Washington University in St. Louis, USA) [23]. HA-tagged Hey-1 construct was a kind gift from Dr. Manfred Gessler (University of Wuerzburg Würzburg, Germany) and the FLAG-tagged HES-1 construct was kindly provided by Dr. Stafano Stifani (McGill University, Montreal, Canada). The region of 4799 bp 5′ upstream of human IDE gene (−4825/−26) cloned into the pGL3-Basic vector (Promega, Madison, USA) was kindly provided by Dr. Wesley Farris (Pittsburgh University, Pennsylvania, USA).
2.6. RNA extraction and quantitative real-time polymerase chain reaction (QRT-PCR)
Total RNA was extracted from cells using RNAspin Mini kit (GE Bioscience, Piscataway, USA) and from tissues using RNeasy Lipid Tissue Mini Kit (Qiagen, Hilden, Germany). SuperScript II Reverse transcriptase (Invitrogen, Carlsbad, USA) was used to synthesize first strand cDNA according to the manufacturer’s instructions. Synthesized cDNA was used in independent PCR reactions containing 6 mM MgCl2, 0.4 mM dNTP mixture, 200 nM each primer, 1 U Platinum Taq Polymerase (Invitrogen. Carlsbad, USA) SybrGreen and ROX reference dye in appropriate buffer conditions. Reactions were run in an Mx3005P Cycler (Stratagene, Santa Clara, USA). Data were analyzed using MxPRO-Mx3005P software. Primers used for IDE amplification were: F: 5′-AAAGAAACTCTCTGCAGA-3′ and R: 5′-TTATGAATCACCTCAGGT-3′; for Hey-1 amplification: Forward: 5′-TGAGCTGAGAAGGCTGGTAC-3′ and R: 5′-ACCCCAAACTCCGATAGTCC-3′; for BACE-1 amplification: F: 5′-GATGGTGGACAACCTGAG-3′ and R: 5′-CTGGTAGTAGCGATGCAG-3′. Primers used for TATA-Binding-Protein (TBP) amplification were: F: 5′-ACCGTGAATCTTGGCTGTAA-3′ and R: 5′-CCGTGGCTCTCTTATTCTCA-3′ and for HPRT were: F: 5′-TGGGAGGCCATCACATTGTAG-3′ and R: 5′-TGTCCCCTGTTGACTGGTCATT-3′. TBP and HPRT were used as normalizing genes.
2.7. Cell cultures, transient transfection and treatments
N2aSW (N2a mouse neuroblastoma cells stably expressing human APP Swedish mutation [24] kindly provided by Dr. Gopal Tinakaran from University of Chicago, USA) were maintained as previously described [25]. Cells were plated at 70–80% confluency 16 h prior to transient transfection using Lipofectamine LTX (Invitrogen, Carlsbad, USA) according to the manufacturer’s instructions. Cells were harvested 24 h after transfection. The γ-secretase inhibitor was used at a final concentration of 20 μM. Cells were incubated with 100 μM JAG-1 peptide for 48 h as previously reported [26].
2.8. SDS-PAGE and Western blots
Aliquots of 50 μg of nuclear fractions, 100 μg of cytosolic fractions or 50 μg of cell homogenates were subjected to electrophoresis on 7.5% SDS-Tris-Tricine polyacrylamide mini-gels, and transferred to polyvinylidene fluoride (PVDF)-membranes (GE Bioscience, Piscataway, USA). Immunoreactivity was detected with HRP-coupled anti-mouse or anti-rabbit IgG and ECL Plus (GE Bioscience, Piscataway, USA). Immunoblots were scanned with STORM 840 and analyzed with ImageQuant 5.1 software (GE Bioscience, Piscataway, USA).
2.9. Luciferase reporter assay
The fragment of 556 bp (−576/−19) upstream of the first translational start site was amplified by PCR using as template the 5′ upstream region of human IDE gene and the following forward (F) and reverse (R) primers: F-5′-GTATGGTACCCACGCCGCACTCGA-3′ and R-5′-GACTAAGCTTGATCACCGCAAACGCT-3′. For directional cloning the restriction enzyme recognition sites KpnI and HindIII were introduced in the F and R primers, respectively. PCR products were cloned into the pGL3-basic (Promega, Madison, USA). The correct sequence and direction was confirmed by sequencing (Macrogen, Seoul, Korea). To generate mutant versions of the 556 bp construct site-directed mutagenesis was performed for the −431/−425 E-box, the −379/−372 Class C1 and the −310/−303 N-box/Class B sites, respectively, using the following set of primers: E-box (Mut1): 5′-CACGAACCTTGTTCCAATGCCTACAGGGTCCGGCACGCT-3′; Class C1 box (Mut2) 5′-GCCTCAGAAAACATAGCTCCGCATGCCCTCACTGGATTTA-3′ and N-box/Class B (Mut3): 5′-ACACGTCGCCACCCTCATGAGTACGGCCGGTGGGATCTA-3′ and their corresponding complementary primers. These primers were used in separate temperature-cycling reactions using the 556 bp of IDE promoter construct as template and Pfx DNA polymerase (Invitrogen). Then, the products were treated with DpnI endonuclease to digest parental DNA template and subsequently introduced into DH5α cells. Plasmid extraction was performed and mutations confirmed by automated sequencing (Macrogen, Seoul, Korea). To generate a deleted version of the 556 bp fragment containing only the N-box/Class B sites a fragment of 354 bp was amplified by PCR using the following forward and reverse primers: F-5′-GTATGGTACC TGCCCTCACTGGATT-3′and R-5′ GACTAAGCTTGATCACCGCAAACGCT-3′. The restriction enzyme recognition sites KpnI and HindIII were introduced in the F and R primers, respectively. PCR products were cloned into the pGL3-basic (Promega, Madison, USA). The correct sequence and direction was confirmed by sequencing (Macrogen, Seoul, Korea). The generated plasmids were designated pGL3-hIDE-556; pGL3-hIDE-Mut1; pGL3-hIDE-Mut2; pGL3-hIDE-Mut3 and pGL3-hIDE-354.
N2aSW cells (4×104) were plated in a 24-well plate 16 h prior to co-transfection using Lipofectamine LTX (Invitrogen, Carlsbad, USA) with 200 ng of the indicated reporter constructs or equimolar amount of empty vector pGL3-Basic and HA-NICD or HA-Hey-1 or FLAG-HES-1 cDNAs, respectively. To control transfection efficiency pRL-TK-Renilla Luciferase plasmid (Promega, UK) was also included in transfections. Forty-eight hours later, cells were lysed using Dual-Luciferase Reporter Assay System (Promega, Madison, USA) and luminescence measured in a Tecan Genius (Männedorf, Switzerland). Values for each experimental condition were calculated as a ratio of luminescence obtained for firefly/renilla. Results are mean values of 4 independent transfected wells performed by duplicate and are expressed as percentage relative to control (100%).
2.10. Quantification of extracellular Aβ levels
Conditioned media were collected 24 h after transfection, spun at 1000×g for 5 min to pellet cell debris and diluted 1:100. Aβ40 levels were quantified by using a commercial ELISA kit (Covance, Princeton, USA). Data for each set duplicate transfection was averaged, the control in each set normalized to 100% and data presented as percentage of control.
