Abstract
We demonstrate an affordable low-noise SPR instrument based on extraordinary optical transmission (EOT) in metallic nanohole arrays and quantify a broad range of antibody-ligand binding kinetics with equilibrium dissociation constants ranging from 200 pM to 40 nM. This nanohole-based SPR instrument is straightforward to construct, align, and operate, since it is built around a standard microscope and a portable fiber-optic spectrometer. The measured refractive index resolution of this platform is 3.1 × 10−6 without on-chip cooling, which is among the lowest reported for SPR sensors based on EOT. This is accomplished via rapid full-spectrum acquisition in 10 milliseconds followed by frame averaging of the EOT spectra, which is made possible by the production of template-stripped gold nanohole arrays with homogeneous optical properties over centimeter-sized areas. Sequential SPR measurements are performed using a 12-channel microfluidic flow cell after optimizing surface modification protocols and antibody injection conditions to minimize mass-transport artifacts. The immobilization of a model ligand, the protective antigen of anthrax on the gold surface, is monitored in real-time with a signal-to-noise ratio of ~860. Subsequently, real-time binding kinetic curves were measured quantitatively between the antigen and a panel of small, 25 kDa single-chain antibodies at concentrations down to 1 nM. These results indicate that nanohole-based SPR instruments have potential for quantitative antibody screening and as a general-purpose platform for integrating SPR sensors with other bioanalytical tools.
Keywords: Surface plasmon resonance, plasmonics, extraordinary optical transmission, nanohole arrays, template stripping, recombinant antibody, microfluidics
With the completion of the human genome project and the advent of rapid, cost-effective nucleic acid sequencing, scientific and technological interest has shifted from analysis of genes to analysis of the corresponding proteins, and, critically, their networks of interactions with other proteins. These protein-protein interactions, from transient, low affinity to stable, high affinity interactions define much of cellular behavior and analysis of their interaction networks helps to assign functions to uncharacterized proteins.1–3 Similarly, during development of therapeutics, such as antibodies, lead molecules are frequently selected based on their binding kinetics and selectivity for a target ligand, as this single parameter is often predictive of activity in in vitro and in vivo assays.4,5 For instance, antibodies that bind and neutralize the protective antigen (PA) of anthrax toxin are protective in animal models and, as a result, are currently undergoing intense clinical development. Analysis of a panel of antibodies neutralizing PA demonstrated that increased affinity for the toxin correlates with enhanced survival in animal models and was the key criterion in selecting a single antibody variant for further development.6
A number of high-throughput approaches have been developed in order to detect and measure protein-protein binding events. To assess equilibrium binding affinities in an in vivo setting, the yeast two hybrid system provides a sensitive but qualitative in vivo assay in which a productive interaction results in expression of a marker protein,7 while immunoprecipitation followed by mass spectrometry provides a more quantitative analysis.8 To more precisely rank equilibrium binding affinities, protein microarrays can measure binding between a protein of interest and a variety of other proteins.8,9
However, for a detailed, kinetic analysis of binding between two proteins, surface plasmon resonance (SPR) instruments have become the accepted standard.10–12 In commercial BiacoreTM SPR instruments, a ligand is immobilized on the gold sensing surface in a flow cell, while buffer containing the analyte flows over the surface. Analyte binding and unbinding to the gold surface modulates the interfacial refractive index, which is measured optically via surface plasmon waves – density fluctuations of conduction electrons propagating along the gold surface. In the SPR sensing scheme, the on-rate (ka) is observed as a temporal increase in interfacial index resulting from analyte-ligand binding, while the off-rate (kd) is detected as signal decay resulting from dissociation and removal of analyte molecules when buffer alone flows over a surface saturated with analyte. Importantly, these kinetic constants are measured without fluorescence or radioactive labeling of the analytes. Furthermore, because the rapid kinetic measurements are performed in a microfluidic flow cell, the analyte consumption can be significantly reduced compared with equilibrium measurements. Because of these key advantages, the growing field of proteomics has concurrently increased the demand for SPR instruments. Commercial SPR instruments, however, suffer from very high cost, low throughput, the difficulty in screening against membrane-bound receptors,13,14 and cannot be readily integrated with other bioanalytical tools.
The emerging field of nanoplasmonics has presented a new generation of optical biosensors. Among these, metallic films perforated with nanoholes have been of particular interest, because they can exhibit plasmon-enhanced extraordinary optical transmission (EOT).15 To generate SPR in BiacoreTM, a flat gold film atop a prism is illuminated at a steep angle in a total internal reflection mode, which complicates optical design, assembly, and alignment. In contrast, nanoholes in gold films can directly launch SPR from normally incident light using a simple illumination setup such as a standard microscope.16–21 Besides acting as a source of SP waves, nanoholes can concurrently function as nanowells to confine supported lipid bilayer membranes,22 a scaffold for pore-spanning lipid membranes,14 nanofluidic channels to transport biomolecules,23,24 or as electrodes for electrochemical sensing.25 Integrating these novel functionalities may justify the added cost of nanofabrication and motivates the development of a general-purpose nanoplasmonic sensing platform. Beyond high-throughput fabrication of metallic nanohole arrays,21,26–28 however, considerable efforts are required to quantify protein binding kinetics accurately and without artifacts, such as optimizing surface modification protocols (immobilization, blocking, and regeneration), microfluidics, low-noise optical measurements, and data analysis. This work reports detailed procedures of constructing a compact, microscope-based SPR instrument using gold nanohole arrays, disposable microfluidic chips, and a portable spectrometer. With rapid data acquisition and frame averaging, this nanohole-based SPR instrument can quantify a wide range of antibody-ligand binding kinetics with a refractive resolution of 3.1 × 10−6.
