Background: HDHB has been implicated in chromosomal replication, but its role has not been determined.
Results: Replication stress recruits HDHB to chromatin in a checkpoint-independent, RPA-dependent manner, and HDHB silencing reduces recovery from replication stress.
Conclusion: HDHB functions in chromosomal replication to relieve replication stress.
Significance: HDHB competition with checkpoint-signaling proteins for binding to RPA may modulate cellular responses to replication stress.
Keywords: DNA Damage, DNA Damage Response, DNA Helicase, DNA Replication, Protein-Protein Interactions, DNA Replication Stress, HDHB, HELB, RPA
Abstract
Maintenance of genomic stability in proliferating cells depends on a network of proteins that coordinate chromosomal replication with DNA damage responses. Human DNA helicase B (HELB or HDHB) has been implicated in chromosomal replication, but its role in this coordinated network remains undefined. Here we report that cellular exposure to UV irradiation, camptothecin, or hydroxyurea induces accumulation of HDHB on chromatin in a dose- and time-dependent manner, preferentially in S phase cells. Replication stress-induced recruitment of HDHB to chromatin is independent of checkpoint signaling but correlates with the level of replication protein A (RPA) recruited to chromatin. We show using purified proteins that HDHB physically interacts with the N-terminal domain of the RPA 70-kDa subunit (RPA70N). NMR spectroscopy and site-directed mutagenesis reveal that HDHB docks on the same RPA70N surface that recruits S phase checkpoint signaling proteins to chromatin. Consistent with this pattern of recruitment, cells depleted of HDHB display reduced recovery from replication stress.
Introduction
DNA helicase activity is a vital component of all DNA transactions that requires separation of the two strands of DNA, including DNA replication, DNA repair, and recombination. The abundant variety of DNA helicases, which greatly exceeds that of DNA polymerases, has hindered efforts to elucidate their functional role in DNA processing pathways, particularly in vertebrates. The conserved vertebrate DNA helicase B (HELB)3 was initially discovered in extracts of a temperature-sensitive mouse cell line as a thermolabile ATPase whose activity depended on single-stranded DNA (ssDNA) (1–3). Subsequent biochemical studies revealed that the ATPase displayed ssDNA-dependent helicase activity with 5′ to 3′ polarity (4–6). More recently, analysis of mouse and human HELB cDNAs revealed their sequence homology and biochemical similarity to several prokaryotic superfamily 1B helicases that unwind DNA with 5′-3′ polarity, e.g. Escherichia coli RecD, Bacillus subtilis YrrC, and bacteriophage T4 Dda (7–9).
A potential role for HELB in chromosomal replication was initially suggested by studies of the mutant mouse cell line expressing temperature-sensitive HELB helicase activity; when shifted to the nonpermissive temperature, the cells accumulated in early S-phase (5). Consistent with this finding, microinjection of purified recombinant human HELB (HDHB) protein with a substitution in the Walker B motif, i.e. helicase-dead, into human cells in G1 inhibited DNA synthesis in up to 70% of the injected cells, whereas injection of the wild type protein did not (9). Also of note, purified mouse and human HELB were found to interact functionally with purified DNA polymerase α-primase, displaying primosome activity on replication protein A (RPA)-coated ssDNA in vitro (4, 9). These activities would be consistent with a role for HELB in initiation of chromosomal replication, in lagging strand synthesis, or possibly in recovery from DNA damage by re-priming the leading strand template downstream of forks stalled at a lesion (10, 11). HDHB has also been identified in proteomic screens as a potential target of the ataxia telangiectasia-mutated (ATM) checkpoint kinase (12) and as part of a mismatch repair complex (13). Taking these findings together, we reasoned that HDHB might function in chromosomal replication, perhaps at the interface of replication with repair, and set out to explore this possibility.
Here we demonstrate that in S phase cells exposed to replication stress, HDHB accumulates on chromatin in a checkpoint signaling-independent, RPA-dependent manner. We identify in detail direct physical interactions of HDHB with RPA, which closely resemble those that recruit S phase checkpoint signaling proteins ATRIP and Rad9 to stalled forks. HDHB depletion does not disrupt activation of S phase checkpoint signaling but instead slightly stimulates it. Moreover, HDHB depletion reduces viability of cells exposed to camptothecin and increases chromosomal breaks and gaps in cells exposed to aphidicolin. Based on these results, we propose that HDHB functions to counteract replication stress.
EXPERIMENTAL PROCEDURES
Cell Culture, Synchronization, and Genotoxin Treatment
Human osteosarcoma U2OS (14, 15), colorectal carcinoma HCT116 (16–18), and HPV18-positive cervical carcinoma HeLa (Ref. 19 and references therein) cell lines were grown as monolayers in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum at 37 °C and 5% CO2. U2OS cells were synchronized at G1/S by incubation for 17 h with 2.5 mm thymidine (Sigma) followed by a 12-h release and another 17 h of incubation with thymidine. Cells were released from the second thymidine incubation for 3 h into S phase and for 9 h into G2/M phase. To enrich U2OS cells in G1 phase, cells were cultured with 30 ng/ml nocodazole (Sigma) for 16 h and then released for 4 h. Cells were irradiated with UV-C at 254 nm in a Stratalinker (Stratagene).
Antibodies against HDHB
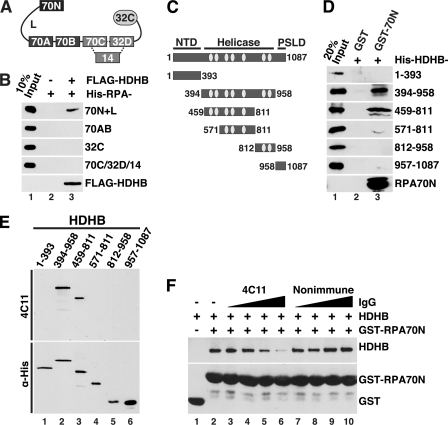
Polyclonal rabbit antibodies were described previously (20). To generate monoclonal antibodies, purified recombinant T7-tagged HDHB protein (9) (50 μg) was injected intraperitoneally and subcutaneously into LOU/C rats using CpG2006 (TIB MOLBIOL) as adjuvant. After 8 weeks, a boost of antigen was given intraperitoneally and subcutaneously. Three days later, fusion of P3X63-Ag8.653 myeloma cells with the rat spleen cells was performed according to standard procedures (21). Hybridoma supernatants were tested in a solid-phase immunoassay using T7-tagged HDHB protein adsorbed to polystyrene microtiter plates. Crude E. coli extract served as a negative control. Hybridoma cells expressing mAb 4C11 and mAb 5C9 were stably subcloned and used to produce antibodies for further analysis (supplemental Fig. S1A and Fig. 4, E and F). Rat IgG was purified using Melon gel IgG purification kit (Pierce) according to the manufacturer's instructions and dialyzed into 25 mm HEPES-KOH, pH 7.5, and 50 mm NaCl. Nonimmune rat IgG was purchased from Jackson ImmunoResearch.
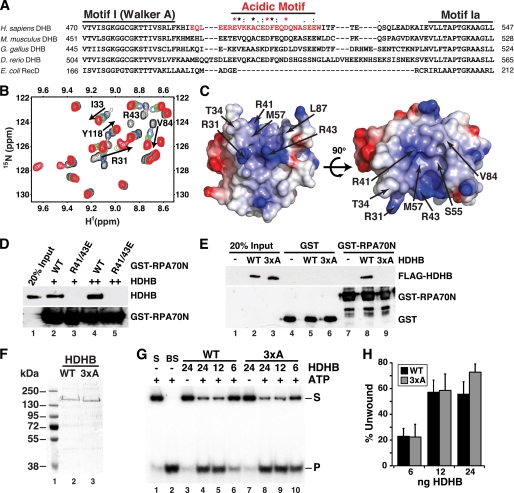
FIGURE 4.
Direct physical interaction of HDHB with RPA. A, modular domain architecture of RPA is shown. The three subunits RPA70, -32, and -14 interact through a three-helix bundle (dotted lines). Compact domains (filled rectangles, oligonucleotide/oligosaccharide binding-fold; filled oval, winged helix) are joined by flexible linkers (Lines) (44–48). The residues at the N and C termini of each domain are listed under “Experimental Procedures.” B, purified His-tagged RPA constructs captured on anti-FLAG antibody beads in the presence (+) or absence (−) of whole cell extract as in Fig. 3C were analyzed by Western blotting with anti-His (top four panels) or anti-FLAG. C, diagram of HDHB domains and truncation constructs. NTD, N-terminal domain; PSLD, phosphorylation regulated subcellular localization domain (20). The seven conserved helicase motifs I, Ia, and II-VI of superfamily I are indicated by light gray ovals. The first and last residue numbers of each construct are indicated. D, glutathione beads prebound to purified GST or His-GST-RPA70N were incubated with purified His-tagged HDHB truncation mutants. Proteins captured on the beads were analyzed by Western blotting with anti-His antibody. E, His-tagged HDHB truncation mutants (E) were separated by SDS-PAGE and visualized by Western blotting performed with HDHB monoclonal antibody 4C11 (top panel) or anti-His antibody (lower panel). F, glutathione beads prebound with GST (lane 1) or GST-RPA70N were incubated with purified HDHB (0.5 μg) in the absence (lanes 1 and 2) or presence of increasing amounts (0.3, 0.6, 1.2, 2.4 μg) of monoclonal IgG 4C11 (lanes 3–6) or of non-immune rat IgG (lanes 7–10).
Cell Fractionation and Western Blotting
To obtain whole cell extract, cells were lysed in radioimmunoprecipitation assay buffer (50 mm Tris-HCl at pH 7.5, 150 mm NaCl, 1% Nonidet P-40, 0.5% deoxycholic acid, 0.1% SDS, 10 mm NaF, 1 mm Na3VO4, 1 mm phenylmethylsulfonyl fluoride, 10 μg/ml aprotinin, 1 μm leupeptin) on ice for 30 min and centrifuged at 12,500 rpm for 15 min. Chromatin fractionation was performed as described (22), except that the final Triton X-100 concentration used for separation of cytoplasmic proteins from nuclei was 0.05% for U2OS or HCT116 and 0.1% for HeLa cells.
