Background: Quinohemoprotein amine dehydrogenase from Gram-negative Proteobacteria undergoes multiple posttranslational modifications.
Results: Disruption of a neighboring gene encoding a subtilisin-like serine protease led to the production of inactive dehydrogenase.
Conclusion: The protease is essential for biogenesis of the dehydrogenase, serving as an unusual cis-acting enzyme without catalytic turnover.
Significance: The protease described here may represent a rare bacterial version of processing enzymes.
Keywords: Enzyme Mechanisms, Enzyme Processing, Posttranslational Modification, Serine Protease, Serpin, Enzyme Maturation, Processing Protease
Abstract
Quinohemoprotein amine dehydrogenase (QHNDH), an αβγ heterotrimer present in the periplasm of several Gram-negative bacteria, catalyzes the oxidative deamination of various aliphatic amines such as n-butylamine for assimilation as carbon and energy sources. The γ subunit of mature QHNDH contains a protein-derived quinone cofactor, cysteine tryptophylquinone, and three intrapeptidyl thioether cross-links between Cys and Asp or Glu residues. In its cytoplasmic nascent form, the γ subunit has a 28-residue N-terminal leader peptide that is necessary for the production of active QHNDH but must be removed in the following maturation process. Here, we describe the role of a subtilisin-like serine protease encoded in the fifth ORF of the n-butylamine-utilizing operon of Paracoccus denitrificans (termed ORF5) in QHNDH biogenesis. ORF5 disruption caused bacterial cell growth inhibition in n-butylamine-containing medium and production of inactive QHNDH, in which the γ subunit retained the leader peptide. Supply of plasmid-encoded ORF5 restored the cell growth and production of active QHNDH, containing the correctly processed γ subunit. ORF5 expressed in Escherichia coli but not its catalytic triad mutant cleaved synthetic peptides surrogating for the γ subunit leader peptide, although extremely slowly. The cleaved leader peptide remained unstably bound to ORF5, most likely as an acyl enzyme intermediate attached to the active-site Ser residue. These results demonstrate that ORF5 is essential for QHNDH biogenesis, serving as a processing protease to cleave the γ subunit leader peptide nearly in a disposable manner.
Introduction
Quinohemoprotein amine dehydrogenase (QHNDH),3 an inducible enzyme present in the periplasm of Gram-negative bacteria such as Paracoccus denitrificans and Pseudomonas putida, catalyzes the oxidative deamination of various aliphatic primary amines, including n-butylamine, for assimilation as carbon and energy sources (1–3). Structurally, it is an αβγ heterotrimer (Fig. 1A). Its α subunit, the largest chain, has a four-domain structure with two hemes in the N-terminal domain that mediate electron transfer from the substrate to an external electron acceptor protein, such as cytochrome c550 and the blue copper protein azurin in P. denitrificans (2) and P. putida (3), respectively. The β subunit, the middle-sized chain, has a seven-bladed β propeller structure that is frequently observed in quinoprotein dehydrogenases. Its γ subunit, the smallest chain, has a particularly unusual structure. It contains a quinone cofactor, cysteine tryptophylquinone (CTQ), posttranslationally derived from Trp and Cys residues, and three intrapeptidyl thioether cross-links between Cys and Glu or Asp residues, as shown schematically in Fig. 1B (4–6). These features clearly indicate that QHNDH must undergo various posttranslational modifications to attain its amine-oxidizing activity in the periplasm of Gram-negative bacteria. Not necessarily in the given order, these modifications are 1) formation of the three intrapeptidyl thioether cross-links and CTQ in the γ subunit, 2) removal of the signal peptides during the periplasmic translocation of the α and β subunits, 3) insertion of the two hemes in the α subunit, and 4) association of the subunits to form active QHNDH. The manner, timing, location, and order of these modifications, which could occur as either independent events in a single subunit or coordinated processes involving dimeric or trimeric subunit complexes, remain unclear. The mechanism of the periplasmic translocation of the γ subunit, without signal peptides, is also unknown.