2.11. IDE and BACE-1 enzymatic activity assay
IDE was immunoprecipitated from conditioned supernatant or hippocampal cytosolic fractions with anti-IDE monoclonal antibodies as described [27] and proteolytic activity was performed by using 125I-insulin as substrate as previously reported [21]. Activity assays for BACE-1 were performed by using synthetic peptide substrate (MCA-Ser-Glu-Val-Asn-Leu-Asp-Ala-Glu-Phe-Arg-Lys-(DNP)-Arg-Arg-NH2) from R&D Systems (Minneapolis, USA) and crude extracts following procedures previously described [11].
2.12. Animals and surgical procedures
A total of 6 mice, ages ranging from 8 to 12 months weighing between 25 and 30 g, were maintained under controlled temperature and light with water and food ad libitum. All animal procedures were performed according to the rules, standards and the regulations of the National Institute of Health, USA. Animal experiments were approved by the Institutional Commission for the Care and Use of the Laboratory Animals of Fundación Instituto Leloir. The stereotaxic coordinates for injection into the hippocampus were antero posterior 2.1 mm from bregma; lateral 1.3 mm; ventral: 1.3 mm. For injections, mice were anesthetized with 100 mg/kg ketamine and 10 mg/kg xylazine intraperitoneally. The skull was immobilized in a stereotaxic apparatus (Stoelting, Illinois, USA). A hole was drilled in the cranium by circular movements with a hand-held drill (Drimer). Mice were injected with 2 μg of JAG-1 (in 1 μl PBS) or vehicle (PBS), using a 50 μm finely stretched glass capillary (Dumont). Animals were sacrificed 3 days post injection.
2.13. Statistical analysis
All experiments were performed in triplicate and values presented as means±SEM. Data were analyzed by Student’s t test, one-way or two-way ANOVA followed by post-hoc Bonferroni tests for comparison among means using GraphPad Prism 3.0 software. Statistical significance was set at p<0.05.
3. Results
3.1. Notch activation is increased in hippocampal formation of SAD patients
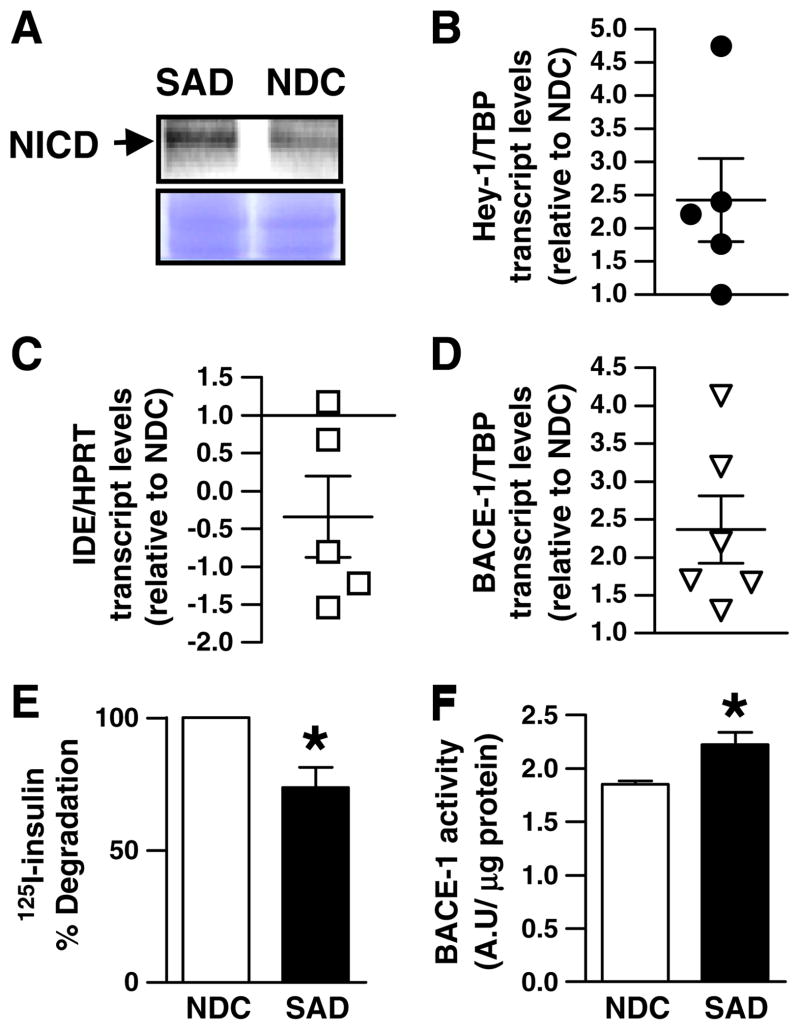
To assess Notch activation in SAD brains, nuclei from frozen hippocampal samples of SAD and non demented control (NDC) were isolated. In nuclear fractions we detected an increase of ~40% in NICD immunoreactivity in SAD as compared to NDC (Fig. 1A, left upper panel). In addition, we isolated total RNA from each hippocampal sample and by QRT-PCR we found an ~2.5-fold increase (2.43± 0.63) in the mRNA of Hey-1, a downstream Notch target gene (Fig. 1A, right panel). In an effort to correlate Notch activation with the gene transcription of relevant enzymes involved in the regulation of Aβ metabolism in SAD brain, we examined IDE and BACE-1 mRNA levels. In accordance with previous reports [11,16] we detected an ~1.5-fold decrease (−0.340±0.54) and an ~2.5-fold increase (2.36± 0.44) in IDE (Fig. 1B, left panel) and BACE-1 (Fig. 1C, left panel) transcript levels, respectively, in SAD as compared to NDC. In addition, human hippocampal homogenates with increased Notch activation were analyzed for IDE and BACE-1 activities. Concurrent with published findings [28], by combining 125I-insulin degradation and trichloroacetic acid (TCA) precipitation, a significant reduction of IDE proteolytic activity (~26%) was found in SAD compared to NDC (Fig. 1B, right panel). BACE-1 activity, as assessed with a fluorogenic peptide substrate, was increased (~20%) in the hippocampal homogenates of SAD as compared to NDC, in agreement with previous reports [11] (Fig. 1C, right panel).
Fig. 1.
Notch pathway activation, IDE and BACE-1 transcript levels and activity in SAD brains. (A) Upper panel, representative Western blot of the expression of NICD protein in nuclear extracts of SAD and control (NDC) hippocampal samples normalized by protein content. Arrow indicates NICD fragment. Lower panel, Coomassie blue staining of the PVDF membrane corresponding to the Western blot described above. (B); (C) and (D) Hey-1, IDE and BACE-1 mRNA levels in SAD hippocampus (n=5). Each point represents the mean value of at least 3 independent experiments performed by triplicate for each sample. The mean±SEM in SAD relative to NDC cases are indicated. (E) Percentage of 125I-insulin degraded by endogenous IDE in NDC and SAD samples as determined by TCA precipitation. (F) BACE-1 activity in NDC and SAD hippocampal extracts. Bars represent the mean±SEM. of four independent experiments performed in triplicate; *p<0.05 compared with NDC. Note that Notch activation, IDE and BACE-1 activities were obtained from the same hippocampal region.
