Background: Bacterial pentameric ion channels provide model systems to delineate structures and mechanisms of eukaryotic Cys-loop receptor channels.
Results: In the bacterial channel GLIC, histidine protonation promotes hydrogen bonding between transmembrane (TM) helices.
Conclusion: Protonation of intramembrane binding site promotes ion channel opening in GLIC.
Significance: Interhelical hydrogen bonds promote concerted tilting of TM helices associated with the open channel conformation. The TM site may contribute to phototaxis in bacteria and allosteric potentiation in eukaryotes.
Keywords: Cys-loop Receptors, Gating, Histidine, Ion Channels, pH Regulation, Allosteric Binding Site, Bacterial Ligand-gated Ion Channel, Histidine Protonation, Hydrogen Bond, Intramembrane Binding Site
Abstract
Prokaryotic orthologs of eukaryotic Cys-loop receptor channels recently emerged as structural and mechanistic surrogates to investigate this superfamily of intercellular signaling proteins. Here, we examine proton activation of the prokaryotic ortholog GLIC using patch clamp electrophysiology, mutagenesis, and molecular dynamics (MD) simulations. Whole-cell current recordings from human embryonic kidney (HEK) 293 cells expressing GLIC show half-maximal activation at pH 6, close to the pKa of histidine, implicating the three native His residues in proton sensing linked to activation. The mutation H235F abolishes proton activation, H277Y is without effect, and all nine mutations of His-127 prevent expression on the cell surface. In the GLIC crystal structure, His-235 on transmembrane (TM) α-helix 2, hydrogen bonds to the main chain carbonyl oxygen of Ile-259 on TM α-helix 3. MD simulations show that when His-235 is protonated, the hydrogen bond persists, and the channel remains in the open conformation, whereas when His-235 is deprotonated, the hydrogen bond dissociates, and the channel closes. Mutations of the proximal Tyr-263, which also links TM α-helices 2 and 3 via a hydrogen bond, alter proton sensitivity over a 1.5 pH unit range. MD simulations show that mutations of Tyr-263 alter the hydrogen bonding capacity of His-235. The overall findings show that His-235 in the TM region of GLIC is a novel proton binding site linked to channel activation.
Introduction
Members of the pentameric Cys-loop receptor channel superfamily bind small molecule ligands to mediate rapid excitatory or inhibitory synaptic transmission (1, 2). For Cys-loop receptors from eukaryotes, both structure determination and mechanistic analyses have been hampered by their inherent structural complexity, including their large size, pentameric structure, hydrophobicity, and post-translational modifications. However, discovery of structurally simpler and readily expressible bacterial orthologs (3, 4) opens the way to high resolution structure determination in parallel with functional studies in heterologous expression systems. The crystal structure of the pentameric ligand-gated ion channel from Erwinia chrysanthemi (ELIC) was determined in the closed state (5), whereas the structure of the channel from Gloeobacter violaceus (GLIC) was determined in the apparent open state (6, 7). Comparison of the two structures suggested local and global rearrangements that mediate channel opening, although neither ELIC nor GLIC could be crystallized in both closed and open states.
A fundamental question is how binding of small molecule ligands elicit opening of Cys-loop receptor channels. For GLIC, the activating ligand appears to be a proton, as step decreases in pH evoke large inward ionic currents (4). By analogy to eukaryotic Cys-loop receptors, a logical candidate for the proton binding site is the subunit interface where multiple extracellular segments of opposing subunits form the principal and complementary faces of the site (8). In the subclass of nicotinic receptors, binding of agonist causes a contraction of the binding site around the ligand through rearrangements of segments from both principal and complementary subunits (9–11). In GLIC, the analogous subunit interface is rich in ionizable residues, yet whether any of these residues mediates proton activation remains unknown. Thus, the first step toward understanding activation of GLIC is to identify a proton binding site linked to channel opening.
We sought to identify a site in GLIC that mediates proton activation of ionic currents through mutagenesis, expression in heterologous cells, and whole-cell patch clamp recording. After identifying a proton binding site linked to channel activation, we used MD2 simulations to characterize structural changes associated with both proton sensing and channel opening. Our findings reveal a novel intramembrane site between the second and third TM α-helices of each subunit that, on protonation or deprotonation, leads to expansion or contraction of the ion translocation pathway.
EXPERIMENTAL PROCEDURES
Construction of GLIC
The GLIC sequence (NCBI Gene ID 2602600) was generated by Invitrogen as a synthetic cDNA with optimized codon usage for Homo sapiens. The signal peptide of the human neuronal α7 acetylcholine receptor (AChR; GenBankTM accession no. X70297) was joined in-frame to the synthetic cDNA, and the completed sequence was subcloned into the CMV-based mammalian expression vector pRBG4 (12). To generate GLIC with a substituted loop C, the sequence KPANFALEDRLESK from GLIC was replaced by the sequence SERFYECCKEPYPD from the α7 AChR. Point mutations were generated using the QuikChange site-directed mutagenesis kit (Stratagene). All constructs were confirmed by sequencing the entire cDNA insert.
