Background: PUF protein RNA recognition is critical for target gene regulation.
Results: A chemically conserved binding pocket in a subset of PUF proteins recognizes cytosine at different positions upstream of the core PUF recognition sequence.
Conclusion: A specialized cytosine-binding pocket introduces qualitative and quantitative differences in RNA recognition by PUF proteins.
Significance: Simple adaptations can diversify PUF protein RNA recognition.
Keywords: Gene Regulation, RNA, RNA Metabolism, RNA-binding Proteins, X-ray Crystallography, PUF Proteins
Abstract
mRNA control networks depend on recognition of specific RNA sequences. Pumilio-fem-3 mRNA binding factor (PUF) RNA-binding proteins achieve that specificity through variations on a conserved scaffold. Saccharomyces cerevisiae Puf3p achieves specificity through an additional binding pocket for a cytosine base upstream of the core RNA recognition site. Here we demonstrate that this chemically simple adaptation is prevalent and contributes to the diversity of RNA specificities among PUF proteins. Bioinformatics analysis shows that mRNAs associated with Caenorhabditis elegans fem-3 mRNA binding factor (FBF)-2 in vivo contain an upstream cytosine required for biological regulation. Crystal structures of FBF-2 and C. elegans PUF-6 reveal binding pockets structurally similar to that of Puf3p, whereas sequence alignments predict a pocket in PUF-11. For Puf3p, FBF-2, PUF-6, and PUF-11, the upstream pockets and a cytosine are required for maximal binding to RNA, but the quantitative impact on binding affinity varies. Furthermore, the position of the upstream cytosine relative to the core PUF recognition site can differ, which in the case of FBF-2 originally masked the identification of this consensus sequence feature. Importantly, other PUF proteins lack the pocket and so do not discriminate upstream bases. A structure-based alignment reveals that these proteins lack key residues that would contact the cytosine, and in some instances, they also present amino acid side chains that interfere with binding. Loss of the pocket requires only substitution of one serine, as appears to have occurred during the evolution of certain fungal species.
Introduction
mRNA control is pervasive. Translation, stability, and localization of many mRNAs are governed by elements in their 3′ untranslated regions (3′UTRs)3 (1). The specificity of interactions between 3′UTRs and regulatory proteins underlie networks of control, enabling coordinate regulation of functionally related mRNAs (2–4). The RNA sequences recognized are often single-stranded, requiring discrimination of a specific series of nucleotides rather than folded structures.
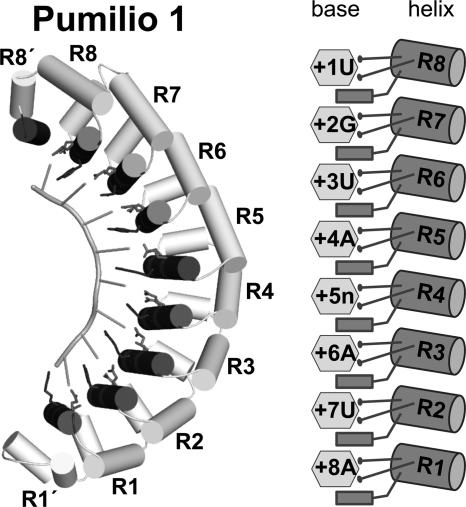
The PUF family of proteins regulates mRNAs using a common polypeptide scaffold. They comprise a series of α-helical repeats arranged along an arc (Fig. 1) (5, 6). A ladder of α-helices, the so-called RNA recognition helices, lie on the concave face of the protein (7). Each helix contacts predominantly one base, using two amino acid side chains to make edge-on contacts and another to stack between adjacent bases (Fig. 1). The simplest condition, exemplified by human Pumilio 1, uses eight helices to recognize eight RNA bases (7).
FIGURE 1.
PUF protein-RNA interaction. Ribbon diagram of human Pumilio 1 in complex with 5′-UGUAUAUA-3′ NRE1 RNA (left panel, PDB code 3Q0P) and schematic representation of the protein-RNA interaction of repeats R1-R8 (right panel). RNA recognition helices are colored dark gray, and RNA-interacting side chains are shown as stick models.
Variations of the PUF scaffold enable different PUF proteins to discriminate unique groups of mRNAs, even though the proteins use very similar sets of atomic contacts. In FBF-2, for example, a distortion in the central region of the protein requires the presence of an “extra” base relative to Pumilio (8). Yeast Puf4p also demands an extra base but at a different location and with distinct structural changes (4, 9). In these cases, the key base does not contact the protein but is solvent-exposed, or “flipped.”
FBF-1 and FBF-2 are two nearly identical proteins with highly overlapping functions, collectively referred to as FBF. The core RNA-binding site for FBF, termed the FBF binding element (FBE), is the 9-mer sequence 5′-UGUDHHAUA-3′, where H is A, U, or C, and D is A, G, or U (10). The FBE, as defined in this report, represents the highest affinity sites. It was elucidated using either purified FBF protein in vitro or through yeast three-hybrid assays (10). In vivo, natural FBF binding sites generally conform to this in vitro FBE but include variations with suboptimal affinity.
PUF protein core RNA-binding sites are recognized by RNA recognition helices R1 to R8 (Fig. 1). PUF proteins contain a C-terminal α-helical region, or pseudo-repeat, that contributes an additional helix to the RNA-binding surface. In yeast Puf3p, that pseudo-repeat (called R8′) combines with parts of repeat 8 to form a pocket that binds a cytosine residue two positions 5′ of the core RNA-binding site (11). The presence of a C at that −2 position is required for tight binding to target mRNAs in vitro and for regulation in vivo (11). Most mRNAs associated with Puf3p in vivo possess a C at this position and encode proteins with mitochondrial functions (4). This extra binding pocket can be viewed as a specificity device that enables Puf3p to bind only its own targets (11).
We sought to understand the appearance and loss of upstream C-binding pockets among PUF proteins at the structural level. Analysis of the RNAs associated with FBF-2 in vivo revealed that they contain an upstream C residue critical for binding. Structural analysis of FBF-2 revealed that it possesses a binding pocket chemically similar to that of yeast Puf3p. Closely related pockets were identified in Caenorhabditis elegans PUF-6 and PUF-11, and in each case, the pocket enhanced binding to RNA, although with a unique quantitative impact. In contrast, other PUF proteins lack the pocket and do not discriminate upstream bases. Structure-based sequence alignments of proteins with and without upstream pockets reveal the diagnostic features of the pocket with a critical serine residue that directly contacts the upstream C. The simplicity of the variations required to gain or lose this specificity suggest an opportunity for rapid evolution of new networks of control.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
The RNA-binding domain of FBF-2 (residues Ser-164-Glu-575) was expressed and purified, and protein-RNA complexes were prepared as reported previously (8).