3.2. Notch signaling pathway activation impacts on Aβ metabolism
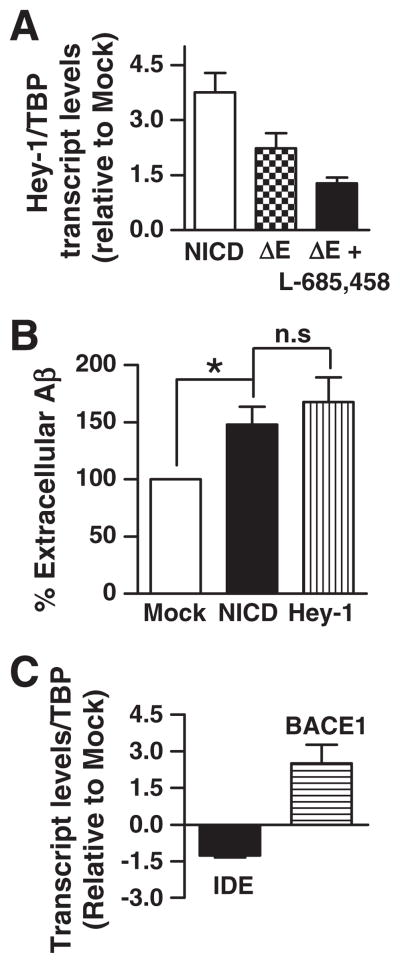
As a first attempt to analyze the effect of Notch signaling activation on Aβ metabolism, a mouse neuroblastoma cell line expressing APP with the Swedish mutation (N2aSW) was transiently transfected with NICD or a constitutively activated form of Notch that is a substrate of γ-secretase, ΔE-Notch. As expected, a 4-fold increase (3.75±0.50; n=7) in Hey-1 mRNA levels were observed in NICD as compared to mock-transfected cells. Transfection with ΔE-Notch resulted in a 2-fold-increase (2.23±0.40; n=3) of Hey-1 transcripts as compared to controls and this effect was partially reverted (1.27±0.16) by the addition of L-685,458, a selective γ-secretase inhibitor known to inhibit Notch signaling in vitro (Fig. 2A). Importantly, in N2aSW cells over-expressing Hey-1 or NICD extracellular steady-state Aβ levels were increased ~48% (148±15.4; n=4; p<0.05) and ~68% (167.5±21.7; n=4; p<0.05), respectively, as compared to mock-transfected cells (100%) (Fig. 2B) suggesting a direct impact of Notch signaling on Aβ production and/or degradation.
Fig. 2.
Effect of Notch signaling pathway activation on extracellular Aβ and on IDE and BACE-1 transcripts levels. (A). Expression of Hey-1 mRNA in N2aSW cells transfected with NICD, or ΔE-Notch-construct in the presence and absence of γ-secretase inhibitor L-685,458. Bars indicate the mean value±SEM of at least 3 independent experiments performed in triplicate in NICD or ΔE-Notch-construct relative to mock-transfected cells. (B) Analysis of Aβ40 in the supernatants of cells transfected with empty vector (mock), NICD, or Hey-1 cDNAs, respectively. Bars indicate the mean value±SEM; *p<0.05 compared with mock-transfected cells; n.s., not significant differences in ex-tracellular Aβ levels between NICD and Hey-1 N2aSW transfected cells. (C) IDE and BACE-1 mRNA levels in cells transfected with NICD construct. Bars indicate the mean value±SEM of at least 3 independent experiments performed in triplicate in NICD construct relative to mock-transfected cells.
3.3. Notch signaling pathway activation impacts on endogenous IDE and BACE-1 mRNA levels
To test if Notch signaling activation may modulate mRNA levels of proteases involved on Aβ metabolism N2aSW cells were transfected with NICD cDNA and mRNA levels of IDE and BACE-1 determined by QRT-PCR. As expected, a 4-fold-increase (4.18±0.45) in Hey-1 mRNA was observed (not shown). Interestingly, a decrease of ~1.5 fold (−1.260±0.07, n=4) on endogenous IDE mRNA levels was confirmed and a 3-fold increase (2.5±0.8; n=3) of endogenous BACE-1 transcripts was detected (Fig. 2C).
3.4. Notch signaling pathway activation impacts on IDE and BACE-1 proteolytic activity
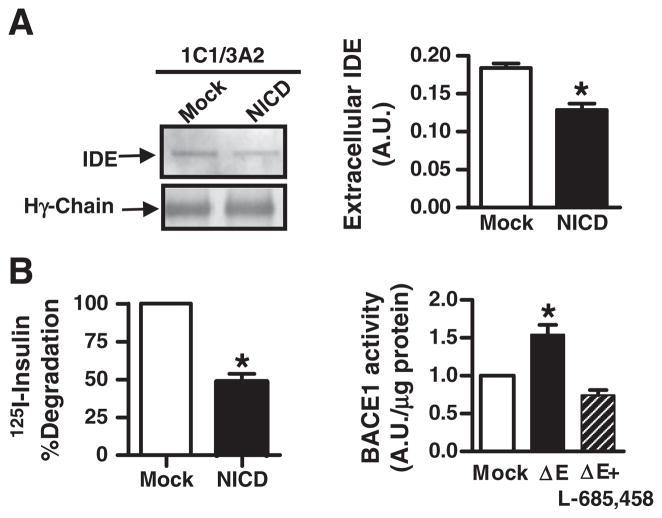
To determine if reduction of IDE and increments of BACE-1 mRNA levels were reflected in IDE and BACE-1 enzymatic activity, IDE was immunoprecipitated from conditioned media of mock- or NICD-transfected N2aSW cells (Fig. 3A, left panel) and a significant reduction in extracellular IDE protein content was detected (Fig. 3A, right panel) (0.18 vs. 0.12; p<0.05) in accordance with a significant reduction of IDE proteolytic activity (48.5%±5.17; p<0.05), as compared to mock-transfected cells (Fig. 3B, left panel). Moreover, a significant increase in the percentage of BACE-1 proteolytic activity was observed in ΔE-Notch as compared to mock-transfected N2aSW cells (154%±0.12) which was reverted by L-685,458 treatment (0.74%±0.60) (Fig. 3B, right panel).
Fig. 3.
Effect of Notch signaling pathway activation on the proteolytic activity of IDE and BACE-1 (A) Left upper panel, Western blot with BC2 of 1C1/3A2 ipp from supernatants of mock-and NICD- N2aSW transfected cells. Left lower panel, Coomassie blue staining of heavy chains of immunoglobulins to show normalization of immunoprecipitated IDE. Right panel, bars show the mean values±SEM of the densitometric quantitation of the immunoreactivity of IDE in the Western blots as described above. (B) Left panel, percentage of 125I-insulin degraded by extracellular IDE in conditionated media of mock- and NICD- transfected cells as determined by TCA precipitation. Right panel, semi-quantitative determination of BACE-1 activity in cellular homogenates of mock- and ΔE-Notch- transfected N2aSW cells in the presence or absence of γ-secretase inhibitor L-685,458. Bars represent the mean value±SEM. of 4 independent experiments performed in triplicate; *p<0.05 compared with mock-transfected cells.