HEK 293 Cell Expression
Cells were maintained in DMEM containing fetal bovine serum (10% v/v) at 37° C until they reached ∼10–30% confluence. GLIC cDNAs were transfected by calcium phosphate precipitation using a final cDNA concentration of 20 μg/ml. For patch clamp recordings, cDNA encoding green fluorescent protein was co-transfected to allow identification of transfected cells. After 8–10 h at 37° C, the medium was replaced with fresh medium, and the cells were incubated 1 to 3 days prior to patch clamp recordings or [125I]α-bungarotoxin binding measurements. We measured binding of [125I]α-bungarotoxin to intact cells expressing GLIC with loop C from the α7 AChR as described (13), using a concentration of 10 nm [125I]α-bungarotoxin and an incubation time of 30 min. Nonspecific binding was defined as that obtained for cells expressing GLIC without the substituted loop C.
Electrophysiology
Patch clamp recordings were made from HEK 293 cells in the whole-cell configuration at 21° C using the Axopatch 200B (Axon Instruments, Inc., Union City, CA). The extracellular solution contained the following: 140 mm NaCl, 2.8 mm KCl, 2 mm CaCl2, 2 mm MgCl2, 10 mm HEPES/NaOH, pH 7.4, or HCl for lower pH values. The patch pipette solution contained 140 mm KCl, 5 mm MgCl2, 5 mm EGTA, 10 mm HEPES/NaOH, pH 7.0. Cells were viewed under fluorescence optics to allow identification of green fluorescing cells indicating heterologous protein expression. Data were low pass-filtered at 10 kHz and recorded to hard disk at 20 kHz using the program pCLAMP 10 (Molecular Devices, Inc., Sunnyvale, CA). Step changes in pH were applied using a second micropipette configured to an eight-channel gravity perfusion system (ALA Scientific, Farmingdale, NY).
Virtual Mutations
We used the comparative protein structural modeling program, MODELLER (version 8.0), to generate homology models containing virtual mutations using the wild type GLIC structure as the template, as described previously (13).
MD Simulations
Methods are as described previously (14–16). Briefly, the GLIC crystal structure was imbedded in a fully solvated lipid 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) bilayer (120 × 120 Å) created using the visual molecular dynamics membrane plug-in. Lipids within 0.8 Å of the protein and those in the pore were removed. The total number of lipids was 298, with 141 on the periplasmic side and 157 on the cytoplasmic side. The membrane-protein complex was then solvated in TIP3 water using the VMD solvate plug-in. To neutralize the net charge, ions were added using the VMD autoionize plug-in, with the salt concentration set at 100 mm NaCl; 60 sodium and 35 chloride ions were added when His-235 was protonated, whereas 63 sodium and 33 chloride atoms were added when His-235 was unprotonated. The combined system comprised 221,283 atoms, including 51,232 TIP3 water molecules. The highly scalable molecular dynamics program NAMD (17), with the CHARMM27 force field, was used for all-atom MD simulations. Four rounds of equilibration were performed after the combined channel/membrane/solvent system was established, as follows: (i) 2000 steps of energy minimization for non-backbone atoms, (ii) five cycles of 500-step energy minimization with decreasing position restraints on protein Cα atoms, (iii) gradual temperature increase from 50 K to 310 K in 10,000 steps of constant volume (canonical ensemble (NVT), in which moles (N), volume (V) and temperature (T) are conserved) simulation with harmonic restraints on protein Cα atoms (force constant 3 kcal/mol/Å2), and (iv) 2-ns constant surface area ensemble MD equilibration with decreasing positional restraints on the Cα atoms. A short cut-off of 9 Å was used for non-bonded interactions, whereas long range electrostatic interactions were incorporated using the particle mesh Ewald method. Langevin dynamics and a Langevin piston algorithm were used to maintain a temperature of 310 K and a pressure of 1 atm. The r-RESPA multiple time step method was employed with a 2-fs time step for bonded, 2 fs for short range non-bonded, and 4 fs for long range electrostatic forces. Bonds between hydrogen and heavy atoms were constrained using the SHAKE algorithm. MD simulations were conducted using a home built supercomputer cluster in the Receptor Biology laboratory at the Mayo Clinic in Rochester and on the Jaguar XT5 supercomputer at the National Center for Computational Science (http://www.nccs.gov/).
BioMOCA Simulations
Detailed procedures for BioMOCA simulations are described elsewhere (16, 18). In brief, we obtained static structural models for each GLIC conformation by extracting snapshots of the structural coordinates before and after MD simulation and added charge and radius of each atom using the Adaptive Poisson-Boltzmann Solver with the CHARMM force field and pH 7.4. The resulting files were then uploaded to a web-based BioMOCA suite for ion conduction simulations. In detail, each GLIC structure was embedded in a rectangular box with dimensions 70 × 70 × 120 Å and wrapped with a 33-Å-thick layer of simplified lipid membrane with a hole of radius 13 Å surrounding the transmembrane domain of the protein. We then transferred all files from the BioMOCA suite to local computers for production simulations. Each simulation was carried out using a single Intel Xeon processor at 3.0 GHz that achieved a 6 ns/day computing speed. To examine current-voltage relationships, we subjected each simulation to external electrostatic potential biases of −200, −100, 0, +100, and +200 mV. The intracellular solution contained the following: 5 mm Na+, 145 mm Cl−, 140 mm K+, and 3 mm Mg2+; and the extracellular solution contained the following: 140 mm Na+, 145 mm Cl−, 5 mm K+, 2 mm Ca2+, and 1 mm Mg2+.