Primary antibodies used for Western blotting were: rabbit anti-HDHB (20), mouse monoclonal anti-RPA 70C, 70B, or 34A (23), anti-Chk1 phospho-S317 (Cell Signaling), anti-RPA32 phospho-S4/S8 (Bethyl), total RPA32 (RPA2, Calbiochem), glyceraldehyde 3-phosphate dehydrogenase (Santa Cruz), mouse anti-PCNA (PC-10, Santa Cruz), mouse anti-tubulin (NeoMarkers or Santa Cruz), mouse anti-histone H1 (Santa Cruz), mouse anti-Chk2 (Upstate), rabbit anti-Chk2 phospho-T68 (Cell Signaling), mouse anti-Chk1 (Santa Cruz), rabbit anti-Chk1 phospho-S345 (Cell Signaling), mouse anti-FLAG antibody (Sigma), mouse anti-His antibody (Genscript). Rabbit polyclonal anti-GST and anti-Orc2 came from Fanning Lab stocks (Vanderbilt University). Western blots were evaluated using chemiluminescence (ECL, PerkinElmer Life Sciences) and exposure to x-ray film for different time periods.
Recombinant Proteins
WT and mutant HDHB were purified from Hi5 insect cells as previously described (9). Mutant 3xA HDHB was generated by site-directed mutagenesis in pFLAG-WT HDHB plasmid (20) using mutagenic primers E499A (AGTTGGAAGAAAGAGCAGTAAAAAAAGCCTG), D506A (AAGCCTGTGAAGCTTTTGAACAAGA), D510A (GAAGATTTTGAACAAGCCCAGAATGCTTCAGAAG). Correct mutagenesis was confirmed by DNA sequencing. The 3xA HDHB coding sequence excised from the pFLAG vector using NotI/SalI was cloned into pFast-Bac HT (Sigma) using NotI/XhoI. Baculovirus for 3xA HDHB expression was generated in Sf9 cells using Bac-to-Bac Baculovirus expression system (Invitrogen).
HDHB truncation mutants were designed based on predicted secondary structure (psipred, predictprotein) (24, 25), disorder (DisEMBL) (26), and sequence conservation among vertebrate DHBs (T-Coffee) (27) and cloned in pET28 for bacterial expression of His-tagged proteins. Plasmids used for bacterial expression of His-tagged RPA truncation mutants were: pET15b-RPA70 (aa 1–120) for RPA70N (28) from Dr. C. Arrowsmith, pET15b-RPA32 (aa 172–270) for RPA32C (29), pET11d-RPA70 (aa 1–168) for RPA70N+L (30) from Dr. M. Wold, pET15b-RPA70 (aa 181–422) for RPA70AB (31), and pET15b-RPA70 (aa 436–616/32 (aa 43–171)/14) for RPA70C/32D/14 (32), both from Dr. A. Bochkarev.
His-tagged RPA and HDHB truncation mutants were expressed in E. coli and purified over a nickel-nitrilotriacetic acid column (Qiagen). GST-tagged WT and R41/43E mutant RPA70-(1–120) constructs, kindly provided by Drs. D. Cortez and X. Xu (33), were expressed in E. coli, then purified over glutathione-Sepharose beads (Sigma). Wild type human RPA was expressed from pET11d-WT RPA (from Dr. M. Wold) in E. coli and purified as described (34).
Co-immunoprecipitation and GST Pulldown Assays
For co-immunoprecipitations using FLAG M2 beads, extracts from cells transfected with FLAG-HDHB or control plasmid were lysed in 50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1 mm EDTA and 1% Triton X-100 and then incubated with FLAG M2 antibody-agarose (Sigma) for 2 h. FLAG-HDHB that co-immunoprecipitated endogenous RPA was washed 3 times with FLAG immunoprecipitation wash buffer (50 mm Tris-HCl, pH 7.4, and 150 mm NaCl), 10 min each, and analyzed by Western blotting. For FLAG co-immunoprecipitations with RPA truncation mutants, FLAG-HDHB-containing cell extracts were bound to FLAG M2 resin as above. The beads were washed with FLAG immunoprecipitation high salt wash buffer (50 mm Tris-HCl pH 7.4 with 800 mm NaCl and then with 1 m NaCl) and incubated with purified His-tagged RPA truncation mutants for 30 min in Tris-HCl, pH 7.4, 150 mm NaCl. Proteins bound to beads were analyzed by Western blotting after three washes with FLAG immunoprecipitation wash buffer.
For co-immunoprecipitation of purified proteins, protein A beads prebound to rabbit anti-HDHB IgG were incubated with purified HDHB for 1 h at 4 °C in binding buffer (30 mm Hepes-KOH, pH 7.8, 10 mm KCl, 7 mm MgCl2) containing 2% milk. Beads were then washed with binding buffer and incubated with purified WT RPA or RPA truncation mutants for 30 min at 4 °C. Alternatively, protein A beads prebound to mouse anti-RPA70C were incubated with RPA for 1 h in binding buffer, washed, and then incubated with HDHB for 1 h in the presence or absence of 250 units of benzonase (Novagen). Beads were washed once with binding buffer, 3 times with wash buffer (30 mm Hepes-KOH, pH 7.8, 75 mm KCl, 7 mm MgCl2, 0.25% inositol, 0.01% Nonidet P-40, 10 μm ZnCl2), and once more with binding buffer, 10 min each, and bound proteins were analyzed by SDS-PAGE and Western blotting.
For GST pulldown assays, purified GST alone or GST-tagged WT or R41/43E RPA70N were allowed to bind to glutathione beads overnight in binding buffer with 2% milk. The beads were then incubated with purified HDHB truncation mutants or with whole cell extracts from cells expressing FLAG-tagged WT- or 3xA-HDHB for 30 min, washed, and analyzed as described above.
Helicase Assay
M13mp18 circular ssDNA (U. S. Biochemical Corp.) annealed to a 33-nucleotide DNA (5′- TCGACTCTAGAGGATCCCCGGGTACCGAGCTCG-3′) 32P-radiolabeled at the 5′ end served as a partial duplex DNA substrate. Helicase reactions (10 μl) contained 20 mm Tris-HCl, pH 7.5, 8 mm DTT, 1 mm MgCl2, 1 mm ATP, 20 mm KCl, 4% (w/v) sucrose, 80 μg/ml BSA, 8 ng of 32P-labeled helicase substrate, and 6–24 ng of HDHB. In negative control reactions, ATP was omitted. Reactions were incubated at 37 °C for 30 min. At the end of incubations, reactions were stopped by the addition of 10 μl of stop buffer (for a final concentration of 0.3% SDS, 10 mm EDTA, 5% glycerol, and 0.03% bromphenol blue). Samples were electrophoresed on 12% native polyacrylamide gels in 89 mm Tris-borate, 2 mm EDTA. The wet gel was wrapped in plastic and exposed to a PhosphorImager screen for visualization and quantification. The average background density determined from no-enzyme and no-ATP samples was subtracted from the unwound product values. The percentage of unwound DNA was calculated with the formula: % unwound = 100 × [product/(remaining DNA substrate + product)].
Isothermal Titration Calorimetry
An HDHB peptide containing residues 493–517 (EQLEEREVKKACEDFEQDQNASEEW) was purchased (Genscript) and further purified by high performance liquid chromatography to >90% purity. RPA70N and HDHB peptide were exchanged into 20 mm Tris, pH 7.2, 75 mm NaCl, and 2 mm β-mercaptoethanol. Binding affinity of RPA70N with HDHB peptide was measured using a MicroCal VP isothermal titration calorimeter. Titration experiments were performed by first injecting 2 μl of 1 mm HDHB peptide into 75 μm RPA70N in the sample cell followed by additional 10-μl injections. Data were analyzed using Origin software. Thermodynamic parameters and binding constant (Kd) were calculated by fitting the data to the best binding model using a nonlinear least squares fitting algorithm.
NMR Spectroscopy
NMR experiments were performed using Bruker DRX 500-MHz or 600-MHz spectrometers equipped with cryoprobes. 15N-1H heteronuclear single-quantum coherence (HSQC) spectra were acquired using 1024 complex points in the 1H dimension and 128 complex points in the 15N dimension. 15N-enriched RPA70N sample was prepared at 100 μm in a buffer containing 20 mm Tris, pH 7.5, 75 mm NaCl and 2 mm DTT. A series of 15N-1H HSQC spectra were collected at RPA70N/HDHB peptide molar ratios of 1:0, 1:0.5, 1:1, 1:2, and 1:4. All spectra were processed by Topspin v2.0 (Bruker, Billerica, MA) and analyzed with Sparky (University of California, San Francisco, CA).
Gene Silencing
The pRetro-Super vector was kindly provided by R. Agami (35). HDHB shRNA (CAGGTGCTTGGTGGAGAGT) and control shRNA (GACCCGCGCCGAGGTGAAG) were cloned into pRetro-Super. The pRetro-Super plasmids were then transfected into the retrovirus packaging cell line Phoenix 293 as described on the Nolan laboratory website (Stanford University) with minor modifications. Briefly, cells were transfected with each pRetro-Super-derived plasmid and selected with 5 μg/ml puromycin (Sigma). To harvest virus, cells at 75% confluence were incubated for 16 h at 37 °C. Collected media were passed through a 0.45-μm syringe filter (Pall Corp.). To obtain stable HDHB knockdown in HCT116, cells were infected with virus stock preincubated with 4 μg/ml Polybrene (Sigma). After overnight incubation, cells were replated in growth medium and selected in 5 μg/ml puromycin for 7–10 days.
For transient HDHB knockdown, HeLa cells were transfected with pGIPZ HDHB 33141 (shRNA1: GCAAGACTGTGATCTAATT) or 33143 (shRNA2: CCAGTTCTCAGTCATCTAA) (Open Biosystems) and selected with 3 μg/ml puromycin. Non-silencing pGIPZ was used as control.