3.5. Notch signaling activation down-regulates IDE transcription
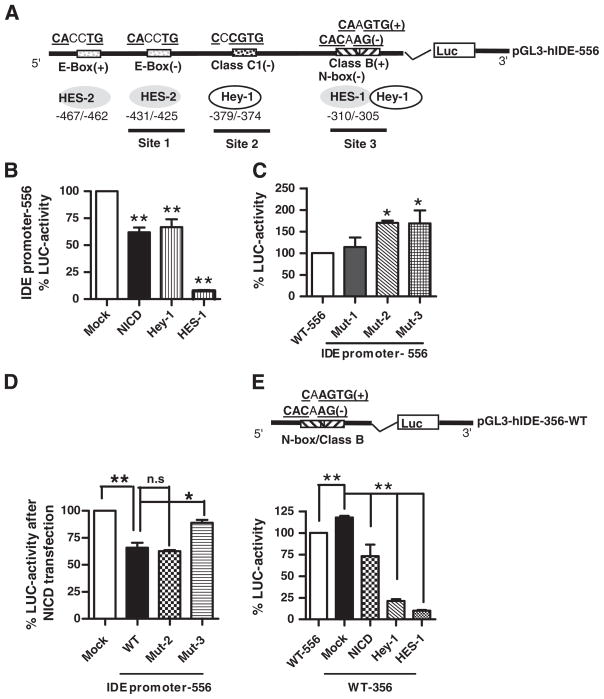
Taking into account that in the −582/−26 promoter region we detected by in silico analysis putative Notch target genes binding sites (schematically depicted in Fig. 4A) and knowing that the canonical Notch signaling is mediated by HES and/or Hey, which are described as transcriptional repressors, we explored whether Notch activation down regulates IDE promoter activity. As a proof of concept we decided to co-transfect N2aSW cells with the IDE 5′ region (556 bp) fused to Luciferase (LUC) and NICD or Hey-1, or HES-1, or empty vector, respectively. There was a significant reduction on IDE promoter activity in NICD (61.99%±4.38; p<0.01), Hey-1 (66.61%±7.37; p<0.01) and HES-1 (7.65±0.81; p<0.01) as compared to mock-transfected cells (100%) (Fig. 4B) suggesting the presence of functional sites that negatively regulate IDE transcription.
Fig. 4.
Effect of Notch activation on IDE promoter. (A) Schematic representation of the 556 bp fragment (−575/−19) corresponding to the IDE proximal promoter showing the location of E-box (site 1), Class C1 (site 2) and the overlapping Class B/N-box (site 3) sites and their putative consensus sequences for HES and Hey-1 binding. (+), foward orientation; (−) reverse orientation. (B) Bars show the mean±SEM of percentage of Luciferase (LUC) activity determined in N2aSW cells co-transfected with the IDE promoter-556 pb and Mock, NICD, Hey-1 and HES-1 constructs, respectively. **p<0.01 compared to mock-transfected N2aSW cells. (C) Bars show the mean±SEM of percentage of Luciferase (LUC) activity determined in N2aSW cells transfected with IDE promoter-556 wild type (WT) or mutated in site 1 (Mut1), site 2 (Mut2) or site 3 (Mut3), respectively. *p<0.05 compared to WT. (D) Bars show the mean±SEM of percentage of Luciferase (LUC) activity determined in N2aSW cells co-transfected with IDE promoter-556 wild type (WT) or mutated in site 2 (Mut2) or site 3 (Mut3), respectively, and NICD construct. n.s; not significant differences between WT and Mut-2 promoters and *p<0.05 significant differences between WT and Mut-3 promoter. (E) Upper panel, schematic representation of the wild type 356 bp fragment (−337/−19) corresponding to the IDE proximal promoter containing only the overlapping Class B/N-box consensus sequence site (site 3); (+), normal orientation; (−) reverse orientation. Lower panel, bars show the mean±SEM of percentage of Luciferase (LUC) activity determined in N2aSW cells transfected with IDE promoter-556 pb wild type (WT-556) or co-transfected with IDE promoter-356 and Mock, NICD, Hey-1 and HES-1 constructs, respectively. **p<0.01 compared to IDE promoter-556 or to mock-transfected N2aSW cells.
When the putative E-box, Class C1 and the overlapping N-box/Class B consensus binding sites on the wild type 556 promoter (WT-556) were mutated (Mut-1, Mut-2 and Mut-3, respectively) the transcriptional activity increased in Mut-2 and Mut-3 but not in Mut-1 (Fig. 4C) suggesting that site 1 lacks an inhibitory effect while sites 2 and 3 function as repressors of IDE transcriptional activity. Consistently, after NICD transfection the negative regulation on the WT-556 promoter was reverted in the Mut-3 but not in the Mut-2 construct (Fig. 4D) indicating that site 3 is a relevant functional site and indeed it may be partially responsible for the Notch-mediated inhibition of the IDE gene expression probably by the direct impact of HES-1 and Hey-1 on the −310/−305 and −308/−303 binding sites. However, disruption of site 2 (in the presence of a functional site 3) is not enough to revert the repression of IDE transcription. In addition, when co-transfection of Hey-1 and Mut-3 construct was done, the inhibition was not reverted (not shown) suggesting the relevance of a functional Hey-1 binding site located at −379/−372 (referred as Class C1) in accordance with data of binding capacity presented in Table 1. Furthermore we prepared a deleted version (356 bp) containing the overlapping N-box/Class B site (Fig. 4E upper panel). Our results showed that the transcriptional activity of the pGL3-hIDE-356 promoter is increased as compared to pGL3-hIDE-556 promoter in accordance with the presence of an inhibitory site (Class C1) located at upstream (−379/−372 bp) in the long construct. Moreover, we detected a significant reduction on pGL3-hIDE-356 promoter activity after NICD, Hey-1 and HES-1 transfection reinforcing the functionality of this site on the inhibition of IDE transcription.