The RNA-binding domain of C. elegans PUF-6 (residues 76–453) was overexpressed in Escherichia coli strain BL21(DE3) with an N-terminal GST tag. Protein expression was induced at 18 °C for 20 h. Cell pellets were lysed in sonication buffer containing 20 mm Tris (pH 8.0), 0.15 m NaCl, 5% (v/v) glycerol, and 1 mm DTT. The fusion protein was purified with glutathione resin, and the GST moiety was removed by tobacco etch virus protease cleavage after elution from the glutathione beads (50 mm Tris (pH 8.0), 50 mm NaCl, 10 mm reduced glutathione). The PUF-6 protein was further purified with a heparin column (20 mm Tris (pH 8.0), 1 mm DTT with a gradient from 0 to 1 m NaCl). For crystallization, the PUF-6–5BE13 RNA (5′-CUCUGUAUCUUGU-3′) complex was purified using a Superdex 200 column (20 mm HEPES (pH 7.4), 0.15 m NaCl, 2 mm DTT).
We also expressed PUF-6 with an N-terminal His6-small ubiquitin-like modifier (SUMO) tag (12). Protein expression was induced at 18 °C for 20 h. Cell pellets were lysed in sonication buffer containing 20 mm Tris (pH 8.0), 0.5 m NaCl, 20 mm imidazole, 5% (v/v) glycerol, and 0.1% (v/v) β-mercaptoethanol. The fusion protein was purified using a nickel-affinity column, and the His6-SUMO tag was removed by Ulp-1 protease cleavage. PUF-6 was further purified with a heparin column and a Superdex 200 column as described above. The final yield from the His6-SUMO-tagged PUF-6 was about 0.5 mg/liter culture, ∼10-fold greater than from the GST fusion.
The RNA-binding domain of C. elegans PUF-11 (residues 118–505) was overexpressed in E. coli strain BL21(DE3) as a fusion protein with an N-terminal His6-SUMO tag. Protein expression was induced at 18 °C for 20 h. Cell pellets were lysed in sonication buffer containing 20 mm HEPES (pH 7.5), 300 mm NaCl, 10 mm imidazole, 10% (v/v) glycerol, and 2 mm DTT. The fusion protein was purified using a nickel-affinity column, and the His6-SUMO tag was removed by Ulp-1 protease cleavage. PUF-11 was further purified with a HiTrap Q HP anion-exchange column (20 mm HEPES (pH 7.5), 50 mm NaCl, 2 mm DTT, 5% (v/v) glycerol, with a gradient from 50 mm to 1 m NaCl) and a Superdex 75 column (20 mm HEPES (pH 7.5), 150 mm NaCl, 2 mm DTT, 5% (v/v) glycerol).
Site-directed mutants, generated using a QuikChange site-directed mutagenesis kit (Agilent), were purified following the same protocols as for wild-type proteins.
Crystallization and Data Collection
Crystals of FBF-2 with wild-type gld-1 FBEa13 RNA (5′-UCAUGUGCCAUAC-3′) were obtained by combining 1 μl of complex (∼2 mg/ml protein concentration) with 1 μl crystallization solution (10% (w/v) PEG 6000, 5% (v/v) (±)-2-methyl-2,4-pentanediol, 0.1 m HEPES (pH 7.5)) at room temperature using the hanging drop vapor diffusion method. Crystals of FBF-2 with the mutant RNA (5′-UACUGUGCCAUAC-3′) were grown with crystallization solution containing 10% (w/v) PEG 8000, 8% (v/v) ethylene glycol, 0.1 m Tris (pH 8.0). Crystals were flash-frozen after being transferred sequentially into modified crystallization solutions containing 5%, 10%, and 20% (v/v) glycerol. Diffraction data were collected at 100 K using a Rigaku Micromax-007HF x-ray generator with Saturn 92 charge-coupled device detector (wavelength 1.5418 Å).
Crystals of PUF-6 with 5BEa13 RNA were grown in hanging drops at 20 °C with crystallization solution (15% (w/v) PEG 3350, 0.1 m succinic acid (pH 7.0)). The crystals were flash-frozen in crystallization solution with 15% (v/v) glycerol. Diffraction data were collected at the Southeast Regional Collaborative Access Team beamline 22-ID (wavelength 1.0 Å) at the Advanced Photon Source, Argonne National Laboratories. Data were indexed and scaled with HKL2000 (13). Data collection and processing statistics are shown in supplemental Table 1.
Structure Determination and Refinement
The crystal structure of the FBF-2-gld-1 FBEa13 complex was determined by molecular replacement using a structure of FBF-2 (PDB code 3K5Y) as the search model. The RNA was excluded from the initial searching and phase calculations, and high-temperature simulated annealing (2500 K) was performed to reduce model bias. The model was refined with CNS (14) and rebuilt manually using O (15). Phenix was employed for addition of water molecules and translation/libration/screw refinement (16). The final FBF-2-gld-1 FBEa13 structure comprises residues Leu-168 to Ser-567 and nucleotides −2C to +9A. The structure of FBF-2 with −1C FBEa13 RNA was determined using the structure of FBF-2-gld-1 FBEa containing nucleotides +1U to +9A as the model and refined as for the complex with gld-1 FBEa13.
The structure of the PUF-6–5BE13 complex was determined by molecular replacement using the protein structure of FBF-2 (3K5Y) as a search model in Phaser (17). Coot (18) and Phenix (16) were used for model building and refinement, respectively. The final model contains PUF-6 residues 82–403 and 409–452 and nucleotides −1C to +6C. All structures were analyzed with MolProbity (19). All torsion angles are within allowed regions of the Ramachandran plot, and ≥ 97% are in the most favored regions. Refinement statistics are presented in supplemental Table 1.