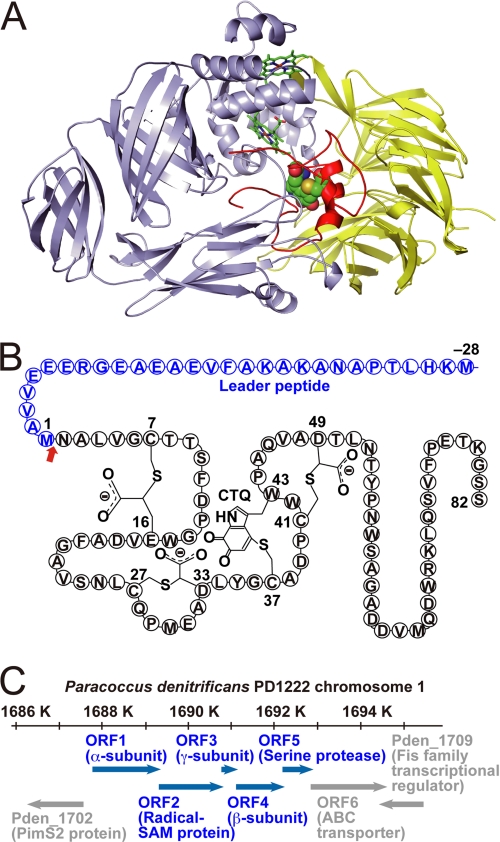
FIGURE 1.
Protein and gene structure of QHNDH. A, ribbon model of QHNDH from P. denitrificans (PDB code 1JJU). The light blue, yellow, and red ribbons represent the α, β, and γ subunits, respectively. The two hemes in the α subunit are indicated as sticks, and CTQ atoms in the γ subunit are depicted as balls. B, schematic of the γ subunit polypeptide with the 28-residue N-terminal leader peptide and intrapeptidyl modifications (three Cys-to-Asp/Glu thioether cross-links and CTQ) at the indicated residue numbers. The red arrow indicates the cleavage site of the leader peptide. C, schematic of the gene structure of QHNDH in chromosome 1 of Pd1222, ORF1-ORF5, and the peripheral genes are shown as blue and gray arrows, respectively. Gene identification numbers from the NCBI genome database are included, with the annotated proteins referred to in parentheses. SAM, AdoMet.
The structural genes encoding the QHNDH subunits constitute an operon harboring at least five ORFs that may be transcribed synchronously by an amine inducer (Fig. 1C). In this operon, the α, β, and γ subunit polypeptides are encoded by ORF1, ORF4, and ORF3, respectively. ORF2, intervening between the genes for the α and γ subunits, encodes a putative [Fe-S] cluster- and S-adenosylmethionine (AdoMet)-binding protein, belonging to the “radical AdoMet superfamily,” which includes various proteins involved in vitamin biosynthesis and posttranslational activation of other enzymes adjacently encoded in the genome (7). We previously demonstrated that ORF2 of P. denitrificans plays an essential role in the posttranslational processing of the γ subunit, most likely by participating in intrapeptidyl thioether cross-link formation via an [Fe-S] cluster- and AdoMet-dependent mechanism (8). Further, in its cytoplasmic nascent form, the γ subunit has a 28-residue N-terminal leader peptide that is necessary for the production of active QHNDH but must be removed during the subsequent maturation process (8). This leader peptide is atypical as a signal peptide for the periplasmic translocation of the γ subunit and instead seems to participate in the cytoplasmic processing of the γ subunit, e.g. serving as a primer or scaffold for intrapeptidyl thioether cross-link formation by ORF2.
ORF5 encodes an ∼22.5 kDa protein belonging to subfamily S8A of peptidase family S8 (subtilisin family), as revealed by a sequence similarity search in a protein database (supplemental Fig. S1). The Asp/His/Ser catalytic triad of subtilisin is fully conserved at Asp-8, His-39, and Ser-177 in P. denitrificans ORF5. To elucidate the role of this protein in the n-butylamine-utilizing operon of P. denitrificans, particularly in relation to the biogenesis of QHNDH, we conducted genetic and biochemical analyses of ORF5. Here, we report its unique properties as a bacterial processing protease.