3.6. Ligand-induced activation of Notch modifies IDE expression levels in vitro and in vivo
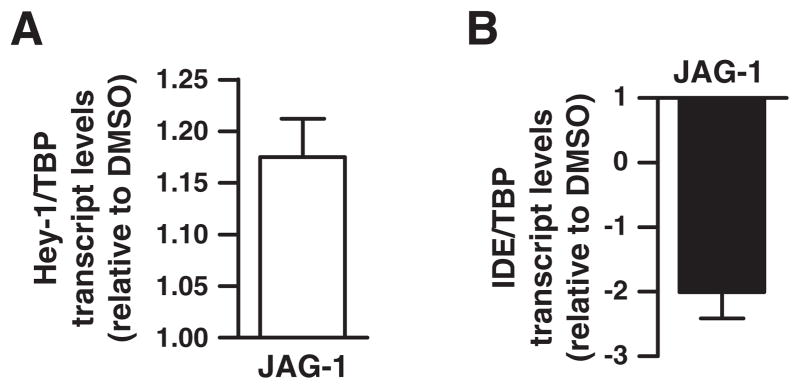
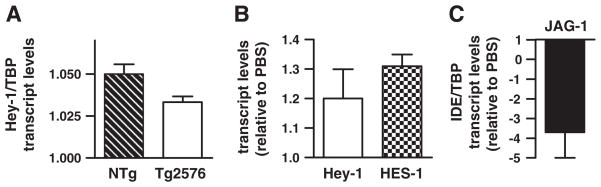
N2aSW cells were exposed to JAG-1 synthetic peptide, homologous to the δ/Serrate/Lag-2 domain of Jagged1 known to activate Notch pathway [29]. After JAG-1 exposure an increase of ~20% (1.21±0.04) in Hey-1 mRNA was detected (Fig. 5A) in accordance with previous reports [30], and ~2-fold reduction (−2.00±0.41; n=4) of IDE transcripts was observed (Fig. 5B). As a preliminary approach to determine the in vivo role of Notch signaling on IDE transcription we aimed to recapitulate an acute Notch pathway activation by performing a single injection of JAG-1 peptide into the hippocampus of Tg2576 mice (9.2±0.94 month). In this well-established AD mouse model the increase in IDE protein observed with aging occurred without any change in mRNA levels [31–32] providing a rational model to evaluate acute Notch-mediated IDE mRNA modulation. To assess the basal activation status of the Notch pathway in Tg2576 mice, Hey-1 mRNA levels were measured by QRT-PCR. No statistically significant differences were detected in Hey-1 transcript levels between non transgenic (NTg) and Tg2576 mice (Fig. 6A) suggesting that Notch activation is not a consequence of Aβ accumulation in this mouse model (32). After 3 days of JAG-1 injection, QRT-PCR showed an increase of ~30% on Hey-1 and HES-1 mRNA levels (Fig. 6B). In contrast, IDE mRNA abundance was reduced almost 4-fold (−3.7±1.3) in JAG-1 injected hippocampus as compared to control (Fig. 6C) suggesting a direct impact of Notch signaling on IDE transcription in vivo.
Fig. 5.
Hey-1 and IDE transcript levels in response to a ligand-induced Notch activation. N2aSW cells were treated with Jagged-1 peptide (JAG-1) and Hey-1 (A) and IDE (B) transcript levels determined by QRT-PCR. Bars show 3 independent determinations performed by triplicate. The mean and the SEM expression levels relative to vehicle are indicated.
Fig. 6.
In vivo effect of Notch signaling pathway activation on IDE gene expression. (A) Hey-1 mRNA levels determined by QRT-PCR in hippocampal extracts from non-transgenic (NTg) and Tg2576 mice. (B) Hey-1 and HES-1 and (C) IDE mRNA levels determined by QRT-PCR in hippocampal extracts from Tg2576 mice treated with JAG-1 relative to vehicle (PBS) injected animals (n=3). Bars show 3 independent determinations performed by triplicate for each sample. The mean±SEM of expression levels in the each case are indicated.
4. Discussion
A range of different studies has produced conflicting results for IDE expression in SAD brains [16,18–19,21,28,33]. In this study, we measured mRNA and enzyme activity for IDE in hippocampus of a series of well-characterized patients and control cases and correlated these findings to Notch signaling activation. We found lower IDE mRNA and proteolytic activity in SAD compared with NDC consistent with findings from the original reports [16,28] and reinforcing the role of IDE as a dominant Aβ protease in regulating the steady-state levels of Aβ in SAD. Moreover, our data provide direct evidence that Notch pathway is activated in the hippocampi of SAD cases as compared to NDC and that such activation may be implicated in the dysregulation of IDE and BACE-1 genes, both critical for Aβ metabolism. This hypothesis was supported experimentally in vitro by transfection of neuroblastoma cells and in vivo by intracranial injections in mice of a Notch ligand.
We showed that in N2aSW cells, over-expression of NICD or Hey-1 resulted in higher steady-state levels of extracellular Aβ. These results were consistent with reduced levels of IDE and increased levels of BACE-1 endogenous mRNA and enzymatic activities, respectively.
In our report we show that IDE is a new and direct target of HES-1 and Hey-1, key downstream targets of Notch signaling. So far, and despite its recognized importance in insulin and Aβ clearance, little is known about how IDE gene expression is regulated. The data reported to date are limited to the description of IDE promoter as a CG-rich, “slippery” and typical of housekeeping genes with several transcription initiation sites [20].
It was recently reported that the regulation of IDE gene expression may be mediated by peroxisome proliferator-activated receptor γ(PPARγ) which promotes gene transcription through binding to functional elements located at −1549/−1530 bp in IDE 5′ DNA region [34]. In addition, it was shown that selective PPARδ agonist (GW742) [35] and noradrenaline precursor L-threo-3,4-dihydroxy-phenylserine (L-DOPS) [36] increase mRNA levels of IDE in 5xFAD mice. Moreover, in vitro analyses in primary cultures of rat hippocampal neurons revealed that 17 beta-estradiol (17beta-E2) increased IDE in both mRNA and protein levels upon activation of estrogen receptor (ER) β and phosphatidylinositol 3-kinase (PI3-K) signaling cascade [37] By contrast, in primary cultures of astrocytes it was shown that glucocoticorticoids (GC) decrease IDE expression through the activation of GC receptors [38].
In our study, we have obtained data suggesting that HES-1 and Hey-1 directly repress IDE transcription in N2aSW cells through binding to the proximal IDE promoter. Our data show that endogenous and exogenous HES-1 and Hey-1 can be recruited to the IDE promoter in N2aSW cells presumably through an E-box suggesting that this proximal consensus binding site in the IDE promoter region is a relevant element in IDE transcription after Notch activation.
Our in vivo experiments strongly support that, by the age at which substantial Aβ is present in Tg2576 mice, Notch signaling is not altered as compared to non-Tg. This indicates that Notch may not be involved in the amyloidogenic pathway in this animal model and supports that Notch activation, as found in SAD is unlikely to be a secondary process due to Aβ accumulation. However, a transient and acute Notch activation in Tg2576 mice impacts on IDE transcription. These findings support that, at least in this animal model, changes in Notch expression do not replicate what is found in human brain. The time-course analysis of Notch pathway components and IDE expression as well as Aβ levels and memory deficit in young Tg2576 mice will be a valuable experimental tool to test the hypothesis that chronic Notch1 activation may accelerate Aβ-associated pathology. The down-regulation of IDE mediated by Notch1 activation in SAD brain may be a late-stage consequence of the pathological process. Alternatively, Notch activation in vulnerable regions such as the hippocampus may be an early event in the disease that promotes the accumulation of Aβ. Although Jagged-1 is overexpressed in fetal DS brain [39], the analysis of Notch pathway components and IDE expression in DS brains at different ages may help to clarify this issue. The function of Notch in aging human brain has not been fully established, yet, it seems likely that Notch signaling is involved in mammalian adult neurogenesis at the subventricular zone (SVZ) and subgranular layer of the hippocampus. More specifically, the role of Notch has been related to the maintenance of the pool of neuronal precursor cells (NPCs) in these neurogenic regions (reviewed in [40]). In addition, it was also speculated that expression of Notch1 protein in post-mitotic neurons from adult brain in areas with high synaptic activity/plasticity, such as hippocampus and cerebellum may play a role in neuronal plasticity, maintenance of complex neurite arborization or synaptic remodeling during memory formation [5]. A very recent report showed that expression of several FAD PS1 mutants in NPCs leads to impaired NPC self-renewal due a reduced γ-secretase cleavage of Notch [41]. If this defect in Notch activation due to FAD PS mutations is widespread in the brain, it is likely that increased amyloidogenesis in FAD is related primarily to the processing of APP. Gene expression and immunohistochemical studies show that Notch is over-expressed in neurogenic and non-neurogenic regions in SAD [4,42] and adult DS brains [3,39]. Together with our results, this evidence suggests that amyloidogenesis in SAD may be favored by a Notch-mediated decrease of IDE expression.