For RPA70 knockdown, RPA70 siRNA (AACACUCUAUCCUCUUUCAUG) and control siRNA (AUGAACGUGAAUUGCUCAA) (Dharmacon) were transfected into HeLa cells exactly as described (33). Transfections were done using Lipofectamine 2000 (Invitrogen) for HeLa cells and FuGENE HD (Roche Applied Science) for U2OS cells according to the manufacturer's protocol. Cells transiently silenced for HDHB or RPA70 were analyzed 72 h post-transfection.
Clonogenic Survival Assay
Stably HDHB- or control-silenced HCT116 cells were seeded in 60-mm dishes (∼800 per dish). Cells were allowed to attach to the dish for 12 h, then treated in triplicate with different concentrations of CPT for 12 h, washed twice with PBS, and incubated in fresh growth medium for 10 days. Cell colonies were fixed and stained with 0.5% crystal violet in 70% ethanol. Visible colonies were counted. Experiments were repeated at least 3 times.
Chromosome Analysis in Metaphase Spreads
HCT116 cells stably silenced for HDHB or control-silenced were cultured in a 100-mm dish to 30% confluence followed by the addition of aphidicolin (0.2 or 0.4 μm) for 24 h and then 100 ng/ml colcemid (Roche Applied Science) for 2 h. Cells were trypsinized, washed once with PBS, resuspended in 10 ml of prewarmed 75 mm KCl, and incubated for 10 min at 37 °C. Cells were collected by centrifugation (5 min, 800 rpm) and resuspended in 0.2 ml of 75 mm KCl. To fix the cells, 5 ml of pre-chilled acetic acid/methanol (1:3) was dropped into the cell suspension while vortexing, mixed immediately, and incubated for at least 30 min on ice. For staining, cells were collected by centrifugation, washed once with cold fresh fixative, then resuspended in fresh fixative (0.5 ml for ∼107 cells) and dropped onto wet cold slides (slides were kept in 70% ethanol at −20 °C) on ice from an ∼10-cm height. Slides were air-dried, baked at 65 °C for 2 h, and stained with 4% Giemsa in 10 mm phosphate buffer for 15 min. Slides were rinsed with water, dried, and mounted on coverslips with Cytoseal 60 (Richard-Allan Scientific). Slides were observed under a bright field microscope, and 100 cells, each with 45–46 chromosomes, per sample were counted.
RESULTS
DNA Damage Induces Accumulation of HDHB on Chromatin
To elucidate potential roles for HDHB in chromosomal replication, we reasoned that variation in its subcellular localization as a function of the cell cycle might correlate with its function. Because HDHB was easily detectable in whole cell extracts of U2OS, HeLa, and HCT116 tumor cells but much less abundant in primary cells (supplemental Fig. S1B), these cell lines were chosen for study.
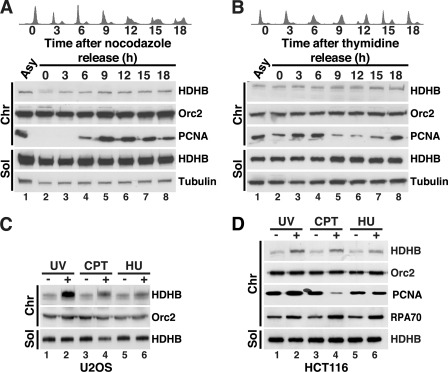
Initially, U2OS cells were biochemically fractionated using an established method (supplemental Fig. S1C). U2OS cells were released from a nocodazole block and fractionated at 3-h intervals, and proteins in each sample were analyzed in Western blots (Fig. 1A). Tubulin was detected in the soluble fraction and Orc2 in the chromatin fraction as expected. Chromatin-bound PCNA was absent in G1 and began to accumulate in early S (6 h after release) as expected. HDHB was found mainly in the soluble fraction throughout the cell cycle, and the level of chromatin-bound HDHB remained very low, with a barely detectable increase in S phase (Fig. 1A, lanes 5–8). Cells released from a double thymidine block displayed a similar subcellular distribution of HDHB (Fig. 1B). The results indicate that unlike the authentic replication fork protein PCNA, the subcellular distribution of HDHB fluctuated little during the cell cycle.
FIGURE 1.
DNA damage induces HDHB accumulation on chromatin. A, U2OS cells were released from a nocodazole block for the indicated times, and cell cycle distribution was characterized by flow cytometry (upper panels). Cells from each time point were separated into soluble (Sol) and chromatin fractions (Chr) (22) and analyzed by Western blot with the indicated antibodies (lower panels). Fractions from asynchronous cells were analyzed in parallel (Asy). B, U2OS cells were released from a double thymidine block for the indicated times, and cell cycle distribution was characterized by flow cytometry (upper panels). Cells from each time point were separated into soluble (Sol) and chromatin fractions (Chr) (22) and analyzed by Western blot with the indicated antibodies (lower panels). Fractions from asynchronous cells were analyzed in parallel (Asy). C and D, asynchronous U2OS (C) or HCT116 (D) cultures treated (+) with 100 J/m2 UV, 10 μm CPT, or 5 mm HU for 2 h or left untreated (−) were fractionated as in A and B and analyzed by Western blotting with the indicated antibodies.
To examine the possibility that the low level of chromatin-bound HDHB might function in DNA repair rather than in bulk DNA replication, asynchronously growing U2OS cells were treated with DNA-damaging agents, biochemically fractionated, and analyzed by Western blot. Exposure to ultraviolet irradiation (UV), the topoisomerase I inhibitor camptothecin (CPT), or the ribonucleotide reductase inhibitor hydroxyurea (HU) induced accumulation of HDHB on chromatin (Fig. 1C). The level of chromatin-bound HDHB correlated with the amount or duration of exposure to each genotoxin (supplemental Fig. S2, A–F). A similar increase in chromatin-bound HDHB was observed in HCT116 cells treated with UV, CPT, or HU (Fig. 1D), indicating that damage-induced HDHB accumulation on chromatin was not a peculiarity of the U2OS cell background. Chromatin-bound PCNA was clearly decreased in the CPT-treated cells (Fig. 1D, lane 4), an observation consistent with the ability of CPT-topoisomerase I cleavage complexes to cause replication fork collapse (36). All three agents induced a modest, but clearly detectable, increase in chromatin-bound RPA, suggestive of replication stress (33, 37, 38) (Fig. 1D, compare lanes 2, 4, and 6 with lanes 1, 3, and 5).
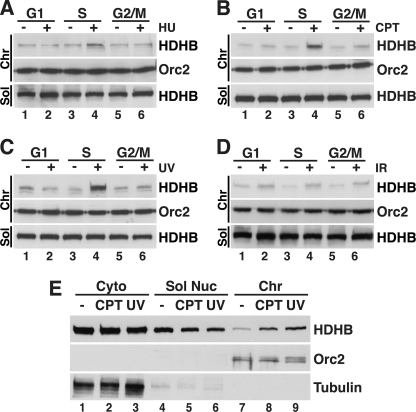
If the damage-induced accumulation of HDHB on chromatin were indeed associated with replication stress, one would expect to observe it preferentially in S phase cells. This prediction was tested by enriching U2OS cells in G1, early-mid S phase, or G2/M, then exposing them to HU, CPT, UV, or ionizing radiation (IR) followed by biochemical fractionation and Western blot analysis. HU, CPT, and UV induced the accumulation of chromatin-bound HDHB almost exclusively in S phase cells, with little or no increase observed in G1 or G2/M cells (Fig. 2, A–C). These findings are consistent with replication stress-induced accumulation of HDHB on chromatin. In addition, cells exposed to IR also displayed a modest induction of chromatin-bound HDHB in G1, S, and G2/M (Fig. 2D, compare lanes 1, 3, and 5 with lanes 2, 4, and 6). IR-induced damage includes DNA double strand breaks, which undergo limited processing by nucleases to generate short stretches of ssDNA that facilitate repair (39–42). Thus, chromatin structures generated in response to IR may display features in common with the extended stretches of RPA-coated ssDNA in replication-stressed chromatin. These findings indicate that HDHB accumulates on chromatin in response to replication stress and to a lesser extent in response to IR-induced damage.
FIGURE 2.
HDHB accumulates on chromatin in response to replication stress. A–D, U2OS cells enriched in G1, S, or G2/M phase as described under “Experimental Procedures” were treated with 5 mm HU (A), 10 μm CPT (B), or 100 J/m2 UV (C), or 20 gray IR (D) or left untreated (−) and analyzed as described in (Fig. 1C). E, U2OS cells synchronized in S phase and treated with UV or CPT as in Fig. 1C or left untreated were extracted using digitonin in isotonic buffer to release cytosolic proteins as described previously (20). Nuclei were extracted to separate soluble nuclear from chromatin-bound proteins. Fractions were analyzed by Western blotting with the indicated antibodies. Sol, soluble fraction; Chr, chromatin fraction.
This DNA damage-induced accumulation of chromatin-bound HDHB could reflect a redistribution of soluble HDHB to chromatin or an increased level of total HDHB after damage. To distinguish between these possibilities, S phase cells that had been treated with CPT, UV, or left untreated were biochemically fractionated with a different protocol to generate separate cytosolic, soluble nuclear, and chromatin-associated protein fractions. We found that the soluble nuclear fractions from CPT- and UV-treated cells contained less HDHB than did that from control cells (Fig. 2E, top row, compare lane 4 with lanes 5 and 6). Conversely, the chromatin fractions from the CPT- and UV-treated cells contained more HDHB than that from untreated cells (Fig. 2E, lanes 7–9). The level of HDHB in the cytosolic fraction of CPT- or UV-treated cells was not detectably different from that of untreated cells (compare lane 1 with lanes 2 and 3). The results are most consistent with a replication stress-induced recruitment of soluble nuclear HDHB to chromatin.