MATERIALS AND METHODS
Fabrication of the Au nanohole arrays and microfluidics
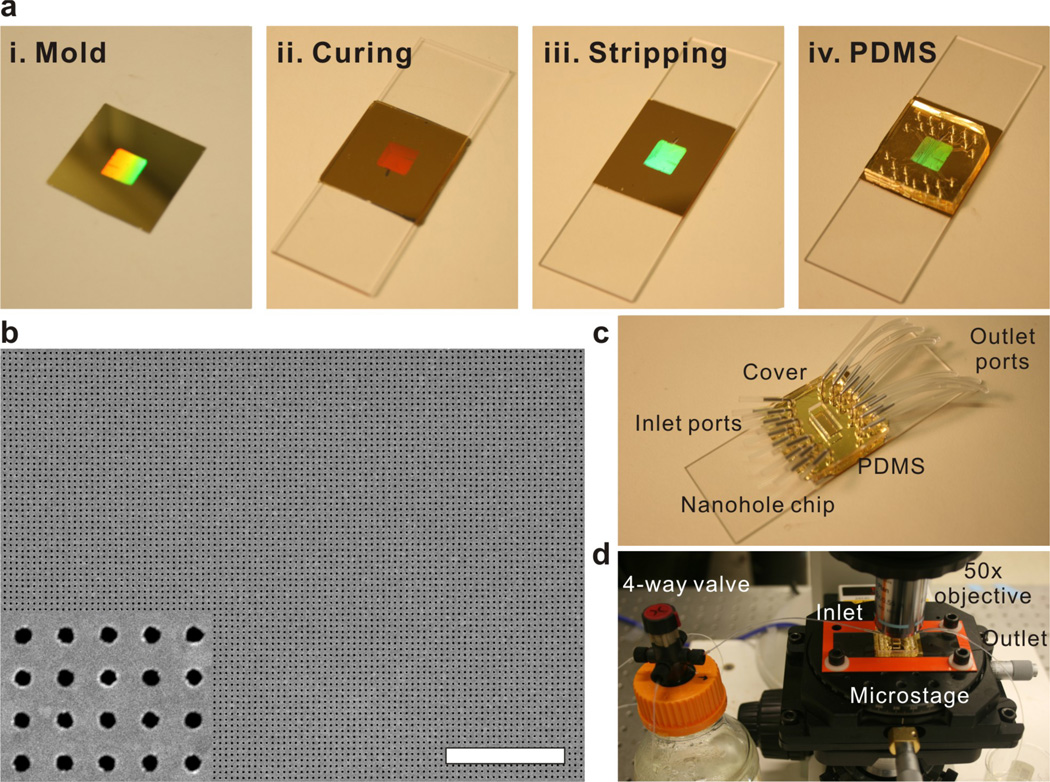
Au nanohole arrays were fabricated using a template-stripping technique,28,29 as shown in Figure 1a. A reusable Si template is patterned with deep circular trenches using nanoimprint lithography followed by deep trench Si etching. After cleaning the Si template with a piranha solution (1:1 mixture of sulfuric acid and hydrogen peroxide), a 200 nm-thick Au film was deposited using electron-beam evaporation, forming Au nanohole arrays (Step 1). The diameter of the nanoholes and periodicity of the array are 200 nm and 500 nm, respectively. An UV-curable optical epoxy (NOA 61, Norland Products) was applied to the Au surface and covered by a microscope slide (Step 2). After UV curing, the Au film, now adhered on the microscope slide with the epoxy, was peeled off (Step 3) and integrated with a polydimethylsiloxane (PDMS, Sylgard 184, Dow Corning) microfluidic chip (Step 4). After template stripping, the Si template was reused after cleaning in Au etchant followed by piranha cleaning. Figure 1b shows a scanning electronic microscope (SEM) image of template-stripped Au nanohole array. The root-mean-square roughness of the template-stripped Au surface, measured with atomic force microscope, was 0.6 nm (data not shown), and the smooth Au surface improves the SPR detection sensitivity, as previously reported.28,29 Figure 1c shows a ready-to-use Au nanohole sensor chip that is attached to a 12-channel PDMS microfluidic chip made via soft lithography.30 The height and width of each channel are 50 µm and 250 µm, respectively. A plastic cover was used to press the PDMS flow cell against the Au sensor chip for sealing, and no leak was observed during experiments. Inlet and outlet ports consisted of Tygon microtubes (Cole-Parmer, IL) and metal tubes (New England Small Tube Corp., NH).
Figure 1.
(a) The procedures for template-stripping periodic Au nanohole array chips: (i) deposit 200 nm-thick Au on a Si template, (ii) cure an UV-curable optical epoxy coated on the Au surface, (iii) peel off the Au film, and (iv) attach a PDMS microfluidic flow cell. (b) Scanning electronic microscope (SEM) image of template-stripped Au nanohole arrays. The diameter of the nanoholes and periodicity of the array are 200 nm and 500 nm, respectively. The scale bar is 10 µm. The inset shows a zoomed-in image. (c) Photograph of a ready-to-use nanohole chip attached to a microfluidic flow cell. (d) Photograph of a time-resolved SPR biosensing setup. A broadband light source illuminates the nanohole array through a 50× objective and transmitted light is collected using a multi-mode optical fiber placed underneath the nanohole chip, which is connected to a portable fiber-optic spectrometer. Either analyte or buffer solution is delivered to a microfluidic flow cell using a four-way valve.
Optical measurement setup and data acquisition
The Au nanohole chip assembled with a PDMS microfluidic flow cell was placed on the stage of an upright microscope (Nikon LV100, USA) as shown in Figure 1d. The microtube connects the flow cell and a four-way valve (Cole-Parmer, USA), which selects either an analyte or a buffer solution for injection via syringe pumps (Harvard Apparatus). The PDMS chip forms 12 parallel microfluidic channels above gold nanohole arrays, thus a single sensor chip can be used to measure 12 separate binding events in a sequential manner without surface regeneration. A standard tungsten-halogen lamp illuminates one channel of the nanohole array through a 50× objective (illumination spot size of ~200 µm) and the transmitted light is collected by a multi-mode optical fiber (200 µm in core diameter) placed underneath the nanohole chip, which is connected to an uncooled fiber-optic spectrometer (USB4000, Ocean Optics). A linear silicon CCD array inside the spectrometer acquires a full EOT spectrum with an acquisition time of 10 msec. A custom MATLABTM script is used to fit a 2nd order polynomial function around the measured EOT peak and automatically calculates the centroid of the peak.