Several reports indicate that Notch signaling is increased in response to ischemic, traumatic or cryo-injury in the adult brain resulting in cell proliferation, variable effects upon neurogenesis, microglia activation and inflammation [43–46]. The inhibition of endogenous Notch signaling during these pathologic processes may help to evaluate the actual functional significance of activated Notch in neurodegenerative diseases.
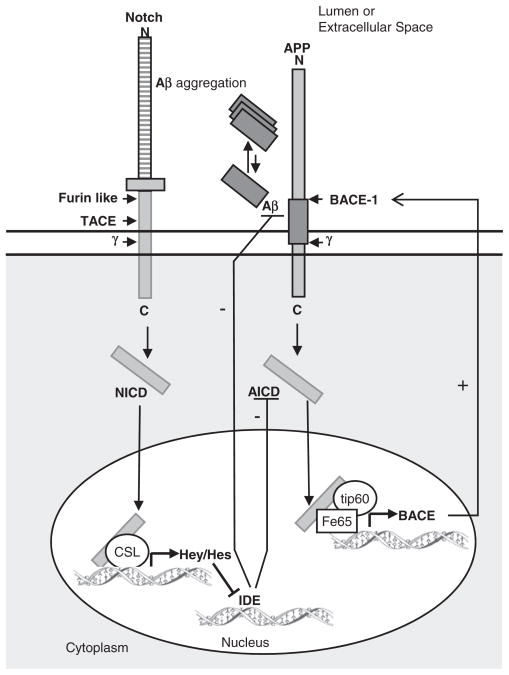
Taken together, we propose that binding of HES-1 and Hey-1 to the functional N-box of the IDE gene promoter may cooperate with a number of factors including other bHLH factors or other transcriptional regulators, already not described, and that the combined output of these interactions determines the decrement of IDE gene transcription in SAD brain. Moreover, taking into account that IDE is capable of degrading AICD [47–48], and that AICD positively regulates BACE-1 expression, Notch-mediated down-regulation of IDE would result in increased BACE1 levels, further enhancing the production of Aβ, as schematically represented in Fig. 7. Due to the multiplicity of gene targets and cellular processes regulated by Notch, it is difficult to speculate about how its activation in SAD may impact upon the disease course. Similarly, several cell-specific pathways regulate the control of IDE expression. The identification that their activities can be modulated by Notch signaling may provide novel insights on the consequences of therapeutic strategies for AD based on γ-secretase pharmacological inhibition.
Fig. 7.
Schematic representation of the effects of the γ-secretase cleavage of Notch and APP on IDE and BACE-1 transcriptional activation. Cleavage of Notch or APP by PS1/γ-secretase system produces NICD and AICD fragments, respectively. NICD translocates to the nucleus, binds the transcriptional cofactor CSL and promotes the transcription of the HES/Hey transcriptional inhibitors which further impact directly on IDE promoter inhibiting IDE mRNA synthesis. AICD interacts with the nuclear adaptor protein Fe65 and promotes transcription of BACE-1 by the histone acethyltransferase Tip60. BACE-1 promotes Aβ generation while IDE is involved in Aβ and AICD degradation. Abbreviations: γ, γ-secretase.
5. Conclusions
The following are the main conclusions of this work:
The decrement of IDE transcript levels observed in the hippocampus of SAD brains as compared to controls may be explained by the increment of Hey-1, a Notch target gene and a family member of transcriptional repressors.
Extracellular Aβ accumulation may be the consequence of local Notch activation and reduction of IDE mRNA and enzymatic activity levels.
The inhibitory effect of Notch activation on IDE transcription is mediated by the direct binding of HES-1 and Hey-1 to both functional sites located at −379/−372 and −310/−303 in the IDE proximal promoter.
Intracranial injection of the Notch ligand JAG-1 in Tg2576 mice induced overexpression of HES-1 and Hey-1 and reduction of IDE mRNA levels, respectively.
Our results support that a Notch-dependent IDE transcriptional modulation may impact on Aβ metabolism providing a functional link between Notch signaling and the amyloidogenic pathway in SAD.
Acknowledgments
The authors would like to acknowledge Dr. Gopal Thinakaran (Department of Neurology, University of Chicago) for providing us with the N2aSW cell line and molecular reagents, and M. I. Farías for her assistance in the histology procedures. This work was supported by grants from the sponsor-id=“gs0005” id=“gts0005”>Agencia Nacional de Promoción Científica y Tecnológica (ANPCyT) PICT38009 (to LM); PICT 2354 (to EMC) and from the sponsor-id=“gs0010” id=“gts0010”>Consejo Nacional de Investigaciones Científicas y Tecnológicas (CONICET)-PIP6168 and PIP693 (to LM); PIP1660 (to ES). The brain tissues were provided by the Harvard Brain Tissue Resource Center, which is supported in part by PHS grant number R24-MH 068855.