Requirements for HDHB Recruitment to Chromatin
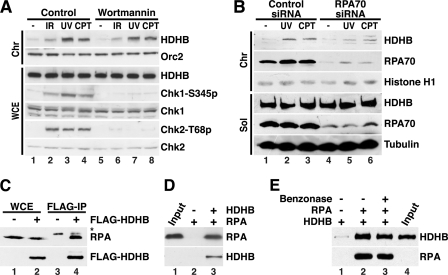
To determine the requirements for HDHB recruitment to chromatin in response to replication stress, we first considered that phosphoinositide 3-kinase-related protein kinases (PIKK) ATM, ATR, and DNA protein kinase activated by DNA damage might recruit HDHB to chromatin. Consistent with this possibility, HDHB contains several predicted PIKK phosphorylation sites and was identified by mass spectrometry as a target for ATM/ATR after DNA damage (12). To test for a possible role for PIKK activity in recruitment of HDHB to chromatin, we briefly treated cells with wortmannin, a broad-spectrum inhibitor of PIKK family kinases (43), before exposing them to DNA damaging agents. As expected, exposure to IR, UV, and CPT resulted in robust phosphorylation of Chk1 serine 345 and Chk2 threonine 68 in control cells, with little effect on total Chk1 or Chk2 (Fig. 3A, compare lane 1 with lanes 2–4), and checkpoint signaling was strongly inhibited in the presence of wortmannin (compare lane 5 with lanes 6–8). Importantly, HDHB recruitment to chromatin after genotoxin treatment was virtually identical in the presence and absence of checkpoint signaling (Fig. 3A, top row, compare lanes 1–4 with lanes 5–8). Thus inhibition of PIKK activity does not reduce DNA damage-induced recruitment of HDHB to chromatin.
FIGURE 3.
Replication stress-induced redistribution of HDHB does not require checkpoint signaling but correlates with the level of RPA on chromatin. A, U2OS cells were pretreated with DMSO (control) or 200 μm wortmannin for 30 min and then exposed to 20-gray IR, 100 J/m2 UV, or 10 μm CPT for 2 h. Whole cell extracts (WCE) or chromatin fractions (Chr) were analyzed by Western blotting with the indicated antibodies. B, HeLa cells transiently transfected with RPA70 siRNA or control siRNA were treated with UV or CPT as in Fig. 1C or left untreated (-) as a control. Soluble (Sol) and chromatin fractions (22) were analyzed by Western blotting with the indicated antibodies. C, whole cell extracts prepared from HeLa cells transiently transfected with pFLAG-HDHB (+) or empty pFLAG (-) were incubated with anti-FLAG-M2 agarose beads. Proteins bound to the beads were analyzed by SDS-PAGE and Western blotting with anti-FLAG or monoclonal anti-RPA antibody 34A as indicated. *, nonspecific band. IP, immunoprecipitation. D, purified WT heterotrimeric RPA was incubated in the absence (−) or presence (+) of purified HDHB as indicated with protein A beads prebound to anti-HDHB antibody. Proteins bound to the beads were analyzed by SDS-PAGE and Western blotting with mouse anti-RPA70C and rabbit anti-HDHB. Input shown is 20%. E, purified HDHB was incubated in the absence (−) or presence (+) of purified RPA trimer as indicated with protein A beads pre-bound to anti-RPA70C IgG. Concurrent incubation with benzonase (lane 3) had no effect on the interaction. Proteins bound to the beads were visualized by Western blotting with monoclonal RPA70B or polyclonal rabbit anti-HDHB. Input shown is 80%.
Replication stress-induced recruitment of S phase checkpoint proteins, e.g. ATRIP, to chromatin depends on their ability to bind to the RPA-coated ssDNA that accumulates at sites of DNA damage (33, 37, 38). To test the possibility that damage-induced recruitment of HDHB may be mediated by RPA, HeLa cells transiently depleted of RPA70 were exposed to UV or CPT and then biochemically fractionated. The level of RPA70 in the soluble and chromatin fractions was substantially lower in RPA-silenced cells than in control-silenced cells, validating the knockdown (Fig. 3B, RPA70 rows, compare lanes 1–3 with lanes 4–6). Consistent with published evidence (33), RPA70 depletion slightly increased the fraction of cells in S phase and reduced S phase checkpoint signaling. Also as expected, the level of RPA70 on chromatin increased in response to UV and CPT both in control-silenced and in RPA-silenced cells (Fig. 3B, row 2, compare lanes 2 and 3 with lane 1 and lanes 5 and 6 with lane 4). RPA70 knockdown did not affect the level of HDHB in the soluble fraction (row 4, compare lanes 1–3 with 4–6), but the level of chromatin-bound HDHB after UV and CPT treatment was lower in RPA70-silenced cells than in control-silenced cells (Fig. 3B, top row, compare lanes 2 and 3 with lanes 5 and 6). Thus HDHB recruitment to chromatin in response to replication stress correlates with the level of chromatin-bound RPA.
The recruitment of soluble nuclear HDHB to chromatin in response to replication stress, the dispensability of PIKK activity for this recruitment, and the correlation of HDHB recruitment with the RPA level on chromatin led us to question whether HDHB recruitment to chromatin might be mediated by RPA. If chromatin-bound RPA were directly responsible for recruiting soluble HDHB to chromatin, one would expect HDHB to interact physically with RPA. Consistent with this prediction, endogenous RPA was co-precipitated with FLAG-HDHB from extracts of cells transiently expressing FLAG-HDHB but not FLAG-vector (Fig. 3C, compare lanes 3 and 4). Purified RPA bound to anti-HDHB antibody beads in the presence of purified recombinant HDHB (Fig. 3D, lane 3), but not in its absence (lane 2), suggesting a direct physical interaction between the two proteins. Reciprocal pulldown assays of purified RPA and HDHB in the presence of the potent Benzonase nuclease did not diminish their physical interaction (Fig. 3E), providing additional evidence for a direct interaction.
HDHB Interacts Specifically with the N-terminal Region of RPA70
Detailed biochemical mapping and structural analysis of the RPA-HDHB interaction was then pursued to fully define the molecular basis for HDHB localization to replication-stressed chromatin. RPA interacts with partner proteins, utilizing four of its seven structural domains: the N-terminal domain of RPA70 (RPA70N), the tandem high affinity ssDNA binding domains A and B of RPA70 (RPA70AB), and the C-terminal domain of RPA32 (RPA32C) (44–48) (Fig. 4A). To map the HDHB binding site(s) in RPA, purified His-tagged RPA domains were added to FLAG antibody beads preincubated with control or FLAG-HDHB extracts, and RPA domains captured on the beads were detected by immunoblotting with anti-His antibody. Under these conditions, HDHB interacted specifically with RPA70N+L but not with RPA70AB, RPA32C, or the trimerization core RPA70C/32D/14 (Fig. 4B).
RPA70N (residues 1–120) serves as a chromatin recruitment domain for several DNA damage response proteins (33, 38), raising the possibility that HDHB might also be recruited to RPA-ssDNA by docking with RPA70N. To search for potential RPA70-interacting regions in HDHB, we first designed HDHB fragments using tools for secondary structure, disorder, and -fold prediction, as described under “Experimental Procedures” (Fig. 4C), and expressed them as His-tagged polypeptides in E. coli. The purified His-tagged HDHB fragments (Fig. 4D, lane 1) were then incubated with glutathione beads bound to GST or GST-RPA70N. HDHB residues 394–958 and 459–811 bound specifically to GST-RPA70N beads but not to GST (Fig. 4D, lanes 2 and 3). Other HDHB polypeptides did not bind to either GST or GST-RPA70N, demonstrating a specific interaction between HDHB 459–811 and RPA70N. Because this same fragment of HDHB was also recognized by the monoclonal antibody 4C11 (Fig. 4E; supplemental Fig. S1A), we asked whether the antibody might compete with RPA70N to bind HDHB. Interestingly, preincubation of HDHB with increasing concentrations of 4C11 IgG inhibited binding of HDHB to GST-RPA70N, whereas non-immune control IgG had no effect (Fig. 4F). We conclude that the HDHB residues 459–811 are sufficient to interact directly and specifically with RPA70N.
A Conserved Acidic Motif in HDHB Interacts Physically with the Basic Cleft of RPA70N
The RPA70N-interacting surfaces of p53, ATRIP, Rad9, and Mre11 were recently mapped at the atomic level to an acidic stretch of residues in each protein (33). We used this information together with the mapping data in Fig. 4 and amino acid sequence alignment of HDHB orthologs and E. coli RecD to search for potential RPA70N binding motifs. The search revealed a phylogenetically conserved acidic peptide, residues 493–517, inserted between the superfamily 1B helicase motifs I and Ia (Fig. 5A). This sequence motif is absent in the corresponding region of E. coli RecD, the prototype member of helicase superfamily 1B, implying that it may not be necessary for helicase activity and might have another role. To investigate the interaction of this region of HDHB with RPA70N, we designed a synthetic peptide (HDHB residues 493–517) that contains the conserved acidic residues of HDHB. Isothermal titration calorimetry experiments were performed to measure the affinity of interaction between RPA70N and the HDHB peptide (supplemental Fig. S3A). The binding isotherm was fit with a single site binding model and resulted in a Kd of 15 ± 0.05 μm, confirming that the peptide interacts physically with RPA70N.
FIGURE 5.
The basic cleft of RPA70N physically interacts with a conserved acidic motif in HDHB. A, shown is the sequence alignment of HELB from four vertebrate species and E. coli RecD. Consensus sequence in the acidic motif is based on all available vertebrate HELB sequences: Star (*), identical; colon (:), conserved; dot (.), semi-conserved residues. Red asterisk, residues substituted by alanine to generate the HDHB 3xA mutant (see E below). Red font, a synthetic HDHB peptide Glu-493–Trp-517 used in NMR (B and C) and isothermal titration calorimetry (supplemental Fig. S3A) experiments. B, shown are overlaid 15N-1H HSQC spectra of RPA70N in the absence (black) and presence of HDHB peptide at 1:0.5 (blue), 1:1 (green), 1:2 (pink), and 1:4 (red) molar ratios. The black arrows show chemical shift perturbations that result from binding of RPA70N with HDHB peptide. C, molecular surface diagram of RPA70N with the significantly perturbed residues labeled. Red, acidic residues; blue, basic residues. D, GST pulldown assays of purified full-length HDHB with wild type (WT) or mutant (R41/43E) GST-RPA70N. Bound proteins were detected by immunoblotting with anti-HDHB (upper panel) or anti-GST (lower panel). E, extracts from HeLa cells transiently expressing FLAG-WT or -3xA HDHB were mixed with glutathione beads prebound to GST or GST-RPA70N. Proteins bound to the beads were analyzed by SDS-PAGE and immunoblotting with anti-FLAG (upper panel) or anti-GST (lower panel) antibodies. F, purified His-tagged WT and 3xA HDHB were analyzed by SDS-PAGE and stained with Coomassie. G and H, helicase activity of purified His-tagged WT and 3xA HDHB proteins was assayed as described (9) using a radiolabeled partial duplex DNA substrate (S) and the indicated amounts of purified HDHB. Helicase activity was visualized by phosphorimaging and quantified (lanes 4–9) after subtraction of radiolabeled background (product band P) detected in the DNA substrate (lane 1) and in the absence of ATP (lanes 3 and 7). BS, boiled substrate. Brackets show S.E. (n ≥ 3).