EXPERIMENTAL PROCEDURES
Materials and Bacterial Strains
All primers used in PCR were designed on the basis of the sequence reported previously (5) (GenBanktm accession no. AB063330) (supplemental Table S1). Plasmid pUC4K containing a kanamycin-resistant (Kmr) gene was provided by the National Institute of Genetics (Mishima, Japan). A suicide vector, pGRPd1 (9), P. denitrificans strain Pd1222, and Escherichia coli strain S17-1 (10) were provided by R. J. van Spanning (Vrije Universiteit, The Netherlands). P. denitrificans mutant strains PdKO4 and PdKO6, in which ORF2 and ORF3 had been disrupted respectively, were reported previously (8).
Molecular Genetics Protocols
Protocols for construction of plasmid vectors, growth conditions, homologous recombination, and site-directed mutagenesis were mostly conventional and are described in detail in the supplemental information.
Purification of His6-tagged ORF5
E. coli strain BL21 (DE3) was transformed with pET-ORF5 (supplemental Fig. S2) and pRARE encoding tRNAs for rare codons of E. coli, and then grown at 37 °C in a Luria-Bertani medium supplemented with 100 μm ampicillin and 34 μm chloramphenicol to a mid-logarithmic phase. After addition of isopropyl-β-d-thiogalactopyranoside (0.15 mm), cells were further cultivated at 30 °C for 4 h, harvested by centrifugation, and washed with 0.85% NaCl. Washed cells were resuspended in 20 mm Tris-HCl (pH 7.5) containing 50 mm imidazole and 0.5 m NaCl (buffer C). After ultrasonic disruption of the cells, the cell extract was collected by centrifugation and loaded onto a 5-ml HisTrap FF crude column (GE Healthcare) equilibrated with buffer C. The column was washed with 20 column volumes of buffer C. Proteins were eluted with a linear gradient of 50–300 mm imidazole. Fractions containing His6-tagged ORF5 were pooled, concentrated, and dialyzed against 10 mm HEPES (pH 6.5), containing 50 mm NaCl and 0.05% (w/v) Triton X-100 (buffer D). Insoluble aggregates were removed by centrifugation, and the supernatant solution was applied to a 5-ml HiTrap SP FF column (GE Healthcare) pre-equilibrated with buffer D. The column was washed with 20 column volumes of buffer D. Proteins were eluted with a linear gradient of 50–500 mm NaCl in buffer D. His6-tagged ORF5 expressed in PdKO7/pRK-H6ORF5 was also purified as described above using a 1-ml Resource PHE column (GE Healthcare) equilibrated with 1.35 m ammonium sulfate in 50 mm potassium phosphate buffer (pH 7.5) instead of the HiTrap SP column. Proteins were eluted with a linear gradient of 1.35–0 m ammonium sulfate in 50 mm potassium phosphate buffer. For both purifications, fractions containing His6-tagged ORF5 were pooled, concentrated, and stored at 4 °C until use.