RESULTS
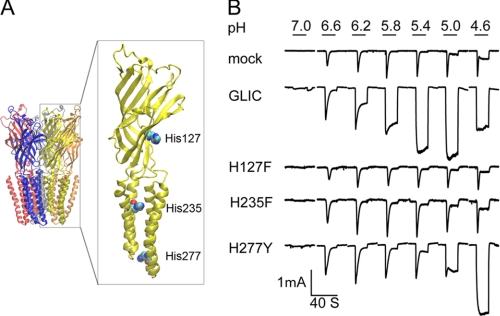
Whole-cell patch clamp recordings from HEK 293 cells expressing GLIC reveal large inward currents in response to step changes in extracellular pH from 7.0 to 4.6 (Fig. 1B). At high pH, the current rises rapidly and decays to a small, steady plateau, whereas at low pH, the current also rises rapidly but does not decay during the 20-s application period. The amplitude of the current increases as the pH is decreased to 5, but as the pH is decreased further, the amplitude decreases. We found that currents evoked by exposure to low pH ran down in successive cycles of exposure and wash-out (data not shown), suggesting the ionic concentration gradients changed. In contrast, in mock-transfected cells, changes in pH elicit small currents, likely arising from pH-sensitive channels endogenous to HEK 293 cells (19). For cells expressing GLIC, half-maximal activation of current occurs at approximately pH 6, which is close to the pKa for histidine.
FIGURE 1.
Ionic currents evoked by step changes in pH for HEK 293 cells expressing GLIC and mutants of the three intrinsic His residues. A, crystal structure of GLIC (7) (PDB code 3EAM) with the three His residues shown in space-filling representation. B, ionic currents versus time evoked by the indicated step changes in pH for mock-transfected cells, wild type GLIC, and the indicated His mutants. Currents with fast response shown in mock-transfected cells are from endogenous pH-sensitive channels.
Each subunit in the GLIC pentamer contains three His residues located in the extracellular, mid-TM, and lower-TM regions (Fig. 1A). To test whether any of these His residues is required for proton-activated ionic currents, we mutated each residue individually, expressed each mutant in HEK 293 cells, and measured whole-cell current in response to step changes in pH. Cells expressing the mutation H235F in the mid-TM region exhibit small, rapidly decaying currents similar to those in mock-transfected cells, as did cells expressing the mutation H127F, suggesting His-235, His-127, or both residues are required for proton activation. By contrast, cells expressing the mutation H277Y in the lower-TM region exhibit proton-activated currents, although proton sensitivity shifts to lower pH. Thus, His-235 and His-127 are candidates for a proton binding site linked to activation of ionic current through the GLIC channel.
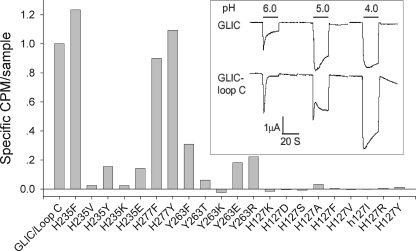
In the GLIC mutants, elimination of pH-activated currents could result from either removal of the pH sensor or lack of GLIC expression on the cell surface. To determine whether the GLIC mutants express on the cell surface, we installed the major recognition element for α-bungarotoxin (20), known as loop C, in place of the equivalent protein segment in GLIC (see “Experimental Procedures”). When GLIC with the modified loop C is transfected into HEK 293 cells, step decreases in pH evoke large inward ionic currents, with similar amplitudes but reduced proton sensitivity compared with wild type GLIC. Moreover, incubation with radio-labeled α-bungarotoxin yields robust binding to the cell surface (Fig. 2). When the mutation H235F is introduced into GLIC containing the modified loop C, α-bungarotoxin binding to the cell surface is robust, similar to that of the control construct, but proton-activated currents are similar to those in mock-transfected cells. By contrast, each of the nine mutations of His-127 eliminate α-bungarotoxin binding. Thus, His-127 appears necessary for protein folding, subunit oligomerization or transport of GLIC to the cell surface, whereas His-235 is a strong candidate for a proton binding site linked to activation of ionic currents through the GLIC channel.
FIGURE 2.
Cell surface expression of GLIC in which loop C was replaced with that from the human α7 AChR. Shown is [125I]α-bungarotoxin bound to intact cells expressing GLIC with loop C modified and the indicated mutants. Inset, pH-activated currents for wild type GLIC and GLIC with loop C modified.