Electrophoretic Mobility Shift Assays
Equilibrium dissociation constants were determined as reported previously (8). Briefly, radiolabeled RNA oligonucleotides (100 pm for FBF-2 and PUF-6, 10 pm for PUF-11) and serially diluted protein were incubated in a buffer containing 10 mm HEPES (pH 7.4), 50 mm NaCl, 0.1 mg/ml BSA, 0.01% (v/v) Tween 20, 0.1 mg/ml yeast tRNA, 1 mm EDTA, and 1 mm DTT. After addition of Ficoll 400 to 2.5% (v/v), reaction mixtures were run on 10% polyacrylamide gels. The apparent dissociation constants, mean, and S.E. were calculated using GraphPad Prism (Graphpad LLC) by fitting data from at least three independent experiments using non-linear regression with a one-site, specific binding model. The percentage of active protein was determined as described previously (20). Wild-type and mutant FBF-2 and PUF-6 were ∼93% active. Wild-type PUF-11 was ∼98 and ∼99% active in different assays, and PUF-11 S491A was ∼89% active. Corresponding adjustments were made to Kd values in supplemental Tables 2–4.
Computational Analyses
C. elegans 3′UTR sequences (WS190) were downloaded from BioMart and analyzed as described in the text.
RESULTS
FBF-2 Target mRNAs in vivo Possess an Upstream −1 or −2C
To define sequences responsible for FBF binding to its target mRNAs in vivo, we identified mRNAs selected from those associated with FBF in C. elegans extracts (21). In that study, FBF transgenes had been created in C. elegans and were used to perform immunoprecipitation (IP) of FBF followed by RNA microarray analysis (RIP chip). These studies identified 1350 putative FBF target mRNAs, defined by their enrichment in the FBF IP compared with the negative control IP.
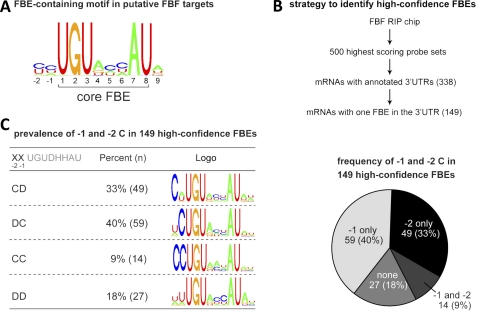
To identify possible FBF binding sites, we first performed an unbiased search for RNA sequence motifs enriched in the 3′UTRs of the 200 mRNAs showing the highest enrichment in the FBF IP versus control IP. The de novo motif-finding tool Cosmo (22) revealed a motif containing the FBE that also suggested a preference for additional nucleotides flanking the core FBE, including an upstream C at positions −1 and −2 (Fig. 2A, see “Experimental Procedures”). To examine the −1 and −2 positions in greater depth, we identified high-confidence FBF binding sites as outlined in Fig. 2B. We focused on mRNAs for the 500 probe sets that were most enriched in the FBF RIP chip study. This group contained several previously validated FBF targets. In addition, this group contained a significant enrichment (p < 10−90) of FBE-bearing transcripts compared with all genes. 82% of these mRNAs contained an FBE in the mRNA 3′UTR compared with only 29% for all C. elegans mRNAs (21). Among these 500, we considered further only the 338 unique, unambiguous mRNAs with annotated 3′UTRs. We eliminated mRNAs that possessed more than one putative FBE, as we could not discern which elements were functional in vivo. This scheme yielded 149 likely FBF targets with single FBEs in their 3′UTRs.
FIGURE 2.
Enrichment of an upstream cytosine in FBF binding sites. A, consensus motif enriched in putative FBF target mRNA 3′UTRs. 3′UTRs for the mRNAs corresponding to the 200 most highly enriched probe sets in the FBF RIP chip (21) were analyzed using MEME. Shown is part of a motif identified using MEME that contains an FBE. B, scheme for identifying high-confidence FBF binding sites. The highest-confidence FBF binding sites likely correspond to FBEs in putative FBF target 3′UTRs with exactly one consensus FBE. The 500 highest scoring probe set in the FBF RIP chip corresponded to 317 unique mRNAs with annotated 3′UTRs. Of these, 149 had only one FBE. C, proportion of high confidence FBEs with Cs at position −1 and/or −2.
The majority (82%) of high-confidence FBF binding sites contained a C at either the −1 or −2 position or both (Fig. 2C). 73% contained a single C, whereas 9% contained a C at both positions. We conclude that high-confidence in vivo FBF binding sites are enriched for a C at either position −1 or −2 upstream of the core PUF binding site. Similarly, mRNAs encoding synaptonemal complex proteins, shown independently to be FBF targets, contain a −2C (23).
To discern any additional sequence patterns, we used Cosmo (22) to determine motifs for four groups of FBEs, those with C only at −2 (CD), those with C only at −1 (DC), those with C at both −1 and −2 (CC), and those with no C at −1 or −2 (DD). The data yielded several patterns (Fig. 2C). All motifs contained a UGU at positions +1 to +3 and an AU dinucleotide at positions +7 and +8, matching the consensus FBE sequence. Of the 63 FBEs with a −2C, 24 have −1A, 21 have −1U, 14 have −1C, and 4 have −1G, indicating that any base may be acceptable, but a preference is observed for −1A, U, or C. RNAs without a −1 or −2C were enriched for a C at position +6 and an A at position +9, as compared with all other FBEs. These observations suggest that the presence of a C at either −1 or −2 is conserved among many FBF target sites. Earlier work that analyzed naturally occurring mutations in fem-3 mRNA noted that a C at what we now understand is the −2 position was required for regulation in vivo (24). In the absence of an upstream C, nucleotides at other positions may compensate to increase affinity, as RNAs with +6C or +9A bind more tightly to FBF-2 than RNAs with other bases at these positions (8).
An Upstream C Is Required for Tight Binding by FBF
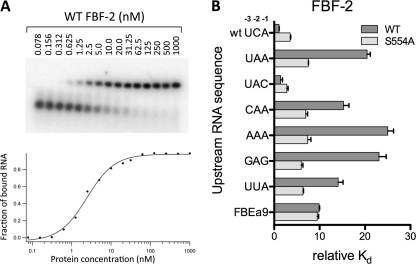
To evaluate the functional significance of an upstream C at the −1 or −2 position, we determined the in vitro binding affinity of FBF-2 for target sequences in the gld-1 3′UTR. The 3′UTR of the gld-1 mRNA contains a well defined FBF binding site with a −2C, termed FBEa (5′-UCAUGUGCCAUAC-3′, −2C is shown in boldface). We measured the binding affinity to 9-mer (+1 to +9) and 13-mer (-3 to +10) RNAs derived from the gld-1 FBEa using electrophoretic mobility shift assays (Fig. 3 and supplemental Table 2). The 9-mer RNA (FBEa9) represents the conserved core FBF binding sequence beginning with a 5′UGU (underlined above), and the 13-mer RNAs (FBEa13) contain three additional nucleotides upstream and one additional nucleotide downstream of the 9-mer core sequences. FBF-2 binds the FBEa13 RNA (WT UCA) ∼9-fold more tightly than the FBEa9 RNA (Kd 3 nm versus 28 nm).