Reagents
Hepes buffered saline (HBS) consists of 10 mM Hepes, 150 mM sodium chloride, buffered to pH 7.2. Recombinant scFv proteins were expressed in the periplasm of E. coli strain BL21, followed by osmotic shock, immobilized metal affinity and S75 size exclusion chromatographic purification steps, as previously reported.6 Micro-bicinchonoinic acid assay (Pierce, Rockford, IL) was used to measure protein concentrations while SDS-PAGE with GelCode Blue stain reagent (Pierce, USA) was used to verify protein preparation homogeneity and purity. The PA ligand was purchased from List Labs (Campbell, USA).
Surface modification
For antibody-ligand binding reactions, the template-stripped Au surface was functionalized using a mixture of 0.5 mM 2-mercaptoundecanoic acids and 0.5 mM 2-mercaptoethanol solutions to form a self-assembled monolayer with a carboxylated functional group for 48 hours at 4 °C. After washing with ethanol, the surface was dried with a stream of nitrogen and assembled with the PDMS microfluidic chip. In order to immobilize PA on the surface, a mixture of 0.4 M 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide) (EDC) and 0.1 M N-Hydroxysuccinimide (NHS) was flowing in the channel for 7 min to active the carboxylate groups. After washing with HBS for 2 min at a flow rate of 30 µm/min, 100 µg/ml of PA was introduced and incubated for 30 min with a slow flow rate of 3 µl/min for covalent binding to the SAM layer. After another washing with HBS for 2 min, the unbound carboxylate groups were quenched with a 1 M ethanolamine solution for 7 min at a flow rate of 30 µl/min. The surface is then washed by HBS about 15 min until the baseline is stabilized before injecting scFv antibody solutions. For binding kinetic measurement of scFv antibodies, after a 3 min baseline with HBS, different concentrations of scFv antibodies ranging from 1 nM to 100 nM were injected for 5 min followed by HBS washing for dissociation curve measurements for 15 min. Between each antibody binding measurement, the surface was regenerated by flowing 3 M MgCl2 for 30 sec and washed by HBS for at least 3 min. During the kinetic measurements, the flow rate of analyte was kept at 30 µl/min.
RESULTS AND DISCUSSION
Optical properties of template-stripped Au nanohole arrays
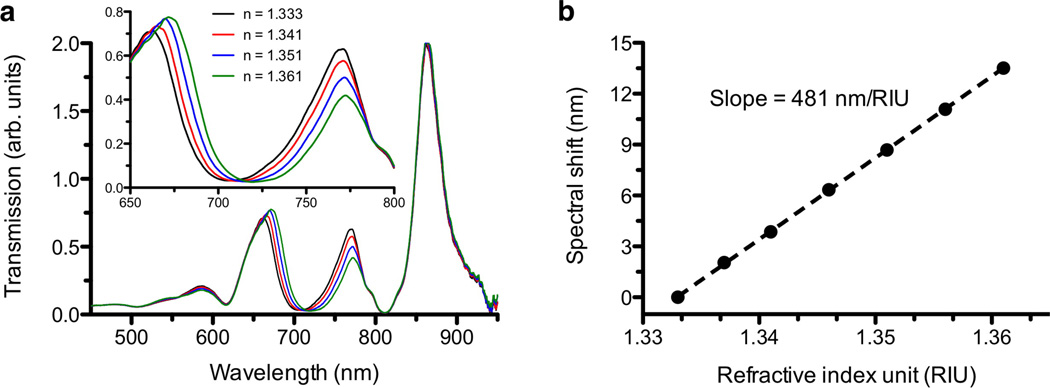
Figure 2a shows optical transmission spectra measured through the Au nanohole array in index-calibrated water-glycerol mixtures with refractive indices (RI) ranging from 1.333 to 1.361. The multiple transmission maxima and minima arise from the interactions between the incident light and the surface plasmons generated on both sides of the Au nanohole film via grating coupling, and their underlying physical mechanisms have been studied by many research groups.31 The wavelengths at which these maxima and minima occur in periodic nanohole arrays depend on the array periodicity, film thickness, hole size, hole shape, as well as the refractive index of the substrate and the surrounding medium. Molecular binding on the Au surface modulates the local refractive index, which in turn shifts the positions of these spectral features. Therefore, binding kinetics can be recorded by continually tracking either the transmission peak19 or the transmission minimum32 in metallic nanohole arrays. As the RI increases, the transmission peaks around 660 nm and 770 nm and a transmission minimum around 710 nm all shift towards longer wavelengths. These spectral features originate from the surface plasmons at the interface between Au and the aqueous solution, thus their spectral shifts can be used to monitor molecular binding to the Au surface. In contrast, the transmission minimum and maximum, near 810 and 880 nm respectively, remain stationary because they are associated with the resonances at the interface between Au and the epoxy substrate, which is not exposed to the solution. For the nanohole design parameters used in this work, the transmission minimum around 710 nm shows the highest sensitivity to the RI changes. As shown in Figure 2b, the measured spectral sensitivity of this transmission minimum, as the bulk RI increases from 1.333 to 1.361, is 481 nm/RIU (refractive index unit).
Figure 2.
(a) Measured optical transmission spectra of the template-stripped nanohole array in a 200 nm-thick Au film (hole diameter: 200 nm, periodicity: 500 nm) in index-calibrated water-glycerol solutions. The refractive index (RI) of the solution increases from 1.333 to 1.361. (b) As the RI increases, the transmission minimum around 710 nm shifts towards longer wavelengths. The measured bulk refractive index sensitivity is 481 nm/RIU.
Optimizing the signal-to-noise ratio
The limit of detection (LOD) is the lowest analyte concentration that can be detected but not necessarily quantitated as an exact value. For direct assays, the LOD is primarily a function of the signal-to-noise ratio in the measurement itself, and is set in relation to statistical variations in response values for blank samples. A commonly used value is 1 × S.D. or 3 × S.D., where S.D. is the standard deviation of replicate measurements on blank samples. Therefore, decreasing a noise level is an appropriate strategy to lower the LOD within the same detection scheme.