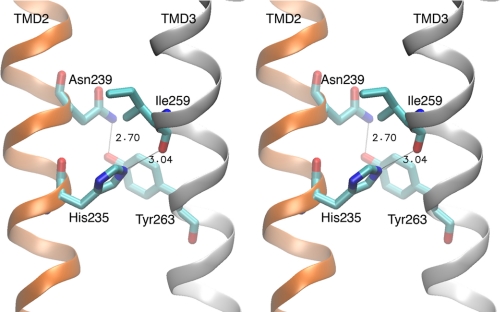
The crystal structure of GLIC, obtained at low pH and with the channel apparently open, shows that His-235, located on the side of TM2 opposite the channel lumen, hydrogen bonds to the main chain carbonyl oxygen of Ile-259 of TM3 (Fig. 3). In a standard α-helix, hydrogen bonds form between main chain carbonyl oxygen atoms and amide groups from successive turns of the helix. However, in the GLIC structure, instead of an intrahelical hydrogen bond to the carbonyl oxygen of Ile-259, an interhelical hydrogen bond is established by His-235. This interhelical hydrogen bond may facilitate association of TMs 2 and 3 and consequently stabilize the open channel conformation.
FIGURE 3.
Stereo view of the interhelical cleft separating TMs 2 and 3 obtained from the GLIC crystal structure (7). Key residues and hydrogen bonds are indicated.
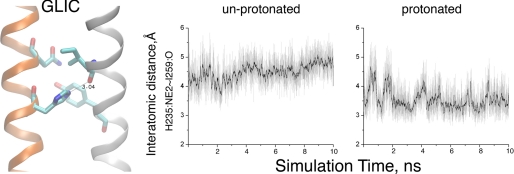
To determine whether this interhelical hydrogen bond depends on protonation of His-235, we carried out all-atom MD simulations of GLIC, embedded in a lipid bilayer and surrounded by explicit water molecules and ions, and monitored the distance between the Nϵ atom of His-235 and the main chain oxygen of Ile-259. When His-235 is protonated, the interatomic distance approaches ∼3.2 Å throughout the 10-ns simulation (Fig. 4). However, when His-235 is unprotonated, the interatomic distance steadily increases as the simulation time increases, approaching a separation distance of 5 Å at 10 ns. Thus, the interhelical linkage mediated by His-235 depends on protonation of its Nϵ atom.
FIGURE 4.
MD simulations of GLIC with His-235 unprotonated and protonated. The left panel shows the hydrogen bond between Nϵ of His-235 and the main chain oxygen atom of Ile-259. The right panels show the interatomic distance versus simulation time for each condition. Black traces indicate the interatomic distances averaged over 40 ps, whereas the light traces show distances measured every 2 ps.
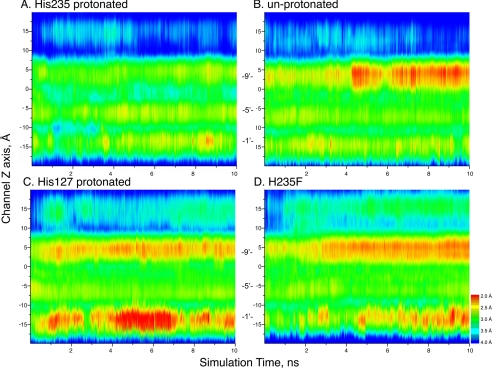
To investigate the relationship between protonation of His-235 and the functional state of the channel, we further analyzed the simulations to assess changes in the channel radius. Toward this end, we constructed a contour plot in which the radius along the channel z axis for each simulated frame is plotted against simulation time, with red hues indicating a small radius and blue hues indicating a large radius (Fig. 5). When His-235 is protonated, the channel radius remains similar to that in the starting, apparently open channel structure. However, when His-235 is unprotonated, the channel constricts approximately halfway through the simulation, suggesting the channel closed. By contrast, when His-127 is protonated or with the virtual mutation H235F, the channel constricts early in the simulation. Thus, protonation of His-235 and maintenance of the interhelical hydrogen bond are associated with the open channel conformation, whereas removal of the proton from His-235 or substitution with Phe eliminate the interhelical hydrogen bond and lead to the closed channel conformation.
FIGURE 5.
Pore radius versus simulation time for the indicated GLIC structures. For each panel, pore radius is indicated by color along an axis passing through the center of the pore (y axis) as a function of simulation time (x axis). Pore radii were calculated using the program HOLE 2.0 (31).
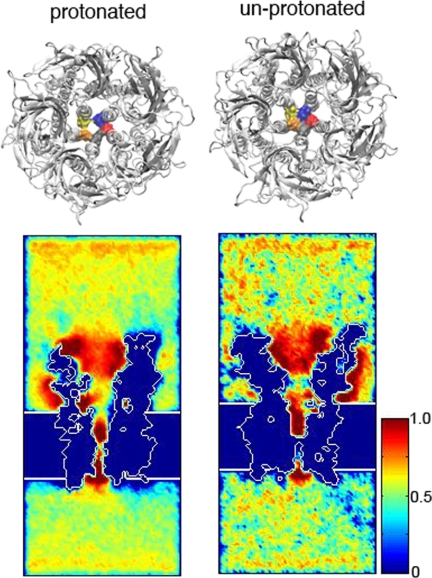
Inspection of the narrowest region of the channel shows an apparently open pathway in the protonated simulation, whereas the pathway appears occluded in the unprotonated simulation (Fig. 6). To assess cation permeability of the two simulated structures, we employed the course-grained method BioMOCA that simulates ion transport in electrolyte environments (16). In BioMOCA simulations, the protein, membrane, and water are modeled as continuum media, with permittivity values computed from the atomic structure, whereas the ions are represented explicitly and are subjected to scattering events and Newtonian physics. The system is then subjected to a specified transmembrane electrostatic potential. In the simulation with His-235 protonated, cation density is appreciable throughout the transmembrane lumen, whereas with His-235 unprotonated cation density is interrupted by a gap midway through the lumen (Fig. 6). Thus, with His-235 protonated the channel is permeable to cations, whereas with His-235 unprotonated cation permeability is immeasurably low.