To map the specific binding surface of the HDHB peptide on RPA70N, NMR chemical shift perturbation experiments were performed on 15N-enriched RPA70N. The series of 15N-1H HSQC spectra acquired with an increasing concentration of unlabeled peptide added into the solution revealed perturbations to a select number of peaks in the spectrum. This observation indicates that the binding interface between RPA70N and the peptide is specific (Fig. 5B). The disappearance of signals at a substoichiometric molar ratio of peptide to RPA70N is consistent with the 15 μm Kd estimated by isothermal titration calorimetry. Analysis of the data showed that, in addition to the RPA70N basic residues (Arg-31, Arg-41, and Arg-43), hydrophobic residues (Ile-33, Tyr-42, Leu-44, Phe-56, Met-57, Leu-58, Ala-59, Val-84, Leu-87, Ile-95, Leu-96, and Leu-99) are involved in binding with HDHB (Fig. 5, B and C; supplemental Fig. S3B). Mapping of these residues onto the structure of RPA70N reveals that HDHB binds to the basic cleft of the oligonucleotide/oligosaccharide binding-fold domain (Fig. 5C). This binding surface in RPA70N resembles that recognized by p53, Rad9, ATRIP, and Mre11 (33, 49, 50).
The importance of the RPA70N basic cleft and the HDHB acidic motif in the binding interaction was then tested in pulldown experiments with wild type and mutant proteins. GST-RPA70N interacted with HDHB as expected, and charge reverse substitutions in the basic cleft of GST-RPA70N (R41/43E) abolished HDHB binding (Fig. 5D). A 3xA mutant form of FLAG-HDHB with alanine substitutions in HDHB acidic residues Glu-499, Asp-506, and Asp-510 was generated to test the role of this motif in binding to RPA70N. Full-length FLAG-HDHB WT was pulled down by GST-RPA70N, but FLAG-HDHB 3xA was not (Fig. 5E). The results confirm the roles of the acidic motif in HDHB and the basic cleft of RPA70N in the interaction.
We purified recombinant WT and 3xA HDHB proteins to assess the specificity of the 3xA substitution on HDHB loss of function (Fig. 5F). The 3xA and WT HDHB displayed comparable helicase activity (Fig. 5, G and H), demonstrating that the 3xA substitution did not perturb the helicase domain stability, overall fold, or activity but specifically impaired the ability of HDHB to bind to RPA70N. Altogether, these results establish a direct physical interaction between the HDHB acidic motif and the RPA70N basic cleft.
HDHB Depletion Impairs Recovery from Replication Stress
The evidence presented above demonstrates a replication stress-induced, RPA-dependent, PIKK activity-independent redistribution of soluble nuclear HDHB to chromatin in two different tumor cell lines as well as a specific, direct physical interaction of a conserved acidic peptide in HDHB with the RPA70N basic cleft. These findings would be consistent with the hypothesis that the unidentified role of HDHB in replication may lie in mitigating replication stress. To establish a basis to address this possibility, shRNAs H1 and H2, targeting two different HDHB sequences, were expressed in HeLa cells and selected for co-expression of puromycin resistance (Fig. 6A). Comparison of HDHB- and control-silenced cells by two-dimensional flow cytometry revealed that the substantial reduction in HDHB levels had little effect on cell cycle distribution in the absence of overt damage (Fig. 6B).
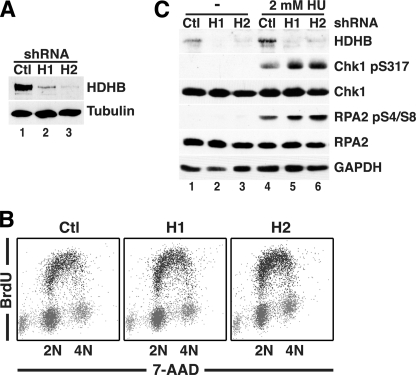
FIGURE 6.
Transient HDHB depletion does not disrupt replication stress-induced checkpoint signaling but impairs recovery of HeLa cells from replication stress. A, whole cell extracts of HeLa cells transiently expressing non-silencing (Ctl) or HDHB-silencing shRNAs (H1 and H2) were analyzed by Western blotting with the indicated antibodies. B, HeLa cells transiently expressing non-silencing (Ctl) or HDHB-silencing shRNAs (H1 and H2) were incubated with 10 μm BrdU for 30 min and stained for BrdU and total DNA and analyzed by flow cytometry. C, HeLa cells transiently expressing non-silencing (Ctl) or HDHB-silencing shRNAs (H1 and H2) as in A were exposed to 2 mm HU for 2 h or left untreated (−) as indicated. Whole cell extracts were then analyzed by SDS-PAGE and Western blotting with antibodies against HDHB, Chk1 phospho-Ser-317, RPA32 phospho-Ser-4, Ser-8, total RPA32 (RPA2), and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) as a loading control.
Based on the correlation of replication stress-induced HDHB recruitment with the level of chromatin-bound RPA (Fig. 3B) and the specific interaction of HDHB with the basic cleft of RPA70N (Figs. 4 and 5), we first considered that HDHB recruitment to chromatin might be important for activation of checkpoint signaling. To assess this possibility, HDHB- or control-silenced HeLa cells were exposed to HU to induce replication stress or left untreated. Analysis of whole cell extracts by Western blotting revealed that in control-silenced cells, HU induced HDHB recruitment to chromatin (Fig. 6C, compare lane 4 with lane 1), just as was observed in HU-treated U2OS and HCT116 cells (Fig. 1, C and D). HDHB knockdown did not induce detectable checkpoint signaling in the absence of HU (Fig. 6C, lanes 1–3). Both HDHB-silenced cultures displayed robust induction of phospho-Chk1 and N-terminally phosphorylated RPA32 in HU-treated extracts (Fig. 6C, compare lanes 5 and 6 with lanes 2 and 3). Unexpectedly, slightly stronger checkpoint signaling was detected in HDHB-silenced extracts than in control-silenced extract (compare lanes 5 and 6 with lane 4). We conclude that HDHB does not function to promote HU-induced activation of S phase checkpoint signaling. On the contrary, HDHB depletion modestly enhanced replication stress signaling, consistent with the hypothesis that HDHB might counteract replication stress.
The role of HDHB in cellular recovery from replication stress was further examined in two different assays using CPT-treated HCT116 cells. HDHB was first stably knocked down using a retroviral shRNA expression vector that targeted a third HDHB sequence. The cell cycle distribution of the stably HDHB-silenced cells was comparable with that of the control-silenced cells (Fig. 7, A and B), consistent with the results observed above in transiently silenced HeLa cells.
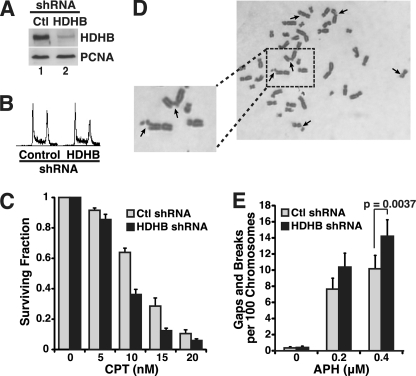
FIGURE 7.
Delayed recovery from replication stress in HCT116 cells stably depleted of HDHB. A, extracts of HCT116 cells stably expressing control (Ctl) or HDHB shRNAs were analyzed by immunoblotting with indicated antibodies. B, flow cytometry of HDHB- and control-depleted HCT116 cells is shown. C, colonies formed by HDHB-depleted (black) or control-depleted (gray) HCT116 cells after exposure to the indicated doses of CPT for 12 h were quantified. The surviving fraction of colonies formed by untreated cells was set to 1. Values for CPT-treated cells represent the average from three independent experiments; brackets indicate S.D. D, images of metaphase chromosomes from HDHB-silenced HCT116 cells exposed to 0.4 μm aphidicolin (APH) for 24 h are shown. Arrows indicate chromosome gaps and breaks. E, shown is quantification of chromosome gaps and breaks in metaphase spreads from control (light gray)- and HDHB (black)-silenced HCT116 cells after exposure to the indicated concentrations of APH for 24 h. Brackets indicate S.D. n ≥ 3. p value was calculated using two-tailed Student's t test.
The role of HDHB in recovery from CPT-induced replication stress was then monitored in clonogenic survival assays. Equal numbers of HDHB- and control-depleted HCT116 cells were cultured in the absence or presence of CPT for 12 h, and colonies formed by surviving cells were counted after 10 days. Exposure to 10, 15, or 20 nm CPT reduced colony formation of HDHB-depleted cells to about half that of control-depleted cells (Fig. 7C), suggesting that HDHB-depletion sensitizes cells to CPT-induced damage.
We also monitored the ability of stably HDHB-silenced HCT116 cells to recover from replication stress induced by exposure to partially inhibitory concentrations of aphidicolin, which uncouples DNA synthesis from DNA unwinding at the fork, resulting in extended stretches of RPA-ssDNA and expression of common fragile sites (51–54). Metaphase chromosomes prepared from HDHB-silenced cells exposed to aphidicolin displayed breaks and gaps (Fig. 7D, arrows), whereas few chromosomal aberrations were observed in either HDHB- or control-silenced cells cultured without aphidicolin (Fig. 7E). Evaluation of chromosomal breaks and gaps in aphidicolin-treated cells revealed a dose-dependent increase in chromosomal instability, with significantly more aberrations in HDHB-silenced than in control-silenced cells (Fig. 7E). These results provide additional evidence for a role of HDHB in recovery from replication stress.