Isolation of γ Subunit Precursor
ΔORF5 strain PdKO7 was cultured in a medium containing 0.5% (w/v) n-butylamine and 20 mm choline·HCl as described in supplemental experimental procedures. The spheroplasts prepared from the cultured cells as described previously (8) were washed twice with PBS and resuspended at a final concentration of 0.1 g/ml in 20 mm Tris-HCl buffer (pH 8.0) containing 6 m urea, 0.1 m dithiothreitol, and 10 mm EDTA·2Na. The spheroplasts were disrupted by sonication, and the supernatant was obtained by centrifugation at 50,000 × g for 60 min. Formic acid was added to the supernatant to a final concentration of 1.5% (v/v), and the precipitated proteins were removed by centrifugation at 50,000 × g for 30 min. CH3CN was added to the supernatant to a final concentration of 10% (v/v), and this solution was applied to the solid-phase extraction column (SiliaBond® C4, 25 ml, 5 g, SiliCycle) pre-equilibrated with 10% CH3CN in 0.1% TFA, followed by washing thoroughly with 0.1% TFA in 20% CH3CN. The polypeptide was eluted with 50% CH3CN in 0.1% TFA. Fractions containing the γ subunit polypeptide were detected by Western blotting (8). The eluate was diluted 2-fold with 20 mm Tris-HCl buffer (pH 8.0) containing 6 m urea, 50 mm dithiothreitol, and 100 mm ammonium bicarbonate. The suspension was then applied to a 1-ml HiTrap Q FF column (GE Healthcare) pre-equilibrated with the same buffer, and the peptide was eluted with a linear gradient of 0–0.5 M NaCl in the buffer. Fractions containing the γ subunit polypeptide were pooled, and formic acid was added to a final concentration of 1.5%. The γ subunit polypeptide was further purified on an HPLC system (Hitachi, L-7100) using a C18 reversed-phase column (Tosoh, TSK-gel Octadecyl-4PW, 4.6 × 150 mm). The sample was loaded onto the column pre-equilibrated with 20% CH3CN containing 0.1% TFA, and the column was washed thoroughly with the same solvent. The γ subunit polypeptide was eluted with a 50-min linear gradient of 20–35% (10 min), 35–50% (30 min), and 50–80% (10 min) CH3CN containing 0.1% TFA at a flow rate of 0.5 ml/min. Fractions containing the γ subunit polypeptide were collected and concentrated with a centrifugal concentrator (Tomy, CC-180). The second HPLC separation was repeated, and the resultant sample was concentrated and used for mass spectrometric analysis.
HPLC Assay of ORF5 Protease Activity with Synthetic Peptides
Two synthetic heptapeptides N- or C-terminally labeled with fluorescein (N- or C-Flc), Flc-CO-EVVAMNA-NH2 (Mr = 1090.18) and Ac-EVVAMNA-NHCH2CH2NHCO-Flc (Mr = 1175.29), were purchased from Sigma-Aldrich Japan. The peptide substrate (20–200 μm) was incubated with the purified ORF5 protein (0.2 mg/ml) at 30 °C in 20 mm HEPES (pH 7.0) containing 5 mm DTT. After 15 h, a 20-μl aliquot of the reaction mixture was applied to the HPLC system equipped with a C4 reverse phase column (5C4-AR-300, 4.6 mm × 150 mm, Nacalai Tesque) and a fluorescence detector (F-1050, Hitachi). Separation was performed using a 25-min (or 20-min) linear gradient of 0–52% (or 0–80%) CH3CN containing 0.1% TFA at a flow rate of 0.5 ml/min, monitored with absorbance at 220 nm and fluorescence at 520 nm upon excitation at 445 nm.
Trypsin Digestion of Cleaved N-Flc Product Bound to ORF5
HPLC fractions containing the cleaved product from N-Flc substrate bound to ORF5 were collected, dried by vacuum centrifugation, and redissolved in 6 m urea and 94 mm Tris-HCl (pH 8.5). The protein was reduced with 4.6 mm Tris(2-carboxyethyl)phosphine and then carboxyamidomethylated with 31 mm iodoacetamide. After addition of excess DTT, the carboxyamidomethylated protein was digested at 37 °C for 16 h with modified trypsin (New England Biolabs). The tryptic digests were applied to the HPLC system equipped with a fluorescence detector as described above. The major fluorescent peaks were collected and dried for mass analysis.