FIGURE 6.
Upper panel, top views of the GLIC structures from the protonated (left) and unprotonated (right) simulations, with Ile-233 at the narrowest constriction of TM 2 highlighted in surface rendering and in different colors in each subunit. Lower panel, cross-sectional view of time-averaged cation distributions. In both structures, cations are concentrated (red) in the upper half of the channel. With His-235 protonated, there is an open pathway with increased cation density, whereas with His-235, the unprotonated the pathway is occluded.
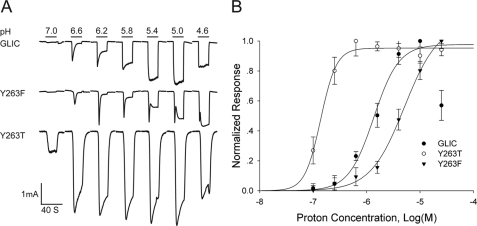
The GLIC crystal structure shows a second interhelical hydrogen bond between Tyr-263 from TM3 and Asn-239 from TM2 (Fig. 3). Although Tyr-263 is not likely a proton binding site, given an expected pKa of ∼10, its proximity to His-235 suggests it may contribute to the proton sensitivity of channel activation. To test whether Tyr-263 contributes to proton sensitivity, we generated the mutations Y263F and Y263T, expressed each mutant in HEK 293 cells, and monitored whole cell current in response to step decreases in pH (Fig. 7). The mutation Y263F retains proton-activated ionic currents, but activation shifts to lower pH, indicating reduced proton sensitivity. The mutation Y263T also retains proton-activated ionic currents, but activation shifts to higher pH, indicating increased proton sensitivity. Thus structural changes at Tyr-263 alter proton sensitivity over a span of 1.5 pH units, suggesting the interhelical link established by Tyr-263 contributes to proton sensing linked to activation.
FIGURE 7.
pH activated ionic currents for mutations of Tyr-263. A, ionic currents versus time in response to the indicated step changes in pH for wild type GLIC and the indicated mutants of Tyr-263. B, relationships between ionic current and pH for GLIC and the indicated mutants. Smooth curves are fits of the Hill equation to the data with the following parameters: for wild type GLIC the EC50 = 1.09 × 10−6 m (pH = 5.96), and the Hill coefficient nH = 2.20 + 0.15 (n = 5); for Y263T the EC50 = 1.38 × 10−7 m (pH = 6.86) and nH = 2.9 + 0.44 (n = 3); for Y263F the EC50 = 5.088 × 10−6 m (pH = 5.3) and nH = 1.36 + 0.15 (n = 3).
The altered proton sensitivity produced by mutations of Tyr-263 may originate from changes in hydrogen bonding between TMs 2 and 3. In wild type GLIC at low pH, the inter-helical hydrogen bond mediated by His-235 comprises a charged hydrogen bond donor and a dipole acceptor. This primary link is supplemented by a secondary inter-helical hydrogen bond mediated by the dipole donor Tyr-263 and the dipole acceptor Asn239. In the Y263F mutant, the secondary hydrogen bond is removed, and the link between TMs 2 and 3 depends primarily on His-235. Thus in the Y263F mutant, the observed decrease in pH sensitivity suggests that more complete protonation of His-235 is necessary to stabilize the link between the TM 2 and 3 α-helices that enables proton activated ionic current.
On the other hand, for the Y263T mutant, the reason for the increase in pH sensitivity is not immediately clear. The side chain of the Thr substitution is less bulky than that of the native Tyr and may enable tighter packing of TMs 2 and 3. Alternatively, the hydroxyl oxygen of the substituted Thr may accept a hydrogen bond from the protonated Nϵ atom of His-235, and thus promote protonation by raising the effective pKa of His-235.
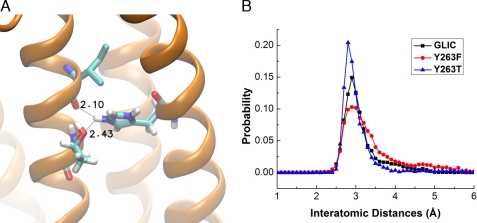
To evaluate these possibilities, we generated homology models of GLIC containing the virtual mutations H263T and H263F, and carried out MD simulations with His-235 in the protonated state. For each frame of the simulation, we measured the distance between the proton on the Nϵ atom of His-235 and the main chain oxygen of Ile-259, and plotted the probability of a given separation distance against the separation distance (Fig. 8B). For simulations of wild type GLIC and the two mutants, the separation distance shows a major peak of probability at ∼3 Å, followed by an asymmetric tail out to longer distances. However, compared with wild type GLIC, the Y263T mutant exhibits a larger peak, indicating a more stable hydrogen bond, whereas the Y263F mutant exhibits a smaller peak, indicating a less stable hydrogen bond. In addition, in the Y263T mutant, the hydroxyl oxygen of the substituted Thr forms a second hydrogen bond with the protonated Nϵ atom of His-235 that may further enhance protonation and stabilize the interhelical interaction (Fig. 8A). Thus, changes in pH sensitivity arising from mutations of Tyr-263 likely originate from changes in interhelical hydrogen bonding involving His-235.