FIGURE 3.
A cytosine base upstream of the FBE core recognition sequence is important for FBF-2 binding affinity. A, representative electrophoretic mobility shift assay. B, relative binding affinities of FBF-2 (wild-type and S554A mutant) for RNAs with varied upstream sequences.
To determine whether this increased binding affinity was due to the presence of the upstream −2C or merely stronger binding to a longer RNA, we tested binding to 13-mer RNAs with mutated sequences in the upstream region. Changing C to A at position −2 (upstream UCA to UAA) decreased affinity ∼19-fold (Fig. 3B). However, binding affinity was restored by insertion of a C at position −1 (upstream UAA to UAC), indicating that a C at either −1 or −2 was sufficient for higher affinity binding and that this higher affinity was not simply due to a longer RNA. A C at position −3 (upstream UAA to CAA) resulted in only a modest increase in affinity (Kd of 43 versus 57 nm). Other RNAs lacking a −1 or −2C with three consecutive purines (upstream AAA or GAG) bound ∼22-fold more weakly than the wild-type RNA. Similarly, an RNA with a −2U (upstream UUA) bound ∼13-fold more weakly than the wild-type RNA. The RNAs lacking a −1 or −2C in the upstream sequence bound more weakly than the 9-mer RNA without an upstream sequence, suggesting that the presence of non-cognate upstream bases interfere with binding to FBF-2.
We also measured binding of FBF-2 to 13-mer natural target sequences in fem-3, fog-1, and a second site in gld-1 (FBEb). These sequences differ from the gld-1 FBEa in upstream sequence as well as other varied positions in the core FBE. All sequences bound with similar affinity as gld-1 FBEa13 (supplemental Table 2). Together these binding data suggest that in different contexts a −1 or −2C promotes tighter binding of FBF-2.
Structure of the FBF-2 Upstream C-binding Pocket
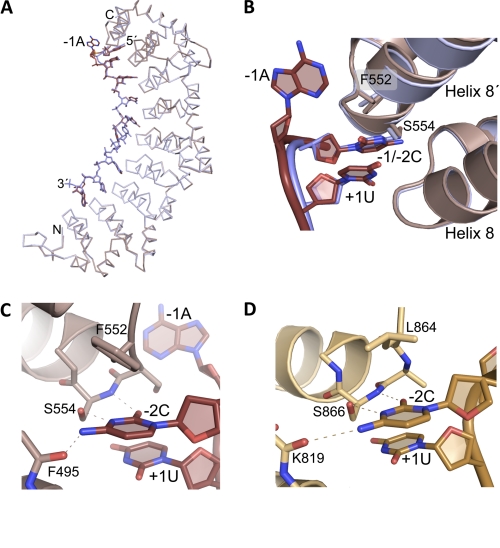
To understand the molecular basis for the interaction of an upstream C with FBF-2, we determined the crystal structures of FBF-2 in complex with a wild-type gld-1 FBEa13 (5′-UCAUGUGCCAUAC-3′) possessing a −2C or a mutant FBEa13 (5′-UACUGUGCCAUAC-3′) with a −1C. The protein structures in these two complexes and in complex with the 9-mer gld-1 FBEa9 (PDB code 3K5Y) are unchanged (root mean square deviation of 0.3–0.7 Å over 394 Cα atoms). The RNA from positions +1 to +9 in these three structures can be superimposed (Fig. 4A). In the structures of FBF-2 in complex with the two 13-mer RNAs, the upstream C, whether at the −1 or −2 position, is bound in a pocket between the last RNA-binding repeat 8 and the C-terminal helix, termed 8′ (Fig. 4B). Modest changes in the RNA backbone conformations allow accommodation of a −1 or −2C. In the structure with wild-type gld-1 FBEa13 RNA with a −2C, the −1A base between the −2C and +1U is not contacted by the protein.
FIGURE 4.
A specific C-binding pocket near the C terminus of FBF-2. A, superposition of Cα traces of structures of FBF-2 in complex with RNAs containing a −2C (mauve) or −1C (blue). B, close-up view of the FBF-2 upstream C-binding pocket. Shown are ribbon diagrams of FBF-2 in complex with −2C or −1C RNAs as shown in A. Nitrogen atoms are blue and oxygen atoms are red. C, interactions of FBF-2 with a −2C. Dashed lines indicate interactions between FBF-2 and −2C. D, interactions of Puf3p with a −2C. Dashed lines indicate interactions between Puf3p and −2C. RNAs are shown in a darker shade for B–D.
FBF-2 makes specific contacts with the Watson-Crick edge of the upstream C through hydrogen bonds with main chain atoms of Phe-495 and Ser-554 and the side chain of Ser-554 (Fig. 4C). To explore the importance of FBF-2 Ser-554 for specific recognition of the upstream C, we mutated Ser-554 to alanine and determined the binding affinity of the mutant protein for RNAs with and without an upstream C. As a control, the mutant protein bound to the 9-mer gld-1 FBEa9 with similar affinity to wild-type protein (Fig. 3B and supplemental Table 2, suggesting that the mutant is properly folded and residue Ser-554 is not involved in interacting with the 9-nucleotide core sequence. The S554A mutant protein bound 2- to 3-fold more weakly than wild-type protein to RNAs with −1 or −2C (Fig. 3B and supplemental Table 2), indicating that Ser-554 contributes to binding to the upstream C. The S554A mutant protein also bound ∼2-fold more weakly to RNAs lacking either a −1 or −2C (Kd 17–21 nm) than to RNAs with a −1 or −2C (Kd 8–10 nm), suggesting that even without Ser-554, the upstream C-binding pocket retains modest base selectivity. However, the selectivity is decreased compared with the 20-fold difference observed for wild-type protein with the same RNAs. Consistent with this, the S554A mutant protein bound ∼3-fold more tightly to RNAs lacking an upstream C (Kd ∼19 nm versus ∼50 nm for wild-type protein). On the basis of the crystal structures, we expect a purine base at the −2 position (as is present in the weaker-binding non-C containing RNA mutants) would clash sterically with the serine side chain of Ser-554 in the C-binding pocket, but not with that of an alanine. Thus S554A allows accommodation of a purine and tighter binding to the mutant RNAs.