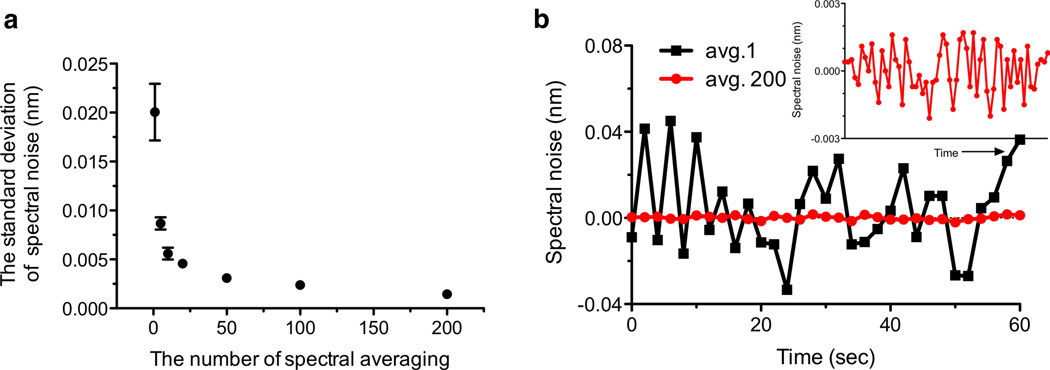
Höök’s group demonstrated that the background noise level in nanohole SPR sensors can be reduced by collecting and averaging many spectra per unit time, dynamically subtracting the dark spectra in real-time, and using robust peak-analysis algorithms.32 The background noise in the nanohole SPR instrument primarily comes from the shot noise in the detector, i.e. statistical variation in the number of collected photons. One strategy to reduce the shot noise in time-resolved SPR sensing is to collect optical transmission spectra using a short acquisition time (10 msec in this work) for frequent temporal averaging. The prerequisite for rapid temporal averaging is large photon flux on each detection pixel, which is provided by large-area template-stripped nanohole arrays in this work. By summing up to 200 frames, the noise level can be reduced by more than 10 fold, as shown in Figure 3. Collecting 200 frames with integration time of 10 msec each corresponds to a temporal resolution of 2 sec, which is still sufficient for measuring the binding kinetics of most antibodies. If a faster temporal resolution is desired, the number of averaged frames can be reduced to 50 or 100 with a moderate increase in the shot noise level (Figure 3a). With 200 averaged spectra, the spectral noise (S.D.) measured over the course of 1 min is 1.5 × 10−3 nm. Using the measured bulk refractive index sensitivity of 481 nm/RIU (Figure 2), the minimum refractive index resolution that can be resolved by the system is 3.1 × 10−6 RIU. The noise level may be further reduced by adding temperature controllers for both chip33 and light source,34 and by using a detector with a higher saturation level (e.g. larger pixel size) and faster frame rates to reduce the shot noise.
Figure 3.
(a) The background noise level with the number of frame averaging. The shot noise is reduced 10 fold by averaging 200 transmission spectra at the interval of 2 sec. (b) Comparison of spectral noise with and without frame averaging. The inset to panel (b) shows a zoomed-in plot for 200 frame averages.
PA-scFv antibody binding system
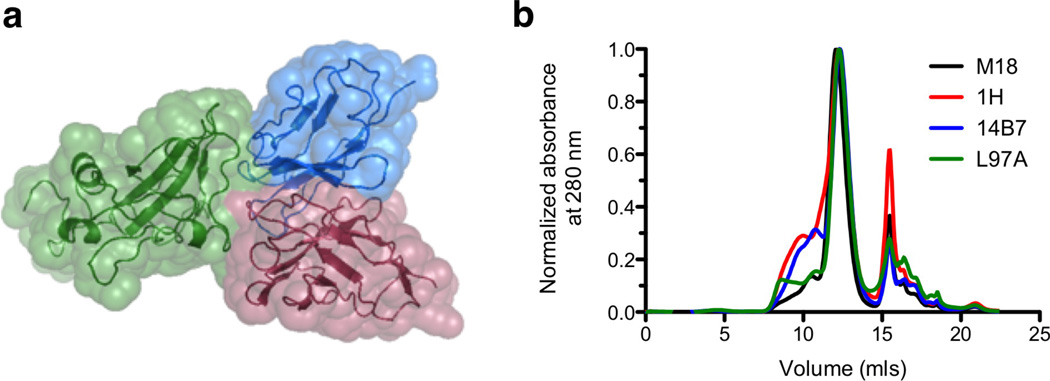
A family of single-chain variable fragment (scFv) antibodies, comprised of variants of the parent 14B7 scFv, were employed to assess the ability of the nanohole SPR instrument to sensitively and quantitatively measure binding kinetics to immobilized PA ligand. These antibodies contain one to ten amino acid substitutions relative to 14B7 and were identified by selection for tighter binding variants from random mutagenesis libraries.6,35 The interaction between these scFvs and PA has been well-characterized by computational, mutational and structural approaches.6,36,37 These data show that the 14B7 family of scFvs has a highly conserved structure in spite of large differences in PA binding affinity.37 Moreover, the complex between the ultra-high affinity M18 variant and PA domain four shows a buried interface of ~1700 Å with the antibody heavy and light chain domains each making similar contributions to PA binding (Figure 4a).37 These proteins were expressed as recombinant antibody fragments in the periplasm of E. coli strain BL21, followed by purification by sequential osmotic shock, immobilized metal affinity and size exclusion chromatographic steps. The chromatographic traces from the final purification step are shown in Figure 4b and indicate that all scFv variants were predominantly expressed in the desired monomeric state. The primary peak, with an elution volume of ~ 12 mls, was collected, the protein concentration quantified with micro-BCA assay and purity by SDS-PAGE and used in all subsequent experiments.
Figure 4.
Characterization of scFv proteins. (a) Structure of the scFv-PA interaction, with domain four of PA shown in green, in complex with the M18 variable light chain in blue and the heavy chain in red. The alpha carbon backbone is shown as a ribbon diagram, with the solvent-accessible surface area indicated by a space-filling model. (From PDB 3ETB; figure prepared with PyMol, www.pymol.org) (b) Traces corresponding to the final scFv purification step, size exclusion chromatography with a GE Healthcare ÅKTA FPLC and Superdex S75 column. The scFv elution peak has an elution volume of 12 mls, corresponding to the expected size of an scFv monomer (25 kDa). The free nickel peak, residual from the metal affinity chromatography step, elutes at 16 mls.