FIGURE 8.
Stability of the hydrogen bond formed by His-235 obtained from MD simulations of GLIC and virtual mutations of Tyr-263. Panel B plots of the probability of the hydrogen bond between the Nϵ atom of His-235 and the main chain oxygen atom of Ile-259 versus the hydrogen bond distance. Panel A shows interhelical hydrogen bonds for the Y263T mutant.
DISCUSSION
We find that the bacterial channel GLIC, from the cyanobacterium Gleobactor violaceus, exhibits half-maximal activation of ionic current at pH 6, close to the pKa of histidine. Among the three His residues in each subunit of the homopentamer, His-235 in the mid-TM region is required for proton mediated activation, whereas His-277 in the lower TM region is not required, whereas His-127 in the extracellular region is necessary for expression of protein on the cell surface. In the GLIC crystal structure, His-235 stems from TM2 and hydrogen bonds to the main chain of TM3, apparently stabilizing the open channel conformation. Our MD simulations support this structural interpretation, showing that protonation of His-235 maintains the interhelical hydrogen bond and is associated with the open channel conformation, whereas deprotonation of His-235 eliminates the hydrogen bond and is associated with the closed channel conformation. In addition, Tyr-263 from TM3 modulates proton sensitivity through a hydrogen bond to Asn-239 on TM2. The mutation Y263F reduces proton sensitivity likely because loss of the aromatic hydroxyl eliminates the hydrogen bond to Asn-239, weakening the inter-helical interaction. Conversely, the mutant Y263T increases proton sensitivity likely because the hydroxyl oxygen of the substituted Thr promotes protonation of His-235.
Both Gleobactor violaceus and its ion channel GLIC are structurally unique. Gleobactor violaceus is the only known cyanobacterium that lacks an intracellular thylakoid membrane so that photosynthesis occurs within the plasma membrane (21). Illumination of cyanobacteria typically evokes flux of protons out of the thylakoid compartment and into the cytoplasm, but illumination of Gleobactor violaceus instead evokes flux of protons out of the cell (22). Proton efflux would locally decrease the extracellular pH, activate GLIC and depolarize the cell, potentially contributing to phototaxis. Although changes in pH have been found to activate several types of bacterial ion channels, including the KcsA potassium (23) and ClC chloride channels (24), the proton sensors in those channels are intracellular rather than in the TM region. In fact, sequence alignments show that among bacterial Cys-loop receptor orthologs identified so far, only GLIC contains His at the position equivalent to His-235 (Table 1). Although His-235 is located in a TM region, it is accessible to extracellular protons through the inter-helical space that extends uninterrupted from the cell surface to deep in the membrane. Furthermore, histidine provides high sensitivity to protons compared with residues with carboxylate side chains. Thus the choice of histidine as a pH sensor in an inter-helical TM cavity seems to be a novel adaptation by Gleobactor violaceus.
TABLE 1.
Although identification of His-235 as the proton sensor eluded previous studies, our findings can be reconciled with those studies. Scanning cysteine substitution along TM2 in GLIC showed that substitution of cysteine for His-235 eliminated pH-activated currents (25), in accord with our findings. However the cysteine substitution studies did not distinguish between loss of function and loss of GLIC expression on the cell surface. By placing the recognition element for α-bungarotoxin binding into the H235F mutant, we find that this mutant expresses on the cell surface in amounts similar to that of the non-mutant counterpart, confirming that H235F eliminates pH sensitivity. Another study generated a chimeria composed of GLIC sequence from the N terminus up to TM1 followed by glycine receptor sequence to the C terminus and found that the chimera exhibited pH activated ionic currents (26) in apparent contrast to our findings. However, the chimera was also active at neutral pH, suggesting that protons may have modulated rather than acted as primary activators. Furthermore, our findings do not exclude the extracellular region as a contributor to pH sensitivity, as mutations of His-127 did not express on the cell surface and thus could not be tested for proton activated ionic currents, and substitution of loop C from the α7 AChR reduced proton sensitivity. Thus, although our findings show that His-235 is required for pH-mediated ionic currents, ionizable residues in the extracellular region may also contribute.
The four TM residues that we implicate in proton sensing congregate within the space between TMs 2 and 3 that is inaccessible from the channel lumen. His-235 corresponds to the 11′ position of TM2, just beyond the central Leu residue at the 9′ position that forms a hydrophobic ring midway along the ion translocation pathway. His-235 extends toward TM3 where it hydrogen bonds to the main chain carbonyl oxygen of Ile-259. This hydrogen bond is unusual because it is mediated by the Nϵ rather than the preferred Nδ nitrogen atom of His-235, but this bonding configuration is present in all of the GLIC crystal structures obtained so far (6, 7, 27). A hydrogen bond in the opposing direction is formed between Tyr-263 of TM3 and Asn-239 at the 15′ position of TM2. Together, this pair of interhelical hydrogen bonds promotes association of TMs 2 and 3.