Conservation of the Upstream C-binding Pocket
Crystal structures of yeast Puf3p in complex with COX17 RNAs have shown a similar binding pocket for a conserved C at the −2 position (11). Interaction between the upstream C and the protein is similar in FBF-2 and Puf3p (Fig. 4D). A serine residue at the beginning of the C-terminal helix (Ser-554 in FBF-2 and Ser-866 in Puf3p) makes specific hydrogen bonds with the upstream C. In addition, both FBF-2 and Puf3p utilize main chain atoms to form hydrogen bonds with the upstream C. Residue Leu-864 in Puf3p forms a stacking interaction with the −2C. Phe-552 in FBF-2 occupies the equivalent position, and its aromatic ring is positioned in a non-parallel orientation relative to the upstream C.
Using the FBF-2 and Puf3p structures, we created a structure-based amino acid sequence alignment of the C-terminal regions to identify equivalent binding pockets in other PUF proteins (Fig. 5A). Earlier sequence-based searches suggested the presence of such a binding pocket in only a limited number of other yeast family PUF proteins (11). The new structure-guided sequence alignment in this region revealed that the C-binding serine is conserved in C. elegans FBF-1/2, PUF-5/6, and PUF-3/11 but not in PUF-8/9. The consensus recognition sequences for these families of PUF proteins suggest a conserved −1C in the 5BE recognized by PUF-5/6 (25) and in some sequences recognized by PUF-11 (26). In yeast Puf4p and human Pumilio 1, the C-binding serine is replaced by a histidine or tyrosine side chain, which occludes this binding pocket (11). On the basis of this sequence alignment, we predict that the bulky side chains in C. elegans PUF-8, yeast Mpt5p, and Arabidopsis PUF proteins also prevent an equivalent upstream C-binding pocket in these proteins. Thus, a small change in amino acid sequence may create a new binding pocket and change specificity.
FIGURE 5.
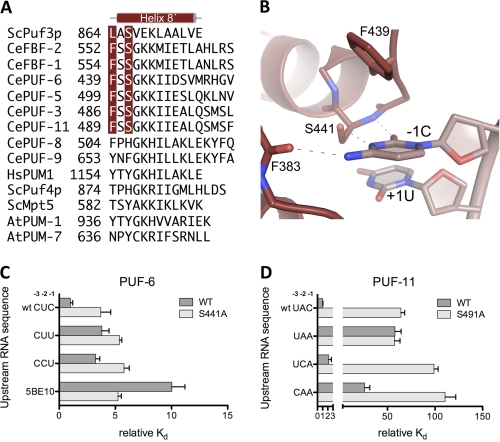
Upstream C-binding pockets in PUF proteins. A, structure-assisted sequence alignment of C-terminal sequences of selected PUF proteins. The aligned crystal structures of S. cerevisiae Puf3p, C. elegans FBF-2, human Pumilio 1, and S. cerevisiae Puf4p were used to generate a sequence alignment. Other PUF protein sequences were aligned manually. Residues in position to interact with upstream C residues are indicated in red. B, close-up view of the upstream C-binding pocket in the crystal structure of C. elegans PUF-6. A ribbon diagram of PUF-6 in complex with −1C RNA is shown. Dashed lines indicate contacts between PUF-6 and −1C. RNA is shown in a lighter shade. C, relative binding affinities of PUF-6 (wild-type and S441A mutant) for RNAs with varied upstream sequences. D, relative binding affinities of PUF-11 (wild-type and S491A mutant) for RNAs with varied upstream sequences.
C. elegans PUF-6 Upstream C-binding Pocket Is Restricted to −1C
To test our prediction of an upstream C-binding pocket in PUF-6, we determined the crystal structure of C. elegans PUF-6 in complex with a 13-mer RNA containing its optimal binding element, 5BE (5BE-13, 5′-CUCUGUAUCUUGU-3′). The overall structure of PUF-6 is similar to that of other PUF protein structures, with RNA bound on the concave surface of the protein (supplemental Fig. 1A). We were able to build a model for bases −1C to +6C of the 5BE RNA, seven of the 13 bases in the RNA sequence. We observed only discontinuous electron density for bases +7U to +10U, indicating disorder in this region. The RNA structure in the PUF-6–5BE complex is similar to that of FBF-2 in the central region (supplemental Fig. 1B). Bases 4–6 stack with each other and turn away from the RNA-binding surface of the protein (supplemental Fig. 1C). However, residue Arg-256 in PUF-6 forms a hydrogen bond with +6C, and the base at position +5 is not contacted by the protein, whereas the corresponding residue Arg-364 in FBF-2 often contacts the +5 base, and the base at position +6 is not contacted (supplemental Fig. 1D). The orientation of RNA-interacting helices in repeats 1–4 of PUF-6 differs from those of FBF-2 (supplemental Fig. 1B), consistent with the different recognition sequences of the two proteins.
As predicted, the crystal structure of PUF-6 confirms the presence of an upstream C-binding pocket. PUF-6 shares the same sequence motif, “FSSGKK,” in the C-terminal helix as FBF-2. Thus the binding pocket for the upstream C in PUF-6 is almost identical to the pocket in FBF-2 (Fig. 5B). The Watson-Crick edge of −1C forms hydrogen bonds with main chain atoms of Phe-383 and Ser-441 and the side chain of Ser-441. Phe-439 also contributes to forming the C-binding pocket.
To probe the importance of the upstream C-binding pocket in PUF-6, we mutated Ser-441 to alanine and determined the RNA-binding activity of the wild-type and mutant proteins. Wild-type PUF-6 bound to 5BE13 RNA with high affinity (Kd = 7.4 nm, Fig. 5C and supplemental Table 3). It bound equally well to 5BE11 RNA, which starts at the −1C (data not shown), indicating that positions −3 and −2 in the RNA sequence make little contribution to the binding. In contrast, PUF-6 bound 10-fold more weakly to 5BE10 RNA, which begins with the 5′UGU and lacks upstream sequences. RNAs with a −2C and/or −3C also bound more weakly than wild-type 5BE13 to PUF-6. Thus, PUF-6 recognizes only an upstream C at position −1. The S441A mutant bound ∼3-fold more weakly than wild-type protein to 5BE13 RNA with a −1C, consistent with the importance of Ser-441 in forming the upstream C-binding pocket.