References
- 1.Citron M, Eckman CB, Diehl TS, Corcoran C, Ostaszewski BL, Xia W, Levesque G, St George Hyslop P, Younkin SG, Selkoe DJ. Additive effects of PS1 and APP mutations on secretion of the 42-residue amyloid beta-protein. Neurobiol Dis. 1998;5:107–116. doi: 10.1006/nbdi.1998.0183. [DOI] [PubMed] [Google Scholar]
- 2.Podlisny MB, Lee G, Selkoe DJ. Gene dosage of the amyloid beta precursor protein in Alzheimer’s disease. Science. 1987;238:669–671. doi: 10.1126/science.2960019. [DOI] [PubMed] [Google Scholar]
- 3.Fischer DF, van Dijk R, Sluijs JA, Nair SM, Racchi M, Levelt CN, van Leeuwen FW, Hol EM. Activation of the Notch pathway in Down syndrome: cross-talk of Notch and APP. FASEB J. 2005;19:1451–1458. doi: 10.1096/fj.04-3395.com. [DOI] [PubMed] [Google Scholar]
- 4.Berezovska O, Xia MQ, Hyman BT. Notch is expressed in adult brain, is coexpressed with presenilin-1, and is altered in Alzheimer disease. J Neuropathol Exp Neurol. 1998;57:738–745. doi: 10.1097/00005072-199808000-00003. [DOI] [PubMed] [Google Scholar]
- 5.Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 6.Sestan N, Artavanis-Tsakonas S, Rakic P. Contact-dependent inhibition of cortical neurite growth mediated by notch signaling. Science. 1999;286:741–746. doi: 10.1126/science.286.5440.741. [DOI] [PubMed] [Google Scholar]
- 7.Poirazi P, Mel BW. Impact of active dendrites and structural plasticity on the memory capacity of neural tissue. Neuron. 2001;29:779–796. doi: 10.1016/s0896-6273(01)00252-5. [DOI] [PubMed] [Google Scholar]
- 8.Fortini ME. Notch signaling: the core pathway and its posttranslational regulation. Dev Cell. 2009;16:633–647. doi: 10.1016/j.devcel.2009.03.010. [DOI] [PubMed] [Google Scholar]
- 9.Iso T, Sartorelli V, Chung G, Shichinohe T, Kedes L, Hamamori Y. HERP, a new primary target of Notch regulated by ligand binding. Mol Cell Biol. 2001;21:6071–6079. doi: 10.1128/MCB.21.17.6071-6079.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tanzi RE, Bertram L. Twenty years of the Alzheimer’s disease amyloid hypothesis: a genetic perspective. Cell. 2005;120:545–555. doi: 10.1016/j.cell.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 11.Li R, Lindholm K, Yang LB, Yue X, Citron M, Yan R, Beach T, Sue L, Sabbagh M, Cai H, Wong P, Price D, Shen Y. Amyloid beta peptide load is correlated with increased beta-secretase activity in sporadic Alzheimer’s disease patients. Proc Natl Acad Sci U S A. 2004;101:3632–3637. doi: 10.1073/pnas.0205689101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Akiyama H, Kondo H, Ikeda K, Kato M, McGeer PL. Immunohistochemical localization of neprilysin in the human cerebral cortex: inverse association with vulnerability to amyloid beta-protein (Abeta) deposition. Brain Res. 2001;902:277–281. doi: 10.1016/s0006-8993(01)02390-3. [DOI] [PubMed] [Google Scholar]
- 13.Eckman EA, Adams SK, Troendle FJ, Stodola BA, Kahn MA, Fauq AH, Xiao HD, Bernstein KE, Eckman CB. Regulation of steady-state beta-amyloid levels in the brain by neprilysin and endothelin-converting enzyme but not angiotensin-converting enzyme. J Biol Chem. 2006;281:30471–30478. doi: 10.1074/jbc.M605827200. [DOI] [PubMed] [Google Scholar]
- 14.Hellstrom-Lindahl E, Ravid R, Nordberg A. Age-dependent decline of neprilysin in Alzheimer’s disease and normal brain: inverse correlation with A beta levels. Neurobiol Aging. 2008;29:210–221. doi: 10.1016/j.neurobiolaging.2006.10.010. [DOI] [PubMed] [Google Scholar]
- 15.Funalot B, Ouimet T, Claperon A, Fallet C, Delacourte A, Epelbaum J, Subkowski T, Leonard N, Codron V, David JP, Amouyel P, Schwartz JC, Helbecque N. Endothelin-converting enzyme-1 is expressed in human cerebral cortex and protects against Alzheimer’s disease. Mol Psychiatry. 2004;9:1122–1128. 1059. doi: 10.1038/sj.mp.4001584. [DOI] [PubMed] [Google Scholar]
- 16.Cook DG, Leverenz JB, McMillan PJ, Kulstad JJ, Ericksen S, Roth RA, Schellenberg GD, Jin LW, Kovacina KS, Craft S. Reduced hippocampal insulin-degrading enzyme in late-onset Alzheimer’s disease is associated with the apolipoprotein E-epsilon4 allele. Am J Pathol. 2003;162:313–319. doi: 10.1016/s0002-9440(10)63822-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miners JS, Baig S, Palmer J, Palmer LE, Kehoe PG, Love S. Abeta-degrading enzymes in Alzheimer’s disease. Brain Pathol. 2008;18:240–252. doi: 10.1111/j.1750-3639.2008.00132.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Caccamo A, Oddo S, Sugarman MC, Akbari Y, LaFerla FM. Age- and region-dependent alterations in Abeta-degrading enzymes: implications for Abeta-induced disorders. Neurobiol Aging. 2005;26:645–654. doi: 10.1016/j.neurobiolaging.2004.06.013. [DOI] [PubMed] [Google Scholar]
- 19.Wang S, Wang R, Chen L, Bennett DA, Dickson DW, Wang DS. Expression and functional profiling of neprilysin, insulin-degrading enzyme, and endothelin-converting enzyme in prospectively studied elderly and Alzheimer’s brain. J Neurochem. 2010;115:47–57. doi: 10.1111/j.1471-4159.2010.06899.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Farris W, Leissring MA, Hemming ML, Chang AY, Selkoe DJ. Alternative splicing of human insulin-degrading enzyme yields a novel isoform with a decreased ability to degrade insulin and amyloid beta-protein. Biochemistry. 2005;44:6513–6525. doi: 10.1021/bi0476578. [DOI] [PubMed] [Google Scholar]
- 21.Perez A, Morelli L, Cresto JC, Castano EM. Degradation of soluble amyloid beta-peptides 1–40, 1–42, and the Dutch variant 1-40Q by insulin degrading enzyme from Alzheimer disease and control brains. Neurochem Res. 2000;25:247–255. doi: 10.1023/a:1007527721160. [DOI] [PubMed] [Google Scholar]
- 22.Schreiber E, Matthias P, Muller MM, Schaffner W. Rapid detection of octamer binding proteins with ‘mini-extracts’, prepared from a small number of cells. Nucleic Acids Res. 1989;17:6419. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kopan R, Schroeter EH, Weintraub H, Nye JS. Signal transduction by activated mNotch: importance of proteolytic processing and its regulation by the extracellular domain. Proc Natl Acad Sci U S A. 1996;93:1683–1688. doi: 10.1073/pnas.93.4.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thinakaran G, Teplow DB, Siman R, Greenberg B, Sisodia SS. Metabolism of the “Swedish” amyloid precursor protein variant in neuro2a (N2a) cells. Evidence that cleavage at the “beta-secretase” site occurs in the golgi apparatus. J Biol Chem. 1996;271:9390–9397. doi: 10.1074/jbc.271.16.9390. [DOI] [PubMed] [Google Scholar]