Crystal structures of GLIC show that the interhelical space encompassing the four key TM residues is accessible to the protein surface through crevices that extend from the periphery of the pentamer to deep within the TM region. In fact, the pathway from the protein surface to the interhelical space forms a site for binding general anesthetics (27), which inhibit proton activated currents in GLIC, likely by impeding access of protons to His-235. Through an analogous structural mechanism, mutations of residues within this pathway alter proton sensitivity of GLIC (26). In the neuronal α7 AChR, residues within the equivalent interhelical space are required for potentiation by the allosteric modulator PNU-120596 (28, 29) and for direct activation by the compound 4BP-TQS (30). Thus, although the interhelical TM cavity has been deemed an allosteric modulatory site, as opposed to the orthosteric site in the extracellular region, the TM site in GLIC may be the primary site for initiating proton-activated ionic currents.
Comparison of the GLIC and ELIC crystal structures shows that although the positions of TMs 2 and 3 differ markedly between the two structures, they can be superimposed by rotation about an axis parallel to the membrane and approximately three helical turns below His-235 (6, 7). Thus, channel opening seems to involve a rigid body rotation of TMs 2 and 3 with little change within either TM α-helix. Our findings suggest that in the closed state, TMs 2 and 3, although associated, are relatively loosely joined, and as a consequence are unable undergo a concerted tilting away from the central axis. However, when His-235 is protonated a strong interhelical hydrogen bond is established between TMs 2 and 3 that enables concerted tilting away from the central axis, opening the channel.
This work was supported, in whole or in part, by National Institutes of Health Grant NS31744 (to S. M. S.) and the Advanced Computing Program, U. S. Department of Energy Grant FWP ERKJE84 (to X. C.). This work also used resources of the National Center for Computational Sciences at Oak Ridge National Laboratory, which is supported by the U. S. Department of Energy.
- MD
- molecular dynamics
- TM
- transmembrane
- AChR
- acetylcholine receptor.
REFERENCES
- 1. Thompson A. J., Lester H. A., Lummis S. C. (2010) The structural basis of function in Cys-loop receptors. Q. Rev. Biophys. 43, 449–499 [DOI] [PubMed] [Google Scholar]
- 2. Sine S. M., Engel A. G. (2006) Recent advances in Cys-loop receptor structure and function. Nature 440, 448–455 [DOI] [PubMed] [Google Scholar]
- 3. Tasneem A., Iyer L. M., Jakobsson E., Aravind L. (2005) Identification of the prokaryotic ligand-gated ion channels and their implications for the mechanisms and origins of animal Cys-loop ion channels. Genome Biol. 6, R4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bocquet N., Prado de Carvalho L., Cartaud J., Neyton J., Le Poupon C., Taly A., Grutter T., Changeux J. P., Corringer P. J. (2007) A prokaryotic proton-gated ion channel from the nicotinic acetylcholine receptor family. Nature 445, 116–119 [DOI] [PubMed] [Google Scholar]
- 5. Hilf R. J., Dutzler R. (2008) X-ray structure of a prokaryotic pentameric ligand-gated ion channel. Nature 452, 375–379 [DOI] [PubMed] [Google Scholar]
- 6. Hilf R. J., Dutzler R. (2009) Structure of a potentially open state of a proton-activated pentameric ligand-gated ion channel. Nature 457, 115–118 [DOI] [PubMed] [Google Scholar]
- 7. Bocquet N., Nury H., Baaden M., Le Poupon C., Changeux J. P., Delarue M., Corringer P. J. (2009) X-ray structure of a pentameric ligand-gated ion channel in an apparently open conformation. Nature 457, 111–114 [DOI] [PubMed] [Google Scholar]
- 8. Sine S. M. (2002) The nicotinic receptor ligand-binding domain. J. Neurobiol. 53, 431–446 [DOI] [PubMed] [Google Scholar]
- 9. Celie P. H., van Rossum-Fikkert S. E., van Dijk W. J., Brejc K., Smit A. B., Sixma T. K. (2004) Nicotine and carbamylcholine binding to nicotinic acetylcholine receptors as studied in AChBP crystal structures. Neuron 41, 907–914 [DOI] [PubMed] [Google Scholar]
- 10. Hansen S. B., Sulzenbacher G., Huxford T., Marchot P., Taylor P., Bourne Y. (2005) Structures of Aplysia AChBP complexes with nicotinic agonists and antagonists reveal distinctive binding interfaces and conformations. EMBO J. 24, 3635–3646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gao F., Bren N., Burghardt T. P., Hansen S., Henchman R. H., Taylor P., McCammon J. A., Sine S. M. (2005) Agonist-mediated conformational changes in acetylcholine-binding protein revealed by simulation and intrinsic tryptophan fluorescence. J. Biol. Chem. 280, 8443–8451 [DOI] [PubMed] [Google Scholar]