DISCUSSION
Previous work on HELB implicated its helicase activity in chromosomal replication, but its functional role remained elusive. Here we show that although the level of HDHB on chromatin is quite constant throughout the cell cycle, additional HDHB is recruited to chromatin in three different tumor cell lines upon exposure to a variety of DNA damaging agents. UV-, CPT-, and HU-induced HDHB recruitment to chromatin is dose-dependent and occurs preferentially in S phase cells. This recruitment does not depend on checkpoint kinase activity but does correlate with the level of RPA on chromatin.
HDHB recruitment thus closely resembles that of S phase checkpoint proteins (37). HDHB interacts directly with the N-terminal domain of the RPA70 subunit, a primary recruitment scaffold for multiple S phase checkpoint proteins (33, 49, 50). On the other hand, HDHB knockdown does not impair S phase checkpoint signaling in response to agents that induce replication stress, arguing against a role for HDHB in checkpoint activation. Nevertheless, HDHB knockdown delays or diminishes cellular recovery from HU- or CPT-induced replication stress. Based on these results, we suggest that the primary role of HDHB in chromosomal replication is to mitigate replication stress. We note that the residual level of HDHB in knockdown cells was sufficient to cope with endogenous replication stress or damage but not with additional stress induced by HU, CPT, or aphidicolin (Figs. 6 and 7). In contrast, mutational inactivation of HDHB helicase activity resulted in early S phase arrest, perhaps due to its inability to overcome endogenous replication stress (5, 8, 9).
Replication Stress-dependent Recruitment of HDHB to Chromatin
The checkpoint-independent, RPA-dependent accumulation of HDHB on chromatin in response to replication stress (Figs. 2 and 3; supplemental Fig. S2) and the direct physical interaction of a conserved acidic peptide in HDHB with the basic cleft of RPA70N (Figs. 4 and 5) are consistent with a role for RPA70N-HDHB interaction in recruiting HDHB to chromatin. Although technical difficulties have so far confounded our efforts to directly demonstrate the importance of these motifs in recruiting HDHB to chromatin, the interaction closely resembles that of RPA70N with ATRIP, Rad9, and Mre11 (28, 33, 50). Qualitatively, acidic peptides from each of these proteins have been shown to bind specifically to the same surface of RPA70N that binds the acidic peptide from HDHB (33). Quantitatively, a substoichiometric molar ratio of the HDHB peptide to RPA70N (0.5:1) was sufficient to induce strong chemical shift perturbations in the NMR spectrum of RPA70N (Fig. 5B; supplementary Fig. S3B). This was unexpected because a 10-fold greater molar ratio of the ATRIP, Rad9, or Mre11 peptide over RPA70N (a peptide:RPA70N ratio of 4–6:1) was needed under very similar experimental conditions to induce a comparable chemical shift perturbation in the RPA70N spectra (33). The comparison implies that the affinity of HDHB interaction with RPA70N is considerably greater than that of ATRIP, Rad9, or Mre11.
It is interesting to consider the potential functional implications of the quantitatively stronger binding of HDHB to RPA70N. The observation that S phase checkpoint kinase activity is not needed to recruit HDHB to chromatin in response to replication stress (Fig. 3A) implies that HDHB is recruited in parallel with the S phase checkpoint signaling proteins. Based on the stronger HDHB-RPA70N interaction, it is possible that HDHB can be more readily recruited than the checkpoint assembly proteins to RPA-coated ssDNA at sites of replication stress, or that a shorter stretch of RPA-ssDNA would be sufficient to attract HDHB. In this case HDHB might act “on the fly” to bypass or otherwise counteract the cause of the replication stress, thereby obviating the need to assemble a checkpoint signaling complex. Should HDHB recruitment fail to promptly relieve the replication stress, longer stretches of RPA-ssDNA would accumulate and serve as the scaffold for recruiting S phase checkpoint proteins. This speculation could provide a plausible explanation for the somewhat more intense checkpoint signaling observed in HDHB-silenced than in control-silenced cells (Fig. 6C). The ability of HDHB to mitigate DNA damage and modulate the intensity of checkpoint signaling may be important in certain tissues, such as thymus and testis, or for tumor cell viability.
How Does HDHB Stimulate Recovery from Replication Stress?
Depletion of HDHB led to increased checkpoint signaling in HU-treated cells, decreased viability of cells exposed to CPT, and increased chromosomal breaks and gaps in cells recovering from aphidicolin (Figs. 6 and 7). The fact that the data were generated in two different cell lines, depleted transiently or stably with three different shRNAs expressed from two different vectors, and evaluated in three different assays suggests that the observed results are not likely to be the consequence of off-target silencing or another experimental peculiarity. We conclude that HDHB has one or more biochemical activities that stimulate cellular recovery from replication stress.
Consistent with the replication-defective mutant phenotypes that led to HELB discovery (5, 9), the helicase activity of HDHB is likely to play a fundamental role in relieving replication stress. Exactly how recruitment of the 5′ to 3′ helicase activity to chromatin might facilitate recovery from replication stress remains unknown. One possible mechanism is based on the ability of superfamily 1 helicases to work as a “cooperative inchworm” capable of generating sufficient force to displace streptavidin from biotin-labeled DNA (55). The superfamily 1B helicase Rrm3, which migrates with progressing replication forks in budding yeast, is thought to use this mechanism to displace stably bound transcription complexes that block replication fork progression (56–58). In a possibly related mechanism observed in prokaryotes, the 3′-5′ superfamily 1 helicase Rep co-migrates with the 5′-3′ hexameric replicative helicase at the fork and serves as an auxiliary helicase to overcome fork stalling (59–61). The HDHB that accumulates upon replication stress may make use of such mechanisms to clear obstacles that impede an advancing fork.
The primosome activity of HELB might also play a role in counteracting replication stress (4, 9). RPA-coated ssDNA is refractory to primer synthesis by DNA polymerase α-primase (62, 63). However, a mediator protein, e.g. the SV40 helicase large T antigen, which interacts physically with both proteins, can displace RPA from the template and in concert load DNA polymerase α-primase on the exposed template (64–66). HDHB interacts with both RPA (Figs. 3–5) and DNA polymerase α-primase (9), suggesting that HDHB recruitment to chromatin might enable it to re-prime the leading strand downstream of stalled forks. Such damage bypass/fork re-priming mechanisms are observed among distantly related bacteria from B. subtilis to E. coli and were recently detected in eukaryotes (10, 11, 61, 67–69).
The sequence similarity of HELB with prokaryotic proteins involved in homology-dependent DNA repair suggests another potential mechanism for HDHB to mitigate replication stress. Of particular interest is the ability of the superfamily 1B helicase Dda from phage T4, in conjunction with the T4 recombinase UvsX, to rescue stalled forks through two sequential template-switching reactions (70, 71). The possibility that HDHB might mitigate replication stress in part through homology-dependent fork recovery mechanisms merits further investigation (72–75). It will be interesting to learn which of these several HDHB activities contribute to its ability to relieve different kinds of replication stress.
HDHB, a Damage Tolerance Protein?
The results of this study demonstrate replication stress-induced recruitment of HDHB to chromatin in a checkpoint-independent and RPA-dependent manner and provide evidence that HDHB functions to relieve replication stress. The molecular features of HDHB interaction with RPA closely resemble those of proteins that initiate the assembly of S phase checkpoint complexes at sites of replication stress, yet we have not detected any HDHB contribution to checkpoint signaling. Instead, HDHB joins a diverse group of damage tolerance proteins that are recruited to sites of replication stress through interactions with one or more surfaces of RPA. Three prominent examples are the PCNA-modifying ubiquitin ligase Rad18, which binds to RPA70AB (76, 77), the DNA translocase Smarcal 1/HARP (Ref. 78 and references therein), which binds to RPA32, and RecQ helicases WRN and BLM, which interact physically and functionally with several regions of RPA70 (79–81). Thus it will be important to elucidate the roles of HDHB in this network of damage tolerance proteins.
Supplementary Material
Acknowledgments
We thank D. Pretto, D. Cortez, X. Xu, C. Lovejoy, J. Gerhardt, M. Wold, M. K. Kenny, A. Bochkarev, R. Agami, J. G. Patton, Y. Dimitrova, M. Karakas, A. Morin, C. Bansbach, E. Nam, and G. Sowd for advice, criticism, reagents, and valuable assistance in assembling the manuscript and K. Zhao and J. Duryea for technical assistance. We are grateful to S. Schaffer and Vanderbilt Cell Imaging Shared Resource for assistance in fluorescence imaging and B. Matlock and D. Flaherty of Vanderbilt Flow Cytometry Core for assistance in flow cytometry.
This work was supported, in whole or in part, by National Institutes of Health Grants GM52948 (to E. F.), GM65484 (to W. J. C.), P30 CA068485 (to the Vanderbilt-Ingram Cancer Center), and P30 ES00267 (to the Center for Molecular Toxicology) and Vanderbilt University.
This work is dedicated to the memory of Arturo Falaschi, M.D., Ph.D., a wonderful colleague whose fascination with DNA helicases was infectious.

This article contains supplemental Figs. S1–S3.
- HELB
- DNA helicase B gene
- aa
- amino acids
- ATM
- ataxia telangiectasia mutated
- ATR
- ATM and Rad3-related
- ATRIP
- ATR interacting protein
- CPT
- camptothecin
- HDHB
- human DNA helicase B
- HSQC
- heteronuclear single-quantum correlation
- HU
- hydroxyurea
- IR
- ionizing radiation
- PIKK
- phosphoinositide 3-kinase-related protein kinases
- RPA
- replication protein A
- RPA70N
- N-terminal domain of replication protein A
- PCNA
- proliferating cell nuclear antigen.