Time-resolved SPR measurements of antibody-antigen binding
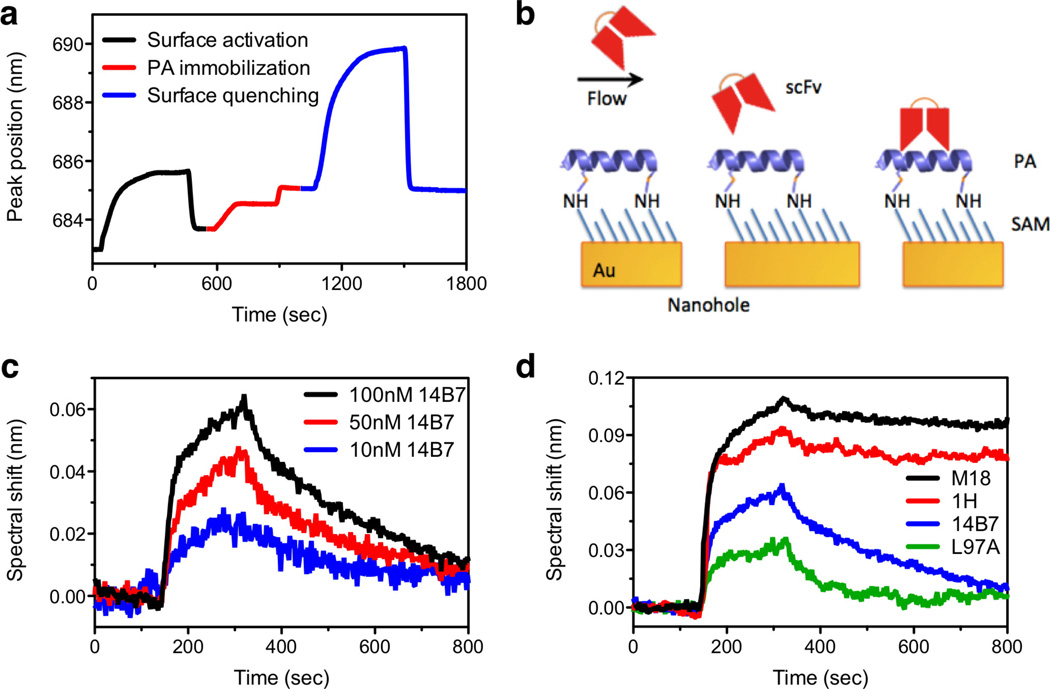
The entire process of surface functionalization, ligand immobilization, as well as the reversible binding and unbinding of receptor-ligand was monitored with the nanohole-based SPR instrument. The first step for biosensing is the immobilization of antigen on the Au nanohole surface coated with a carboxylated SAM using surface chemistry based on amine coupling, as described in the Materials and Methods section. This entire antigen (PA) immobilization process is monitored from spectral shifts of transmission peaks to quantitatively control and analyze the amount of PA immobilized on the surface as shown in Figure 5a. To minimize mass-transport effects, it is crucial to minimize the number of antigen ligands presented on the surface. Otherwise rebinding of dissociated antibodies can result in measurement of an artifactually slow dissociation rate. However, this low immobilized ligand density also confers a low maximal SPR response, even when the antigen surface is saturated with antibody. The immobilization of PA (83 kDa), which is designed to cover at most about 50% of the gold surface, shifts the SPR spectrum by 1.3 nm (Figure 5a). The signal-to-noise ratio of this measurement was over ~860, balancing sensitive detection with minimized re-binding.
Figure 5.
(a) Kinetic responses of spectral peak position during the immobilization of protective antigen (PA). (b) Schematic of scFv binding to PA immobilized on the Au nanohole surface via amine coupling. (c) Binding kinetics curves of 14B7 scFv antibody to PA with various concentrations. (d) Binding kinetic curves of 100 nM scFv antibodies to PA. These scFv molecules having the same molecular weight, but different dissociation constants with the PA in the range of around 10 pM to 100 nM are sequentially injected. Small noise peaks just prior to dissociation event are likely caused by the operation of the four-way mechanical valve, but they do not affect the calculation of binding kinetics.
After immobilizing the PA on the sensing surface, a family of single-chain variable fragment (scFv) antibodies binding the same PA epitope with affinities ranging from 10 pM to 100 nM were injected through the channel (Figure 5b). Figure 5c shows kinetic curves of 14B7 antibody binding with various concentrations to the PA immobilized on the surface, while Figure 5d compares the measured binding kinetic curves between the four different scFv molecules and PA. These antibodies are only 25 kDa in size, one-sixth the size of an IgG and one-thirtieth that of a pentameric IgM antibody. Since the SPR response is proportional to the density of the protein layer, molecules with smaller sizes result in smaller SPR responses on a per molecule basis than larger molecules.38 Because of the smaller molecular weight, the increased distance from the Au surface, and the relatively low concentration, the spectral shifts induced due to their binding to the surface are in the level of 0.1 nm. However, binding kinetics of these smaller scFv molecules can be still measured with a signal-to-noise ratio of ~60 and a sufficient temporal resolution to record and analyze their fast binding reactions (Figure 5c).
Antibody-antigen binding rates,39 i.e. the dissociation rate constant (kd), association rate constant (ka), and equilibrium dissociation constant (KD), are calculated from the kinetic curves shown in Figure 5c and 5d and summarized in Table 1. The nanohole SPR system was able to finely discriminate between antibodies with similar affinities for the same PA ligand, observing the identical affinity ranking and similar quantitative rate data, within an order of magnitude, to that previously measured using a BIAcore3000 system.6,35 While the observed association rates, (ka) are similar for all four scFv antibodies, typical for related antibodies, they exhibit a wide range of dissociation rate constants (kd) from 10−2 to 10−4 s−1. This data verifies that nanohole-based SPR sensors are capable of measuring binding kinetic curves and calculating kinetic parameters accurately even for small molecules. Moreover, many lead molecules identified during development of protein therapeutic show kd values within the range, indicating that the nanohole SPR system is suitable for use in drug discovery programs.13,40
Table 1.