C. elegans PUF-11 Binds to an Upstream C at the −1 or −2 Position
Among C. elegans PUF proteins, PUF-11 is unusual in its flexibility to recognize three distinct classes of core consensus sequence using at least two different binding modes (26). PUF-11 binds with a Kd of 0.05 nm to a model class I PUF-11 binding sequence (11BE I-1, 5′-UACUGUGAAUAGG-3′) (Fig. 5D and supplemental Table 4). Mutation of the −1C in this sequence to A (upstream UAC to UAA) decreases affinity nearly 60-fold (Kd = 2.9 nm). A −2C (upstream UCA) restores binding affinity similar to that with −1C (Kd = 0.11 nm), but a −3C (upstream CAA) binds with an affinity similar to having no upstream C (Kd = 1.3 nm). Yeast 3-hybrid RNA selection experiments identified 66 unique sequences that associate with PUF-11 (26). Of these 66 sequences, 86% contain a −1 or −2C.
Mutation of the putative C-binding pocket of PUF-11 (S491A) resulted in a protein that bound 11BE I-1 RNA 64-fold weaker than wild-type PUF-11 (Fig. 5D and supplemental Table 4). PUF-11 S491A bound to RNA with no upstream C (upstream UAC to UAA) with an affinity similar to wild-type PUF-11 for the same RNA (Kd = 2.9 nm), and the affinity of the mutant protein for RNA with a −2C or −3C was reduced ∼100-fold below the affinity of wild-type PUF-11 for 11BE I-1 RNA. These data suggest that PUF-11 recognizes an upstream C at position −1 or −2 and that Ser-491 is essential in forming the upstream C-binding pocket of PUF-11.
Divergence of the Upstream C-binding Pocket during Evolution
To examine the evolution of the upstream C-binding pocket, we prepared an alignment of amino acid sequences of Puf3p homologues in 23 fungal species for which Puf3p orthologues have been identified (Fig. 6 and supplemental Fig. 2) (27). The regions predicted to contain R8′ helices are shown in Fig. 6, and longer regions predicted to comprise RNA-binding helices R7, R8, and R8′ are shown in supplemental Fig. 2. Most species possess the equivalent of Ser-866 of S. cerevisiae. However, five species diverge. Three of these species lack a Puf3p with the critical serine within a predicted R8′ helix. These include the fission yeast Schizosaccharomyces japonicus with two duplicated Puf3p genes and Aspergillus nidulans and Neurospora crassa with single Puf3p genes. The two fission yeasts, Schizosaccharomyces octosoporus and Schizosaccharomyces pombe, which also possess duplicated Puf3p genes, encode one copy predicted to have an upstream C-binding pocket and a second (designated “B”) lacking the critical serine. Although the sequence alignment might not detect the appropriate serine residue, a protein fold recognition threading program used to guide alignments near the R8′ helix also did not detect a conserved serine. The simplest hypothesis is that the Puf3p protein ancestral to all five of these species possessed the pocket, which was selectively lost in a minority of descendants.
FIGURE 6.
Sequence alignment of predicted R8′ helices of Puf3p homologues in fungal species. Residues predicted to interact with upstream C residues are indicated in red. Dark and light gray denote 80% identical and 50% similar amino acids, respectively. The sequences of Puf3p species lacking a conserved serine residue were analyzed using structural prediction from GenThreader (35), and the residue that aligns with the C-binding serine is boxed in red. Extended alignments are presented in supplemental Fig. 2.
DISCUSSION
Our identification of high-confidence FBF binding sites in the 3′UTRs of associated RNAs has led to the discovery of an upstream C-binding pocket at the C terminus of FBF, adding an additional element of specificity for this well studied PUF protein. The FBF upstream C-binding pocket shares recognition features with a −2C binding pocket in yeast Puf3p. The upstream C-binding pocket of FBF can recognize either a −1C or −2C with only a modest change in RNA structure. High-confidence FBEs in C. elegans mRNAs contain an upstream C at either position or both. In vitro RNA binding to RNAs without a −1C or −2C is reduced ∼20-fold.
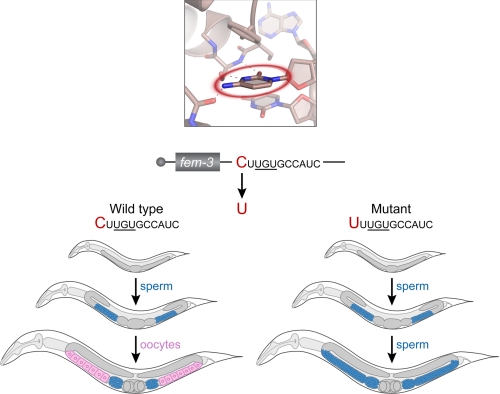
The importance of the upstream pocket in vivo is emphasized by C. elegans mutants obtained through unbiased genetic selections, performed long before the discovery of PUF proteins or upstream pockets (28). Normally C. elegans hermaphrodites first make sperm and then switch to making oocytes (Fig. 7, left panel). Dominant mutants were isolated that prevented the switch to oogenesis (28), and their molecular lesions identified by sequencing (24). These point mutations lay in the 3′UTR of fem-3 mRNA in a region that we now know is the FBF binding element (Fig. 7, right panel). Five independently isolated alleles substituted the C at the −2 position of the binding site with a U. Others altered the G of the UGU sequence (24). This single base substitution at the −2 position prevented FBF repression of fem-3 with dramatic biological consequences: a failure to produce oocytes, and sterility.
FIGURE 7.
fem-3 mutants demonstrate the biological impact of the upstream pocket in C. elegans. Top, the FBF-2 upstream C-binding pocket with the −2C base circled in red. Sequences shown correspond to the now-established FBF binding element in the 3′UTR of fem-3 mRNA, which encodes a protein critical in the switch from spermatogenesis to oogenesis. This regulatory element was first identified through genetic selections (28). Bottom, C. elegans worms at three stages of development. Worms with the wild-type C residue in the upstream site develop normally and switch from making sperm (blue) to making oocytes (pink). Worms with a −2U substitution at the −2 position develop normally but are defective in the switch and make sperm incessantly. Gray indicates undifferentiated germ line cells.