- 12. Lee N. G., Yamaguchi J., Subramanian K. N. (1991) Efficient replication of plasmids containing the SV40 origin in N-myc overexpressing human neuroblastoma cells. Oncogene 6, 1161–1169 [PubMed] [Google Scholar]
- 13. Sine S. M., Wang H. L., Bren N. (2002) Lysine scanning mutagenesis delineates structural model of the nicotinic receptor ligand-binding domain. J. Biol. Chem. 277, 29210–29223 [DOI] [PubMed] [Google Scholar]
- 14. Cheng X., Lu B., Grant B., Law R. J., McCammon J. A. (2006) Channel opening motion of α7 nicotinic acetylcholine receptor as suggested by normal mode analysis. J. Mol. Biol. 355, 310–324 [DOI] [PubMed] [Google Scholar]
- 15. Wang H. L., Cheng X., Taylor P., McCammon J. A., Sine S. M. (2008) Control of cation permeation through the nicotinic receptor channel. PLoS Comp. Biol. 4, e41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang H. L., Toghraee R., Papke D., Cheng X. L., McCammon J. A., Ravaioli U., Sine S. M. (2009) Single-channel current through nicotinic receptor produced by closure of binding site C-loop. Biophys. J. 96, 3582–3590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Phillips J. C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., Chipot C., Skeel R. D., Kalé L., Schulten K. (2005) Scalable molecular dynamics with NAMD. J. Comput Chem. 26, 1781–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. van der Straaten T. A., Kathawala G., Trellakis A., Eisenberg R. S., Ravaioli U. (2005) BioMOCA–a Boltzmann transport Monte Carlo model for ion channel simulation. Mol. Sim. 31, 151–171 [Google Scholar]
- 19. Ma Z. Y., Zhang W., Chen L., Wang R., Kan X. H., Sun G. Z., Liu C. X., Li L., Zhang Y. (2008) A proton-activated, outwardly rectifying chloride channel in human umbilical vein endothelial cells. Biochem. Biophys. Res. Commun. 371, 437–440 [DOI] [PubMed] [Google Scholar]
- 20. Wilson P. T., Lentz T. L., Hawrot E. (1985) Determination of the primary amino acid sequence specifying the α-bungarotoxin binding site on the α subunit of the acetylcholine receptor from Torpedo californica. Proc. Natl. Acad. Sci. U.S.A. 82, 8790–8794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bhaya D. (2004) Light matters: phototaxis and signal transduction in unicellular cyanobacteria. Mol. Microbiol. 53, 745–754 [DOI] [PubMed] [Google Scholar]
- 22. Belkin S., Mehlhorn R. J., Packer L. (1987) Proton gradients in intact cyanobacteria. Plant Physiol. 84, 25–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Heginbotham L., LeMasurier M., Kolmakova-Partensky L., Miller C. (1999) Single Streptomyces lividans K(+) channels: functional asymmetries and sidedness of proton activation. J. Gen. Physiol. 114, 551–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Iyer R., Iverson T. M., Accardi A., Miller C. (2002) A biological role for prokaryotic ClC chloride channels. Nature 419, 715–718 [DOI] [PubMed] [Google Scholar]
- 25. Parikh R. B., Bali M., Akabas M. H. (2011) Structure of the M2 transmembrane segment of GLIC, a prokaryotic Cys-loop receptor homologue from Gloeobacter violaceus, probed by substituted cysteine accessibility. J. Biol. Chem. 286, 14098–14109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Howard R. J., Murail S., Ondricek K. E., Corringer P. J., Lindahl E., Trudell J. R., Harris R. A. (2011) Structural basis for alcohol modulation of a pentameric ligand-gated ion channel. Proc. Natl. Acad. Sci. U.S.A. 108, 12149–12154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nury H., Van Renterghem C., Weng Y., Tran A., Baaden M., Dufresne V., Changeux J. P., Sonner J. M., Delarue M., Corringer P. J. (2011) X-ray structures of general anaesthetics bound to a pentameric ligand-gated ion channel. Nature 469, 428–431 [DOI] [PubMed] [Google Scholar]
- 28. Young G. T., Zwart R., Walker A. S., Sher E., Millar N. S. (2008) Potentiation of α7 nicotinic acetylcholine receptors via an allosteric transmembrane site. Proc. Natl. Acad. Sci. U.S.A. 105, 14686–14691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. daCosta C. J., Free C. R., Corradi J., Bouzat C., Sine S. M. (2011) Single-channel and structural foundations of neuronal α7 acetylcholine receptor potentiation. J. Neurosci. 31, 13870–13879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gill J. K., Savolainen M., Young G. T., Zwart R., Sher E., Millar N. S. (2011) Agonist activation of α7 nicotinic acetylcholine receptors via an allosteric transmembrane site. Proc. Natl. Acad. Sci. U.S.A. 108, 5867–5872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Smart O. S., Neduvelil J. G., Wang X., Wallace B. A., Sansom M. S. (1996) HOLE: a program for the analysis of the pore dimensions of ion channel structural models. J. Mol. Graph. 14, 354–360 [DOI] [PubMed] [Google Scholar]