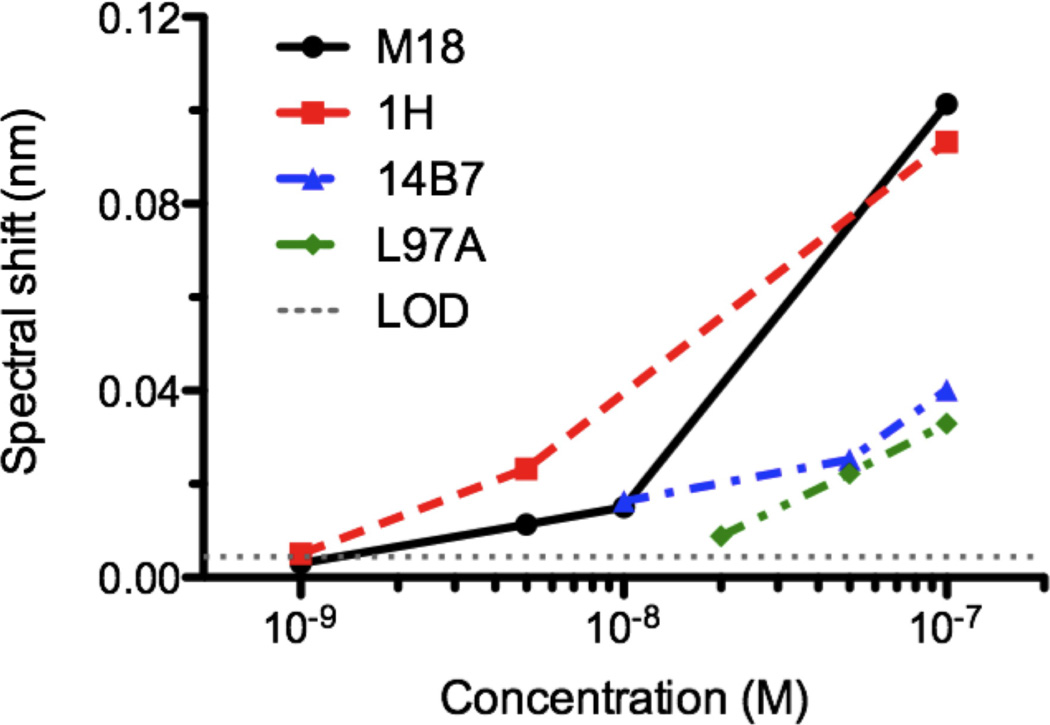
Summary of kinetic values for 14B7 scFv family measured by the nanohole SPR system. The kinetic parameters (kd, ka, and Kd) are calculated from kinetic curves shown in Figure 5. The error terms in kd and ka are from variations in curve fitting. The mass action numbers (Ma#), defined as Kd/LOD concentration, are calculated based on the spectral response to a concentration assay shown in Figure 6.
|
kd (× 10−4 s−1) |
ka (× 105 M−1 s−1) |
Kd (nM) |
LOD (nM) |
Ma# | |
|---|---|---|---|---|---|
| L97A | 101 ± 30 | 2.76 ± 0.43 | 36.7 | 20 | 1.9 |
| 14B7 | 34 ± 0.7 | 2.92 ± 0.23 | 11.7 | 10 | 1.1 |
| 1H | 1.6 ± 0.1 | 5.47 ± 0.37 | 0.29 | 1 | 0.3 |
| M18 | 1.1 ± 0.1 | 5.97 ± 0.38 | 0.19 | 1 | 0.2 |
For SPR systems, device signal (monitored here as spectral shift) depends on the amount of the analyte bound to the sensor surface (Figure 6). Since antibody-antigen interactions are driven by reversible mass action kinetics, the concentration of scFv analyte bound to the surface at equilibrium can be calculated as a function of the injected antibody concentration, the concentration of immobilized antigen and the equilibrium dissociation constant: [Ab:Ag] = [Ab][Ag]/Kd. The observed LOD concentration, or lowest concentration of a particular antibody which can be detected with the SPR instrument, captures the combined effects of antibody molecular size, immobilized antigen concentration and binding affinity. To describe the sensitivity of the SPR sensor to these factors, we define a dimensionless “mass action” number, Ma# = Kd/ LOD concentration. For the nanohole SPR instrument and scFv-PA interactions described here, the Ma# is approximately one (Table 1), with error attributed to uncertainty in measuring the LOD concentration. This Ma# can be used to guide experimental design for analysis of other pairs of binding molecules. For instance, if we next wanted to measure binding of an EE peptide-specific scFv41 with an expected affinity of 76 nM, an Ma# = 1 suggests that the LOD for this new system will be near 76 nM and thus the antibody should be prepared at concentrations ranging from 76 to 760 nM. Similarly, if we wanted to next analyze the 150 kDa 14B7 IgG, we would expect the LOD to increase roughly in proportion to the six-fold increase in molecular size, with a new Ma# = 6. Since the 14B7 IgG also has an affinity of ~11 nM, to determine binding rates with the nanohole SPR sensor, 14B7 concentrations as low as 2 nM would be indicated. This LOD value is at least an order of magnitude lower than previously reported values from EOT-based SPR sensors.20 Furthermore, while the kd value at the 10−4 sec−1 level could not be determined in the previous work because of the baseline drift20, here the problem is addressed by improving the instrumental resolution with the use of large-area gold nanohole arrays and rapid temporal averaging of the EOT spectra.
Figure 6.
Spectral response to a concentration assay of four scFv antibodies, which bind to PA with different dissociation rate constant (kd) values in various concentrations ranging from 1 nM to 100 nM.