The upstream pocket also guides different PUF proteins exclusively to their own correct mRNA targets in vivo. S. cerevisiae Puf3p binds to more than 150 mRNAs with mitochondrial functions (4). Most (88%) of these mRNAs, and others associated with Puf3p, possess a C at the −2 position (11). Mutations in the −2 position of Puf3p targets disrupt regulation by Puf3p in vivo (11). In contrast, Puf4p lacks an upstream pocket, and the mRNAs it binds in vivo lack a −2C (92%) (4, 11). Thus, the presence of a −2C in mitochondrial mRNAs dictates that they will be regulated by Puf3p, whereas the absence of a −2C from the targets of Puf4p help exclude it (11).
Our analysis emphasizes that simple derivation of consensus motifs can miss essential features of true RNA binding sites. The consensus FBF binding site, deduced by computational analysis of FBF targets in vivo, showed only a modest preference for a C at either positions −1 and −2. Yet FBF-2 has a strong requirement for a C but at either of two positions. Simple consensus motifs combine the two classes of RNAs and so do not capture the requirement. Consensus motifs of certain DNA-binding proteins, such as Gata-4 and homeodomain protein Nkx-2.5, have similar limitations (29) and emphasize the need for other modes of analysis.
Using a sequence alignment on the basis of the crystal structures of the FBF-2 and Puf3p binding pockets, we identified additional C. elegans PUF proteins predicted to possess an upstream C-binding pocket. In vitro binding assays confirmed the importance of an upstream C for PUF-6 and PUF-11 RNA recognition, and a crystal structure of PUF-6 revealed conservation of the upstream C-binding pocket. Thus, what appeared initially to be a yeast PUF protein specialization is instead utilized more broadly.
Although the structures of the binding pockets are conserved, as is the chemical role of a key serine, the pockets of different proteins vary in two important ways. First, the preferred position of the upstream C relative to the 5′UGU motif in the core PUF recognition element varies. FBF-2 and PUF-11 accept a C at either −1 or 2, PUF-6 at only −1, and Puf3p at −2. Second, the quantitative effects of the upstream pocket differ substantially among the proteins. For the C. elegans PUF proteins, mutations that change the upstream C decrease in vitro affinity from 3-fold (PUF-6) to 20-fold (FBF-2) to 60-fold (PUF-11). The general features of these binding pockets are structurally conserved (main chain and serine/hydrophobic side chain interactions), but the specific contacts and conformation of the C-terminal helices of the PUF proteins may be responsible for these differences in affinity and specificity. The structure of FBF-2 is constant when bound to either −1 or −2C RNA.
The upstream pockets described here invariably recognize cytosine residues. Mutant PUF proteins that also recognize a cytosine in the core recognition region have recently been selected with the yeast three-hybrid system (30, 31). However, the chemical and structural basis of cytosine recognition is completely different in those cases versus the upstream pockets described here. The mutant proteins possess alterations within the RNA recognition side chains of a typical core recognition helix, with recognition dominated by interaction with an arginine side chain (31). Although core C-recognizing helices do appear to occur in nature, they are rare (31). In contrast, upstream C pockets appear to be widespread and utilize different chemical interactions to discriminate the cytosine.
Taken together with previous studies, the specificity of each PUF protein is determined by the fusion of RNA recognition features. The central element is a characteristic core recognition sequence. The interaction of this core sequence with PUF repeats 1–8 may be highly sequence-specific (e.g. Pumilio 1) or may contain conserved motifs and binding patterns whose sequence and spacing are critical for specificity (e.g. FBF-2). The presence or absence of additional recognition features, like the upstream C-binding pockets described here, modify specificity. Other factors that contribute to regulatory activity include the binding affinity and selectivity associated with these recognition features and the localization and level of expression of the PUF protein.
Multiple chemical features of the protein-RNA interface act in concert to provide selectivity. Their combinatorial nature provides a foundation for understanding PUF control networks and their evolution. The changes required to modify RNA recognition are surprisingly simple. In the recognition helices, substitutions in one or two amino acids can expand or switch specificity. In the upstream pocket, a single mutation in the serine or cytosine alters affinity (Refs. 7, 20, 30–34 and this paper). Variations in the affinity of a particular PUF protein for a set of target mRNAs could cause their differential regulation, whereas overlaps in specificity between two PUF proteins could allow either their interference or coregulation. In some instances, a minimal affinity threshold might be required for activity, producing a regulatory switch. C. elegans fem-3 mRNA is exemplary. A single nucleotide change in −2 position of its FBF binding site has profound consequences for the animal, including sterility. The identification of in vivo RNA targets and the analysis of the structural basis of selectivity are prerequisites for understanding how new specificities arise, disappear, and create new circuits of control.
Supplementary Material
Acknowledgments
We thank Cary Valley for discussions and creating and testing mutant constructs; Maria Wooten for assistance with site-directed mutagenesis; Kathryne Hawthorne for assistance with PUF-6 protein purification, crystallization, and RNA-binding analyses; and Dr. Lars Pedersen and the staff at the Southeast Regional Collaborative Access Team beamline for help with x-ray data collection. Data were collected at Southeast Regional Collaborative Access Team 22-ID beamline at the Advanced Photon Source, Argonne National Laboratory. Use of the Advanced Photon Source was supported by the United States Department of Energy, Office of Science, Office of Basic Energy Sciences, under contract no. W-31–109-Eng-38.
This work was supported, in whole or in part, by National Institutes of Health Grants GM50942 (to M. W.) and HG003747 (to S. K.). This work was also supported by a Howard Hughes Medical Institute grant (to J. K.) and by the Intramural Research Program of the National Institute of Environmental Health Sciences (to T. M. T. H.).

This article contains supplemental Figs. 1 and 2 and Tables 1–4.
The atomic coordinates and structure factors (codes 3V74, 3V6Y, and 3V71) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- UTR
- untranslated region
- PUF
- Pumilio-fem-3 mRNA binding factor
- FBF
- fem-3 mRNA binding factor
- FBE
- fem-3 mRNA binding factor binding element
- IP
- immunoprecipitation
- RIP chip
- ribonucleoprotein immunoprecipitation microarray analysis
- SUMO
- small ubiquitin-like modifier.