REFERENCES
- 1. Seki M., Enomoto T., Hanaoka F., Yamada M. (1987) DNA-dependent adenosinetriphosphatase B from mouse FM3A cells has DNA helicase activity. Biochemistry 26, 2924–2928 [DOI] [PubMed] [Google Scholar]
- 2. Seki M., Enomoto T., Watanabe Y., Tawaragi Y., Kawasaki K., Hanaoka F., Yamada M. (1986) Purification and characterization of a deoxyribonucleic acid-dependent adenosinetriphosphatase from mouse FM3A cells. Effects of ribonucleoside triphosphates on the interaction of the enzyme with single-stranded DNA. Biochemistry 25, 3239–3245 [DOI] [PubMed] [Google Scholar]
- 3. Seki M., Enomoto T., Yanagisawa J., Hanaoka F., Ui M. (1988) Further characterization of DNA helicase activity of mouse DNA-dependent adenosinetriphosphatase B (DNA helicase B). Biochemistry 27, 1766–1771 [DOI] [PubMed] [Google Scholar]
- 4. Saitoh A., Tada S., Katada T., Enomoto T. (1995) Stimulation of mouse DNA primase-catalyzed oligoribonucleotide synthesis by mouse DNA helicase B. Nucleic Acids Res. 23, 2014–2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Seki M., Kohda T., Yano T., Tada S., Yanagisawa J., Eki T., Ui M., Enomoto T. (1995) Characterization of DNA synthesis and DNA-dependent ATPase activity at a restrictive temperature in temperature-sensitive tsFT848 cells with thermolabile DNA helicase B. Mol. Cell. Biol. 15, 165–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Matsumoto K., Seki M., Masutani C., Tada S., Enomoto T., Ishimi Y. (1995) Stimulation of DNA synthesis by mouse DNA helicase B in a DNA replication system containing eukaryotic replication origins. Biochemistry 34, 7913–7922 [DOI] [PubMed] [Google Scholar]
- 7. Singleton M. R., Dillingham M. S., Wigley D. B. (2007) Structure and mechanism of helicases and nucleic acid translocases. Annu. Rev. Biochem. 76, 23–50 [DOI] [PubMed] [Google Scholar]
- 8. Tada S., Kobayashi T., Omori A., Kusa Y., Okumura N., Kodaira H., Ishimi Y., Seki M., Enomoto T. (2001) Molecular cloning of a cDNA encoding mouse DNA helicase B, which has homology to Escherichia coli RecD protein, and identification of a mutation in the DNA helicase B from tsFT848 temperature-sensitive DNA replication mutant cells. Nucleic Acids Res. 29, 3835–3840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Taneja P., Gu J., Peng R., Carrick R., Uchiumi F., Ott R. D., Gustafson E., Podust V. N., Fanning E. (2002) A dominant-negative mutant of human DNA helicase B blocks the onset of chromosomal DNA replication. J. Biol. Chem. 277, 40853–40861 [DOI] [PubMed] [Google Scholar]
- 10. Elvers I., Johansson F., Groth P., Erixon K., Helleday T. (2011) UV stalled replication forks restart by re-priming in human fibroblasts. Nucleic Acids Res. 39, 7049–7057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Heller R. C., Marians K. J. (2006) Replisome assembly and the direct restart of stalled replication forks. Nat. Rev. Mol. Cell Biol. 7, 932–943 [DOI] [PubMed] [Google Scholar]
- 12. Matsuoka S., Ballif B. A., Smogorzewska A., McDonald E. R., 3rd, Hurov K. E., Luo J., Bakalarski C. E., Zhao Z., Solimini N., Lerenthal Y., Shiloh Y., Gygi S. P., Elledge S. J. (2007) ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 316, 1160–1166 [DOI] [PubMed] [Google Scholar]
- 13. Cannavo E., Gerrits B., Marra G., Schlapbach R., Jiricny J. (2007) Characterization of the interactome of the human MutL homologues MLH1, PMS1, and PMS2. J. Biol. Chem. 282, 2976–2986 [DOI] [PubMed] [Google Scholar]
- 14. Diller L., Kassel J., Nelson C. E., Gryka M. A., Litwak G., Gebhardt M., Bressac B., Ozturk M., Baker S. J., Vogelstein B. (1990) p53 functions as a cell cycle control protein in osteosarcomas. Mol. Cell. Biol. 10, 5772–5781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Stott F. J., Bates S., James M. C., McConnell B. B., Starborg M., Brookes S., Palmero I., Ryan K., Hara E., Vousden K. H., Peters G. (1998) The alternative product from the human CDKN2A locus, p14(ARF), participates in a regulatory feedback loop with p53 and MDM2. EMBO J. 17, 5001–5014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Brattain M. G., Fine W. D., Khaled F. M., Thompson J., Brattain D. E. (1981) Heterogeneity of malignant cells from a human colonic carcinoma. Cancer Res. 41, 1751–1756 [PubMed] [Google Scholar]
- 17. Papadopoulos N., Nicolaides N. C., Wei Y. F., Ruben S. M., Carter K. C., Rosen C. A., Haseltine W. A., Fleischmann R. D., Fraser C. M., Adams M. D. (1994) Mutation of a mutL homolog in hereditary colon cancer. Science 263, 1625–1629 [DOI] [PubMed] [Google Scholar]
- 18. Parsons R., Li G. M., Longley M. J., Fang W. H., Papadopoulos N., Jen J., de la Chapelle A., Kinzler K. W., Vogelstein B., Modrich P. (1993) Hypermutability and mismatch repair deficiency in RER+ tumor cells. Cell 75, 1227–1236 [DOI] [PubMed] [Google Scholar]
- 19. Goodwin E. C., DiMaio D. (2000) Repression of human Papillomavirus oncogenes in HeLa cervical carcinoma cells causes the orderly reactivation of dormant tumor suppressor pathways. Proc. Natl. Acad. Sci. U.S.A. 97, 12513–12518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gu J., Xia X., Yan P., Liu H., Podust V. N., Reynolds A. B., Fanning E. (2004) Cell cycle-dependent regulation of a human DNA helicase that localizes in DNA damage foci. Mol. Biol. Cell 15, 3320–3332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kremmer E., Kranz B. R., Hille A., Klein K., Eulitz M., Hoffmann-Fezer G., Feiden W., Herrmann K., Delecluse H. J., Delsol G., Bornkamm G. W., Mueller-Lantzsch N., Grässert F. A. (1995) Rat monoclonal antibodies differentiating between the Epstein-Barr virus nuclear antigens 2A (EBNA2A) and 2B (EBNA2B).Virology 208, 336–342 [DOI] [PubMed] [Google Scholar]
- 22. Méndez J., Stillman B. (2000) Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle. Assembly of prereplication complexes in late mitosis. Mol. Cell. Biol. 20, 8602–8612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kenny M. K., Schlegel U., Furneaux H., Hurwitz J. (1990) The role of human single-stranded DNA-binding protein and its individual subunits in simian virus 40 DNA replication. J. Biol. Chem. 265, 7693–7700 [PubMed] [Google Scholar]
- 24. Jones D. T. (1999) Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292, 195–202 [DOI] [PubMed] [Google Scholar]
- 25. Rost B., Liu J. (2003) The PredictProtein server. Nucleic Acids Res. 31, 3300–3304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Linding R., Jensen L. J., Diella F., Bork P., Gibson T. J., Russell R. B. (2003) Protein disorder prediction. Implications for structural proteomics. Structure 11, 1453–1459 [DOI] [PubMed] [Google Scholar]
- 27. Notredame C., Higgins D. G., Heringa J. (2000) T-Coffee. A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302, 205–217 [DOI] [PubMed] [Google Scholar]
- 28. Ball H. L., Ehrhardt M. R., Mordes D. A., Glick G. G., Chazin W. J., Cortez D. (2007) Function of a conserved checkpoint recruitment domain in ATRIP proteins. Mol. Cell. Biol. 27, 3367–3377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mer G., Bochkarev A., Gupta R., Bochkareva E., Frappier L., Ingles C. J., Edwards A. M., Chazin W. J. (2000) Structural basis for the recognition of DNA repair proteins UNG2, XPA, and RAD52 by replication factor RPA. Cell 103, 449–456 [DOI] [PubMed] [Google Scholar]
- 30. Gomes X. V., Wold M. S. (1995) Structural analysis of human replication protein A. Mapping functional domains of the 70-kDa subunit. J. Biol. Chem. 270, 4534–4543 [DOI] [PubMed] [Google Scholar]
- 31. Pfuetzner R. A., Bochkarev A., Frappier L., Edwards A. M. (1997) Replication protein A. Characterization and crystallization of the DNA binding domain. J. Biol. Chem. 272, 430–434 [DOI] [PubMed] [Google Scholar]
- 32. Bochkareva E., Frappier L., Edwards A. M., Bochkarev A. (1998) The RPA32 subunit of human replication protein A contains a single-stranded DNA binding domain. J. Biol. Chem. 273, 3932–3936 [DOI] [PubMed] [Google Scholar]
- 33. Xu X., Vaithiyalingam S., Glick G. G., Mordes D. A., Chazin W. J., Cortez D. (2008) The basic cleft of RPA70N binds multiple checkpoint proteins, including RAD9, to regulate ATR signaling. Mol. Cell. Biol. 28, 7345–7353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Henricksen L. A., Umbricht C. B., Wold M. S. (1994) Recombinant replication protein A. Expression, complex formation, and functional characterization. J. Biol. Chem. 269, 11121–11132 [PubMed] [Google Scholar]
- 35. Brummelkamp T. R., Bernards R., Agami R. (2002) Stable suppression of tumorigenicity by virus-mediated RNA interference. Cancer cell 2, 243–247 [DOI] [PubMed] [Google Scholar]
- 36. Pommier Y. (2006) Topoisomerase I inhibitors. Camptothecins and beyond. Nat. Rev. Cancer 6, 789–802 [DOI] [PubMed] [Google Scholar]
- 37. Cimprich K. A., Cortez D. (2008) ATR. An essential regulator of genome integrity. Nat. Rev. Mol. Cell Biol. 9, 616–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zou L., Elledge S. J. (2003) Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300, 1542–1548 [DOI] [PubMed] [Google Scholar]
- 39. Ciccia A., Elledge S. J. (2010) The DNA damage response. Making it safe to play with knives. Mol. Cell 40, 179–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Inagaki A., van Cappellen W. A., van der Laan R., Houtsmuller A. B., Hoeijmakers J. H., Grootegoed J. A., Baarends W. M. (2009) Dynamic localization of human RAD18 during the cell cycle and a functional connection with DNA double-strand break repair. DNA Repair 8, 190–201 [DOI] [PubMed] [Google Scholar]