CONCLUSION
We have demonstrated a nanohole-based SPR instrument and quantified kinetic parameters for scFv antibodies binding to their target ligand. Despite its simple optical and mechanical design based on a microscope, an off-the-shelf, uncooled portable spectrometer, and home-built PDMS microfluidic systems, this SPR instrument can resolve refractive index changes down to 3.1 × 10−6 RIU via rapid frame averaging without requiring a temperature-controlled chip or light source. A prerequisite for the fast data acquisition is the production of large-area gold nanohole arrays with homogeneous optical properties, which is accomplished via template stripping. Surface modification protocols and antibody injection conditions have been optimized to minimize mass-transport artifacts during kinetic measurements. Surface immobilization of PA molecules (83 kDa) was measured in real time with the signal-to-noise ratio of 860. Furthermore, this instrument is capable of quantifying binding kinetics of small antibody fragments (25 kDa) at concentrations as low as 1 nM and measuring kinetic parameters with dissociation constants ranging from 200 pM to 40 nM and dissociation rate constants ranging from 10−2 to 10−4 sec−1. Because this SPR instrument is built around a standard microscope and off-the-shelf optical components, it can be readily duplicated by other researchers and provides flexibility to integrate with a variety of sensor chips and optics, including periodic nanoholes, random array of nanoholes, gratings, as well as with high-resolution SPR imaging techniques.42–46 Furthermore, the nanohole systems enable facile integration of membrane-bound receptors using supported lipid membranes,22 pore-spanning lipid membranes,14,47 tethered lipid vesicles,48 or natural cell membranes,49 making them suitable for membrane receptor screening – an important task for new drug discovery.
ACKNOWLEDGMENT
This work was supported by grants to J.A.M. and S.-H.O. from the National Science Foundation (DBI-0964216 and DBI-0964137). J.A.M. also received support from the NIH (1R01GM095638). S.-H.O. also acknowledges the NSF CAREER Award, the Office of Naval Research (ONR) Young Investigator Award, and NIH R01 GM 092993. H.I. acknowledges a 3M Science and Technology Fellowship. Device fabrication was performed at the U. of Minnesota Nanofabrication Center (NFC), which receives support from NSF through the National Nanotechnology Infrastructure Network.
REFERENCES AND NOTES
- 1.Uetz P, et al. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 2.Phizicky E, Bastiaens PIH, Zhu H, Snyder M, Fields S. Nature. 2003;422:208–215. doi: 10.1038/nature01512. [DOI] [PubMed] [Google Scholar]
- 3.Wang PI, Marcotte EM. J. Proteomics. 2010;73:2277–2289. doi: 10.1016/j.jprot.2010.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Steckbeck JD, Orlov I, Chow A, Grieser H, Miller K, Bruno J, Robinson JE, Montelaro RC, Cole KS. J. Virol. 2005;79:12311–12320. doi: 10.1128/JVI.79.19.12311-12320.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu H, Pfarr DS, Johnson S, Brewah YA, Woods RM, Patel NK, White WI, Young JF, Kiener PA. J. Mol. Biol. 2007;368:652–665. doi: 10.1016/j.jmb.2007.02.024. [DOI] [PubMed] [Google Scholar]
- 6.Maynard JA, Maassen CBM, Leppla SH, Brasky K, Patterson JL, Iverson BL, Georgiou G. Nat. Biotechnol. 2002;20:597–601. doi: 10.1038/nbt0602-597. [DOI] [PubMed] [Google Scholar]
- 7.Phizicky EM, Fields S. Microbiol. Rev. 1995;59:94–123. doi: 10.1128/mr.59.1.94-123.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ramachandran N, Larson DN, Stark PRH, Hainsworth E, LaBaer J. FEBS J. 2005;272:5412–5425. doi: 10.1111/j.1742-4658.2005.04971.x. [DOI] [PubMed] [Google Scholar]
- 9.Chan SM, Ermann J, Su L, Fathman CG, Utz PJ. Nat. Med. 2004;10:1390–1396. doi: 10.1038/nm1139. [DOI] [PubMed] [Google Scholar]
- 10.Homola J. Chem. Rev. 2008;108:462–493. doi: 10.1021/cr068107d. [DOI] [PubMed] [Google Scholar]
- 11.Wassaf D, Kuang G, Kopacz K, Wu Q-L, Nguyen Q, Toews M, Cosic J, Jacques J, Wiltshire S, Lambert J, Pazmany CC, Hogan S, Ladner RC, Nixon AE, Sexton DJ. Anal. Biochem. 2006;351:241–253. doi: 10.1016/j.ab.2006.01.043. [DOI] [PubMed] [Google Scholar]
- 12.Rich RL, Myszka DG. Anal. Biochem. 2007;361:1–6. doi: 10.1016/j.ab.2006.10.040. [DOI] [PubMed] [Google Scholar]
- 13.Cooper MA. J. Mol. Recognit. 2004;17:286–315. doi: 10.1002/jmr.675. [DOI] [PubMed] [Google Scholar]
- 14.Im H, Wittenberg NJ, Lesuffleur A, Lindquist NC, Oh S-H. Chem. Sci. 2010;1:688–696. doi: 10.1039/C0SC00365D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ebbesen TW, Lezec HJ, Ghaemi HF, Thio T, Wolff PA. Nature. 1998;391:667–669. [Google Scholar]
- 16.Brolo AG, Gordon R, Leathem B, Kavanagh KL. Langmuir. 2004;20:4813–4815. doi: 10.1021/la0493621. [DOI] [PubMed] [Google Scholar]