REFERENCES
- 1. Thompson B., Wickens M., Kimble J. (2007) in Translational Control in Biology and Medicine (Mathews M., Sonenberg N., Hershey J. W. B., eds) pp. 507–544, Cold Spring Harbor Laboratory Press, New York [Google Scholar]
- 2. Keene J. D. (2007) RNA regulons. Coordination of post-transcriptional events. Nat. Rev. Genet. 8, 533–543 [DOI] [PubMed] [Google Scholar]
- 3. Ule J., Darnell R. B. (2006) RNA binding proteins and the regulation of neuronal synaptic plasticity. Curr. Opin. Neurobiol. 16, 102–110 [DOI] [PubMed] [Google Scholar]
- 4. Gerber A. P., Herschlag D., Brown P. O. (2004) Extensive association of functionally and cytotopically related mRNAs with Puf family RNA-binding proteins in yeast. PLoS Biol. 2, E79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Edwards T. A., Pyle S. E., Wharton R. P., Aggarwal A. K. (2001) Structure of Pumilio reveals similarity between RNA and peptide binding motifs. Cell 105, 281–289 [DOI] [PubMed] [Google Scholar]
- 6. Wang X., Zamore P. D., Hall T. M. (2001) Crystal structure of a Pumilio homology domain. Mol. Cell 7, 855–865 [DOI] [PubMed] [Google Scholar]
- 7. Wang X., McLachlan J., Zamore P. D., Hall T. M. (2002) Modular recognition of RNA by a human pumilio-homology domain. Cell 110, 501–512 [DOI] [PubMed] [Google Scholar]
- 8. Wang Y., Opperman L., Wickens M., Hall T. M. (2009) Structural basis for specific recognition of multiple mRNA targets by a PUF regulatory protein. Proc. Natl. Acad. Sci. U.S.A. 106, 20186–20191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Miller M. T., Higgin J. J., Hall T. M. (2008) Basis of altered RNA-binding specificity by PUF proteins revealed by crystal structures of yeast Puf4p. Nat. Struct. Mol. Biol. 15, 397–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bernstein D., Hook B., Hajarnavis A., Opperman L., Wickens M. (2005) Binding specificity and mRNA targets of a C. elegans PUF protein, FBF-1. RNA 11, 447–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhu D., Stumpf C. R., Krahn J. M., Wickens M., Hall T. M. (2009) A 5′ cytosine binding pocket in Puf3p specifies regulation of mitochondrial mRNAs. Proc. Natl. Acad. Sci. U.S.A. 106, 20192–20197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mossessova E., Lima C. D. (2000) Ulp1-SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Mol. Cell 5, 865–876 [DOI] [PubMed] [Google Scholar]
- 13. Otwinowski Z., Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 14. Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Crystallography and NMR system. A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 15. Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 [DOI] [PubMed] [Google Scholar]
- 16. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX. A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Emsley P., Cowtan K. (2004) Coot. Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 19. Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. (2007) MolProbity. All-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cheong C. G., Hall T. M. (2006) Engineering RNA sequence specificity of Pumilio repeats. Proc. Natl. Acad. Sci. U.S.A. 103, 13635–13639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kershner A. M., Kimble J. (2010) Genome-wide analysis of mRNA targets for Caenorhabditis elegans FBF, a conserved stem cell regulator. Proc. Natl. Acad. Sci. U.S.A. 107, 3936–3941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bembom O., Keles S., van der Laan M. J. (2007) Supervised detection of conserved motifs in DNA sequences with cosmo. Stat. Appl. Genet. Mol. Biol. 6, 1–49 [DOI] [PubMed] [Google Scholar]
- 23. Merritt C., Seydoux G. (2010) The Puf RNA-binding proteins FBF-1 and FBF-2 inhibit the expression of synaptonemal complex proteins in germ line stem cells. Development 137, 1787–1798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ahringer J., Kimble J. (1991) Control of the sperm-oocyte switch in Caenorhabditis elegans hermaphrodites by the fem-3 3′ untranslated region. Nature 349, 346–348 [DOI] [PubMed] [Google Scholar]
- 25. Stumpf C. R., Kimble J., Wickens M. (2008) A Caenorhabditis elegans PUF protein family with distinct RNA binding specificity. RNA 14, 1550–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Koh Y. Y., Opperman L., Stumpf C., Mandan A., Keles S., Wickens M. (2009) A single C. elegans PUF protein binds RNA in multiple modes. RNA 15, 1090–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wapinski I., Pfeffer A., Friedman N., Regev A. (2007) Natural history and evolutionary principles of gene duplication in fungi. Nature 449, 54–61 [DOI] [PubMed] [Google Scholar]
- 28. Barton M. K., Schedl T. B., Kimble J. (1987) Gain-of-function mutations of fem-3, a sex-determination gene in Caenorhabditis elegans. Genetics 115, 107–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Carlson C. D., Warren C. L., Hauschild K. E., Ozers M. S., Qadir N., Bhimsaria D., Lee Y., Cerrina F., Ansari A. Z. (2010) Specificity landscapes of DNA binding molecules elucidate biological function. Proc. Natl. Acad. Sci. U.S.A. 107, 4544–4549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Filipovska A., Razif M. F., Nygård K. K., Rackham O. (2011) A universal code for RNA recognition by PUF proteins. Nat Chem Biol 7, 425–427 [DOI] [PubMed] [Google Scholar]
- 31. Dong S., Wang Y., Cassidy-Amstutz C., Lu G., Bigler R., Jezyk M. R., Li C., Hall T. M., Wang Z. (2011) Specific and modular binding code for cytosine recognition in Pumilio/FBF (PUF) RNA-binding domains. J. Biol. Chem. 286, 26732–26742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Opperman L., Hook B., DeFino M., Bernstein D. S., Wickens M. (2005) A single spacer nucleotide determines the specificities of two mRNA regulatory proteins. Nat. Struct. Mol. Biol. 12, 945–951 [DOI] [PubMed] [Google Scholar]
- 33. Koh Y. Y., Wang Y., Qiu C., Opperman L., Gross L., Tanaka Hall T. M., Wickens M. (2011) Stacking interactions in PUF-RNA complexes. RNA 17, 718–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang Y., Cheong C. G., Hall T. M., Wang Z. (2009) Engineering splicing factors with designed specificities. Nat. Methods 6, 825–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jones D. T. (1999) GenTHREADER. An efficient and reliable protein fold recognition method for genomic sequences. J. Mol. Biol. 287, 797–815 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.