- 25.Bulloj A, Leal MC, Surace EI, Zhang X, Xu H, Ledesma MD, Castano EM, Morelli L. Detergent resistant membrane-associated IDE in brain tissue and cultured cells: relevance to Abeta and insulin degradation. Mol Neurodegener. 2008;3:22. doi: 10.1186/1750-1326-3-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nickoloff BJ, Qin JZ, Chaturvedi V, Denning MF, Bonish B, Miele L. Jagged-1 mediated activation of notch signaling induces complete maturation of human keratinocytes through NF-kappaB and PPARgamma. Cell Death Differ. 2002;9:842–855. doi: 10.1038/sj.cdd.4401036. [DOI] [PubMed] [Google Scholar]
- 27.Morelli L, Llovera RE, Mathov I, Lue LF, Frangione B, Ghiso J, Castano EM. Insulin-degrading enzyme in brain microvessels: proteolysis of amyloid beta vasculotropic variants and reduced activity in cerebral amyloid angiopathy. J Biol Chem. 2004;279:56004–56013. doi: 10.1074/jbc.M407283200. [DOI] [PubMed] [Google Scholar]
- 28.Zhao Z, Xiang Z, Haroutunian V, Buxbaum JD, Stetka B, Pasinetti GM. Insulin degrading enzyme activity selectively decreases in the hippocampal formation of cases at high risk to develop Alzheimer’s disease. Neurobiol Aging. 2007;28:824–830. doi: 10.1016/j.neurobiolaging.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 29.Sainson RC, Aoto J, Nakatsu MN, Holderfield M, Conn E, Koller E, Hughes CC. Cell-autonomous notch signaling regulates endothelial cell branching and proliferation during vascular tubulogenesis. FASEB J. 2005;19:1027–1029. doi: 10.1096/fj.04-3172fje. [DOI] [PubMed] [Google Scholar]
- 30.Young Oh S, Chen CD, Abraham CR. Cell-type dependent modulation of Notch signaling by the amyloid precursor protein. J Neurochem. 2010;113:262–274. doi: 10.1111/j.1471-4159.2010.06603.x. [DOI] [PubMed] [Google Scholar]
- 31.Leal MC, Dorfman VB, Gamba AF, Frangione B, Wisniewski T, Castano EM, Sigurdsson EM, Morelli L. Plaque-associated overexpression of insulin-degrading enzyme in the cerebral cortex of aged transgenic tg2576 mice with Alzheimer pathology. J Neuropathol Exp Neurol. 2006;65:976–987. doi: 10.1097/01.jnen.0000235853.70092.ba. [DOI] [PubMed] [Google Scholar]
- 32.Zhao L, Teter B, Morihara T, Lim GP, Ambegaokar SS, Ubeda OJ, Frautschy SA, Cole GM. Insulin-degrading enzyme as a downstream target of insulin receptor signaling cascade: implications for Alzheimer’s disease intervention. J Neurosci. 2004;24:11120–11126. doi: 10.1523/JNEUROSCI.2860-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dorfman VB, Pasquini L, Riudavets M, Lopez-Costa JJ, Villegas A, Troncoso JC, Lopera F, Castano EM, Morelli L. Differential cerebral deposition of IDE and NEP in sporadic and familial Alzheimer’s disease. Neurobiol Aging. 2010;10:743–174357. doi: 10.1016/j.neurobiolaging.2008.09.016. Epub 2008 Nov 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Du J, Zhang L, Liu S, Zhang C, Huang X, Li J, Zhao N, Wang Z. PPARgamma transcriptionally regulates the expression of insulin-degrading enzyme in primary neurons. Biochem Biophys Res Commun. 2009;383:485–490. doi: 10.1016/j.bbrc.2009.04.047. [DOI] [PubMed] [Google Scholar]
- 35.Kalinin S, Richardson JC, Feinstein DL. A PPARdelta agonist reduces amyloid burden and brain inflammation in a transgenic mouse model of Alzheimer’s disease. Curr Alzheimer Res. 2009;5:431–437. doi: 10.2174/156720509789207949. [DOI] [PubMed] [Google Scholar]
- 36.Kalinin S, Polak PE, Lin SX, Sakharkar AJ, Pandey SC, Feinstein DL. The noradrenaline precursor L-DOPS reduces pathology in a mouse model of Alzheimer’s disease. Neurobiol Aging. 2011 Jun 24; doi: 10.1016/j.neurobiolaging.2011.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao L, Yao J, Mao Z, Chen S, Wang Y, Brinton RD. 17beta-Estradiol regulates insulin-degrading enzyme expression via an ERbeta/PI3-K pathway in hippocampus: relevance to Alzheimer’s prevention. Neurobiol Aging. 2010 Jan 4; doi: 10.1016/j.neurobiolaging.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Y, Li YM, Tang J, Song M, Xu X, Xiong J, Li J, Bai Y. Glucocorticoids facilitate astrocytic Amyloid-{beta}sition by increasing the expression of APP and BACE1 and decreasing the expression of Amyloid-{beta}-degrading proteases. Endocrinology. 2011;152:2704–2715. doi: 10.1210/en.2011-0145. [DOI] [PubMed] [Google Scholar]
- 39.Lockstone HE, Harris LW, Swatton JE, Wayland MT, Holland AJ, Bahn S. Gene expression profiling in the adult Down syndrome brain. Genomics. 2007;90:647–660. doi: 10.1016/j.ygeno.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 40.Lazarov O, Marr RA. Neurogenesis and Alzheimer’s disease: at the crossroads. Exp Neurol. 2010;223:267–281. doi: 10.1016/j.expneurol.2009.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Veeraraghavalu K, Choi SH, Zhang X, Sisodia SS. Presenilin 1 mutants impair the self-renewal and differentiation of adult murine subventricular zone-neuronal progenitors via cell-autonomous mechanisms involving notch signaling. J Neurosci. 2010;30:6903–6915. doi: 10.1523/JNEUROSCI.0527-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nagarsheth MH, Viehman A, Lippa SM, Lippa CF. Notch-1 immunoexpression is increased in Alzheimer’s and Pick’s disease. J Neurol Sci. 2006;244:111–116. doi: 10.1016/j.jns.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 43.Scholzke MN, Schwaninger M. Transcriptional regulation of neurogenesis: potential mechanisms in cerebral ischemia. J Mol Med. 2007;85:577–588. doi: 10.1007/s00109-007-0196-z. [DOI] [PubMed] [Google Scholar]
- 44.Arumugam TV, Chan SL, Jo DG, Yilmaz G, Tang SC, Cheng A, Gleichmann M, Okun E, Dixit VD, Chigurupati S, Mughal MR, Ouyang X, Miele L, Magnus T, Poosala S, Granger DN, Mattson MP. Gamma secretase-mediated Notch signaling worsens brain damage and functional outcome in ischemic stroke. Nat Med. 2006;12:621–623. doi: 10.1038/nm1403. [DOI] [PubMed] [Google Scholar]
- 45.Tatsumi K, Okuda H, Makinodan M, Yamauchi T, Makinodan E, Matsuyoshi H, Manabe T, Wanaka A. Transient activation of Notch signaling in the injured adult brain. J Chem Neuroanat. 2010;39:15–19. doi: 10.1016/j.jchemneu.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 46.Wang X, Mao X, Xie L, Greenberg DA, Jin K. Involvement of Notch1 signaling in neurogenesis in the subventricular zone of normal and ischemic rat brain in vivo. J Cereb Blood Flow Metab. 2009;29:1644–1654. doi: 10.1038/jcbfm.2009.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bernstein HG, Ansorge S, Riederer P, Reiser M, Frolich L, Bogerts B. Insulin-degrading enzyme in the Alzheimer’s disease brain: prominent localization in neurons and senile plaques. Neurosci Lett. 1999;263:161–164. doi: 10.1016/s0304-3940(99)00135-4. [DOI] [PubMed] [Google Scholar]
- 48.Edbauer D, Willem M, Lammich S, Steiner H, Haass C. Insulin-degrading enzyme rapidly removes the beta-amyloid precursor protein intracellular domain (AICD) J Biol Chem. 2002;277:13389–13393. doi: 10.1074/jbc.M111571200. [DOI] [PubMed] [Google Scholar]