- 17.Stark PRH, Halleck AE, Larson DN. Methods. 2005;37:37–47. doi: 10.1016/j.ymeth.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 18.Tetz Ka, Pang L, Fainman Y. Opt. Lett. 2006;31:1528–1530. doi: 10.1364/ol.31.001528. [DOI] [PubMed] [Google Scholar]
- 19.Lesuffleur A, Im H, Lindquist NC, Oh S-H. Appl. Phys. Lett. 2007;90:243110. [Google Scholar]
- 20.Yang J-C, Ji J, Hogle JM, Larson DN. Nano Lett. 2008;8:2718–2724. doi: 10.1021/nl801043t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Menezes JW, Ferreira J, Santos MJL, Cescato L, Brolo AG. Adv. Funct. Mater. 2010;20:3918–3924. [Google Scholar]
- 22.Dahlin A, Zäch M, Rindzevicius T, Käll M, Sutherland DS, Höök F. J. Am. Chem. Soc. 2005;127:5043–5048. doi: 10.1021/ja043672o. [DOI] [PubMed] [Google Scholar]
- 23.Eftekhari F, Escobedo C, Ferreira J, Duan X, Girotto EM, Brolo AG, Gordon R, Sinton D. Anal. Chem. 2009;81:4308–4311. doi: 10.1021/ac900221y. [DOI] [PubMed] [Google Scholar]
- 24.Jonsson MP, Dahlin AB, Feuz L, Petronis S, Höök F. Anal. Chem. 2010;82:2087–2094. doi: 10.1021/ac902925e. [DOI] [PubMed] [Google Scholar]
- 25.Höök F, Stengel G, Dahlin AB, Gunnarsson A, Jonsson MP, Jönsson P, Reimhult E, Simonsson L, Svedhem S. Biointerphases. 2008;3:FA108. doi: 10.1116/1.2948313. [DOI] [PubMed] [Google Scholar]
- 26.Stewart ME, Mack NH, Malyarchuk V, Soares JANT, Lee T-W, Gray SK, Nuzzo RG, Rogers JA. Proc Natl Acad Sci USA. 2006;103:17143–17148. doi: 10.1073/pnas.0606216103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Henzie J, Lee MH, Odom TW. Nat. Nanotechnol. 2007;2:549–554. doi: 10.1038/nnano.2007.252. [DOI] [PubMed] [Google Scholar]
- 28.Im H, Lee SH, Wittenberg NJ, Johnson TW, Lindquist NC, Nagpal P, Norris DJ, Oh S-H. ACS Nano. 2011;5:6244–6253. doi: 10.1021/nn202013v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nagpal P, Lindquist NC, Oh S-H, Norris DJ. Science. 2009;325:594–597. doi: 10.1126/science.1174655. [DOI] [PubMed] [Google Scholar]
- 30.Xia Y, Whitesides GM. Angew. Chem. Int. Ed. 1998;37:550–575. doi: 10.1002/(SICI)1521-3773(19980316)37:5<550::AID-ANIE550>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 31.Garcia-Vidal FJ, Martin-Moreno L, Ebbesen TW, Kuipers L. Rev. Mod. Phys. 2010;82:729–787. [Google Scholar]
- 32.Dahlin AB, Tegenfeldt JO, Höök F. Anal. Chem. 2006;78:4416–4423. doi: 10.1021/ac0601967. [DOI] [PubMed] [Google Scholar]
- 33.Kowalski GJ, Talakoub A, Ji J, O’Connell JG, Sen M, Larson D. Opt. Eng. 2009;48:104402. [Google Scholar]
- 34.Hall WP, Anker JN, Lin Y, Modica J, Mrksich M, Van Duyne RP. J. Am. Chem. Soc. 2008;130:5836–5837. doi: 10.1021/ja7109037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harvey BR, Georgiou G, Hayhurst A, Jeong KJ, Iverson BL, Rogers GK. Proc. Natl. Acad. Sci. U.S.A. 2004;101:9193–9198. doi: 10.1073/pnas.0400187101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sivasubramanian A, Maynard JA, Gray JJ. Proteins. 2008;70:218–230. doi: 10.1002/prot.21595. [DOI] [PubMed] [Google Scholar]
- 37.Leysath CE, Monzingo AF, Maynard Ja, Barnett J, Georgiou G, Iverson BL, Robertus JD. J. Mol. Biol. 2009;387:680–693. doi: 10.1016/j.jmb.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jung LS, Campbell CT, Chinowsky TM, Mar MN, Yee SS. Langmuir. 1998;14:5636–5648. [Google Scholar]
- 39.O’Shannessy DJ, Brigham-Burke M, Soneson KK, Hensley P, Brooks I. Anal. Biochem. 1993;212:457–468. doi: 10.1006/abio.1993.1355. [DOI] [PubMed] [Google Scholar]
- 40.Wu H, Pfarr DS, Tang Y, An L-L, Patel NK, Watkins JD, Huse WD, Kiener PA, Young JF. J. Mol. Biol. 2005;350:126–144. doi: 10.1016/j.jmb.2005.04.049. [DOI] [PubMed] [Google Scholar]
- 41.Pai JC, Culver JA, Drury JE, Motani RS, Lieberman RL, Maynard JA. Protein. 2011;24:419–428. doi: 10.1093/protein/gzq120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith EA, Corn RM. Appl. Spectrosc. 2003;57:320A–332A. doi: 10.1366/000370203322554446. [DOI] [PubMed] [Google Scholar]
- 43.Shumaker-Parry JS, Aebersold R, Campbell CT. Anal. Chem. 2004;76:2071–2082. doi: 10.1021/ac035159j. [DOI] [PubMed] [Google Scholar]
- 44.Ji J, O’Connell JG, Carter DJD, Larson DN. Anal. Chem. 2008;80:2491–2498. doi: 10.1021/ac7023206. [DOI] [PubMed] [Google Scholar]
- 45.Im H, Lesuffleur A, Lindquist NC, Oh S-H. Anal. Chem. 2009;81:2854–2859. doi: 10.1021/ac802276x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lindquist NC, Lesuffleur A, Im H, Oh S-H. Lab Chip. 2009;9:382–387. doi: 10.1039/b816735d. [DOI] [PubMed] [Google Scholar]
- 47.Lazzara TD, Carnarius C, Kocun M, Janshoff A, Steinem C. ACS Nano. 2011;5:6935–6944. doi: 10.1021/nn201266e. [DOI] [PubMed] [Google Scholar]
- 48.Dahlin AB, Jonsson MP, Höök F. Adv. Mater. 2008;20:1436–1442. [Google Scholar]
- 49.Wittenberg NJ, Im H, Johnson TW, Xu X, Warrington AE, Rodriguez M, Oh S-H. ACS Nano. 2011;5:7555–7564. doi: 10.1021/nn202554t. [DOI] [PMC free article] [PubMed] [Google Scholar]