- 41. Lukas J., Lukas C., Bartek J. (2011) More than just a focus. The chromatin response to DNA damage and its role in genome integrity maintenance. Nat. Cell Biol. 13, 1161–1169 [DOI] [PubMed] [Google Scholar]
- 42. Mimitou E. P., Symington L. S. (2009) Nucleases and helicases take center stage in homologous recombination. Trends Biochem. Sci. 34, 264–272 [DOI] [PubMed] [Google Scholar]
- 43. Sarkaria J. N., Tibbetts R. S., Busby E. C., Kennedy A. P., Hill D. E., Abraham R. T. (1998) Inhibition of phosphoinositide 3-kinase-related kinases by the radiosensitizing agent wortmannin. Cancer Res. 58, 4375–4382 [PubMed] [Google Scholar]
- 44. Fanning E., Klimovich V., Nager A. R. (2006) A dynamic model for replication protein A (RPA) function in DNA processing pathways. Nucleic Acids Res. 34, 4126–4137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Iftode C., Daniely Y., Borowiec J. A. (1999) Replication protein A (RPA). The eukaryotic SSB. Crit. Rev. Biochem. Mol. Biol. 34, 141–180 [DOI] [PubMed] [Google Scholar]
- 46. Stauffer M. E., Chazin W. J. (2004) Structural mechanisms of DNA replication, repair, and recombination. J. Biol. Chem. 279, 30915–30918 [DOI] [PubMed] [Google Scholar]
- 47. Wold M. S. (1997) Replication protein A. A heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu. Rev. Biochem. 66, 61–92 [DOI] [PubMed] [Google Scholar]
- 48. Zou Y., Liu Y., Wu X., Shell S. M. (2006) Functions of human replication protein A (RPA). From DNA replication to DNA damage and stress responses. J. Cell. Physiol. 208, 267–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bochkareva E., Kaustov L., Ayed A., Yi G. S., Lu Y., Pineda-Lucena A., Liao J. C., Okorokov A. L., Milner J., Arrowsmith C. H., Bochkarev A. (2005) Single-stranded DNA mimicry in the p53 transactivation domain interaction with replication protein A. Proc. Natl. Acad. Sci. U.S.A. 102, 15412–15417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Olson E., Nievera C. J., Liu E., Lee A. Y., Chen L., Wu X. (2007) The Mre11 complex mediates the S-phase checkpoint through an interaction with replication protein A. Mol. Cell. Biol. 27, 6053–6067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Byun T. S., Pacek M., Yee M. C., Walter J. C., Cimprich K. A. (2005) Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev. 19, 1040–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Durkin S. G., Glover T. W. (2007) Chromosome fragile sites. Annu. Rev. Genet. 41, 169–192 [DOI] [PubMed] [Google Scholar]
- 53. Letessier A., Millot G. A., Koundrioukoff S., Lachagès A. M., Vogt N., Hansen R. S., Malfoy B., Brison O., Debatisse M. (2011) Cell-type-specific replication initiation programs set fragility of the FRA3B fragile site. Nature 470, 120–123 [DOI] [PubMed] [Google Scholar]
- 54. Michael W. M., Ott R., Fanning E., Newport J. (2000) Activation of the DNA replication checkpoint through RNA synthesis by primase. Science 289, 2133–2137 [DOI] [PubMed] [Google Scholar]
- 55. Mackintosh S. G., Raney K. D. (2006) DNA unwinding and protein displacement by superfamily 1 and superfamily 2 helicases. Nucleic Acids Res. 34, 4154–4159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Azvolinsky A., Dunaway S., Torres J. Z., Bessler J. B., Zakian V. A. (2006) The S. cerevisiae Rrm3p DNA helicase moves with the replication fork and affects replication of all yeast chromosomes. Genes Dev. 20, 3104–3116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Azvolinsky A., Giresi P. G., Lieb J. D., Zakian V. A. (2009) Highly transcribed RNA polymerase II genes are impediments to replication fork progression in Saccharomyces cerevisiae. Mol. Cell 34, 722–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Schmidt K. H., Kolodner R. D. (2004) Requirement of Rrm3 helicase for repair of spontaneous DNA lesions in cells lacking Srs2 or Sgs1 helicase. Mol. Cell. Biol. 24, 3213–3226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Atkinson J., Gupta M. K., McGlynn P. (2011) Interaction of Rep and DnaB on DNA. Nucleic Acids Res. 39, 1351–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Guy C. P., Atkinson J., Gupta M. K., Mahdi A. A., Gwynn E. J., Rudolph C. J., Moon P. B., van Knippenberg I. C., Cadman C. J., Dillingham M. S., Lloyd R. G., McGlynn P. (2009) Rep provides a second motor at the replisome to promote duplication of protein-bound DNA. Mol. Cell 36, 654–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lecointe F., Sérèna C., Velten M., Costes A., McGovern S., Meile J. C., Errington J., Ehrlich S. D., Noirot P., Polard P. (2007) Anticipating chromosomal replication fork arrest. SSB targets repair DNA helicases to active forks. EMBO J. 26, 4239–4251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Collins K. L., Kelly T. J. (1991) Effects of T antigen and replication protein A on the initiation of DNA synthesis by DNA polymerase α-primase. Mol. Cell. Biol. 11, 2108–2115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Melendy T., Stillman B. (1993) An interaction between replication protein A and SV40 T antigen appears essential for primosome assembly during SV40 DNA replication. J. Biol. Chem. 268, 3389–3395 [PubMed] [Google Scholar]
- 64. Arunkumar A. I., Klimovich V., Jiang X., Ott R. D., Mizoue L., Fanning E., Chazin W. J. (2005) Insights into hRPA32 C-terminal domain–mediated assembly of the simian virus 40 replisome. Nat. Struct. Mol. Biol. 12, 332–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Huang H., Weiner B. E., Zhang H., Fuller B. E., Gao Y., Wile B. M., Zhao K., Arnett D. R., Chazin W. J., Fanning E. (2010) Structure of a DNA polymerase α-primase domain that docks on the SV40 helicase and activates the viral primosome. J. Biol. Chem. 285, 17112–17122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Huang H., Zhao K., Arnett D. R., Fanning E. (2010) A specific docking site for DNA polymerase {α}-primase on the SV40 helicase is required for viral primosome activity, but helicase activity is dispensable. J. Biol. Chem. 285, 33475–33484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Lopes M., Foiani M., Sogo J. M. (2006) Multiple mechanisms control chromosome integrity after replication fork uncoupling and restart at irreparable UV lesions. Mol. Cell 21, 15–27 [DOI] [PubMed] [Google Scholar]
- 68. Sogo J. M., Lopes M., Foiani M. (2002) Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science 297, 599–602 [DOI] [PubMed] [Google Scholar]
- 69. Cordeiro-Stone M., Makhov A. M., Zaritskaya L. S., Griffith J. D. (1999) Analysis of DNA replication forks encountering a pyrimidine dimer in the template to the leading strand. J. Mol. Biol. 289, 1207–1218 [DOI] [PubMed] [Google Scholar]
- 70. Kadyrov F. A., Drake J. W. (2004) UvsX recombinase and Dda helicase rescue stalled bacteriophage T4 DNA replication forks in vitro. J. Biol. Chem. 279, 35735–35740 [DOI] [PubMed] [Google Scholar]
- 71. Liu J., Morrical S. W. (2010) Assembly and dynamics of the bacteriophage T4 homologous recombination machinery. Virology J. 7, 357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Branzei D., Foiani M. (2010) Maintaining genome stability at the replication fork. Nat. Rev. Mol. Cell Biol. 11, 208–219 [DOI] [PubMed] [Google Scholar]
- 73. Petermann E., Helleday T. (2010) Pathways of mammalian replication fork restart. Nat. Rev. Mol. Cell Biol. 11, 683–687 [DOI] [PubMed] [Google Scholar]
- 74. Vanoli F., Fumasoni M., Szakal B., Maloisel L., Branzei D. (2010) Replication and recombination factors contributing to recombination-dependent bypass of DNA lesions by template switch. PLoS Genet. 6, e1001205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Saleh-Gohari N., Bryant H. E., Schultz N., Parker K. M., Cassel T. N., Helleday T. (2005) Spontaneous homologous recombination is induced by collapsed replication forks that are caused by endogenous DNA single-strand breaks. Mol. Cell. Biol. 25, 7158–7169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Davies A. A., Huttner D., Daigaku Y., Chen S., Ulrich H. D. (2008) Activation of ubiquitin-dependent DNA damage bypass is mediated by replication protein A. Mol. Cell 29, 625–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Motegi A., Liaw H. J., Lee K. Y., Roest H. P., Maas A., Wu X., Moinova H., Markowitz S. D., Ding H., Hoeijmakers J. H., Myung K. (2008) Polyubiquitination of proliferating cell nuclear antigen by HLTF and SHPRH prevents genomic instability from stalled replication forks. Proc. Natl. Acad. Sci. U.S.A. 105, 12411–12416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Driscoll R., Cimprich K. A. (2009) HARPing on about the DNA damage response during replication. Genes Dev. 23, 2359–2365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Doherty K. M., Sommers J. A., Gray M. D., Lee J. W., von Kobbe C., Thoma N. H., Kureekattil R. P., Kenny M. K., Brosh R. M. (2005) Physical and functional mapping of the replication protein a interaction domain of the Werner and Bloom syndrome helicases. J. Biol. Chem. 280, 29494–29505 [DOI] [PubMed] [Google Scholar]
- 80. Shen J. C., Lao Y., Kamath-Loeb A., Wold M. S., Loeb L. A. (2003) The N-terminal domain of the large subunit of human replication protein A binds to Werner syndrome protein and stimulates helicase activity. Mech. Ageing Dev. 124, 921–930 [DOI] [PubMed] [Google Scholar]
- 81. Machwe A., Lozada E., Wold M. S., Li G. M., Orren D. K. (2011) Molecular cooperation between the Werner syndrome protein and replication protein A in relation to replication fork blockage. J. Biol. Chem. 286, 3497–3508 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.