Mass Spectrometric Analyses
MALDI-TOF mass spectra were obtained with an Autoflex MALDI-TOF mass spectrometer (Bruker) equipped with a 337-nm nitrogen laser lamp. The samples were prepared on an MTP AnchorChipTM var/384 TF plate (Bruker) according to the manufacturer's instructions. Briefly, 0.5–1 μl of sample solution was applied onto the target and mixed with 1–2 μl of α-cyano-4-hydroxycinnamic acid (0.3 mg/ml in ethanol:acetone, 2:1) or sinapinic acid (1 mg/ml in CH3CN:0.1% TFA, 9:1) as matrices. All samples were dried and recrystallized by dissolving in 1 μl of ethanol:acetone:0.1% TFA, 6:3:1 (for α-cyano-4-hydroxycinnamic acid) or CH3CN:0.1% TFA, 9:1 (for sinapinic acid). Measurements were performed in the positive-ion linear mode with delayed extraction. All MALDI spectra were initially calibrated with a peptide or protein calibration standard I kit (Bruker). Electrospray ionization MS was carried out in a positive ion mode on a SYNAPT G1 mass spectrometer equipped with a nanoACQUITY HPLC system (Waters) and a Mass PREP Micro desalting column (Waters, 2.1 × 5 mm). The elution system consisted of 0.1% formic acid in water (solvent A) and 0.1% formic acid in CH3CN (solvent B) from 0% to 90% B at a flow rate of 40 μl/min.
RESULTS
n-Butylamine-induced Expression of ORF5
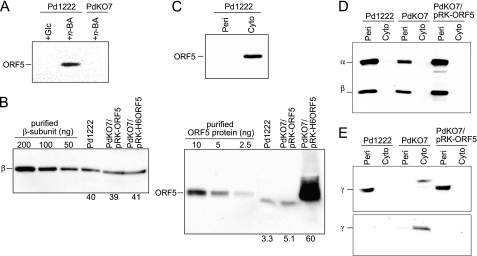
By Western blot analysis with an antiserum raised against ORF5 expressed in E. coli, we confirmed ORF5 expression in WT P. denitrificans Pd1222 cells grown in n-butylamine-containing medium but not in cells grown in medium containing glucose as the energy and carbon source (Fig. 2A). This finding indicates that ORF5 is a gene in the n-butylamine-utilizing operon of P. denitrificans and its expression is controlled under the same promoter as the structural genes for the QHNDH subunits. However, the amount of ORF5 in Pd1222 was estimated to be only about 8% of the QHNDH β subunit (∼12% by molar amount) by quantitative Western blot analysis (Fig. 2B). Presumably, because of an N-terminal sequence that follows the “N-end rule” (11, 12) (supplemental Fig. S1), ORF5 may be rapidly degraded even if its expression is induced to a similar amount as in the QHNDH subunits. Indeed, the amount of ORF5 was also very small when it was expressed through broad host-range plasmid pRK415 with multiple copies (13), in marked contrast to the abundant N-terminal His6-tagged protein (N-His6-ORF5) expressed through the same plasmid (Fig. 2B), which avoided degradation because of the N-end rule. Consistent with the absence of a sequence characteristic of a signal peptide in the N-terminal region (see GenBanktm AB063330), ORF5 was detected only in the cytoplasmic fraction of Pd1222 (Fig. 2C). Therefore, ORF5 probably functions in the cytoplasm similarly to the ORF2-encoded [Fe-S] cluster/AdoMet-binding protein (8) and unlike QHNDH, which occurs in the periplasm (1, 3).
FIGURE 2.
Expression and subcellular localization of ORF5 and QHNDH subunits. A, expression of ORF5 was examined by Western blotting with an antibody against ORF5 for Pd1222 and ΔORF5 strain PdKO7 grown in the medium containing glucose (+ Glc) or n-butylamine (+ n-BA). Total proteins of ∼20 μg from the cell-free extracts were applied in each lane. B, expression levels of ORF5 and the QHNDH β subunit were estimated by Western blotting for Pd1222, PdKO7/pRK-ORF5, and PdKO7/pRK-H6ORF5 grown in the medium containing n-butylamine for 24 h. Three different amounts of β subunit (left panel) or ORF5 (right panel), as calibration standards, were applied in the three left lanes. Total proteins of periplasmic (left panel) or cytoplasmic (right panel) fractions extracted from 5 mg of wet cells were applied in each of the three right lanes. Shown below each lane is a protein amount (ng) calculated from a calibration curve generated by measuring the band intensities of the calibration standards. Band intensities were quantified using Quantity One software (Bio-Rad). C, subcellular localization of ORF5 was analyzed by Western blotting. Total proteins of ∼10 μg from the periplasmic (Peri) and cytoplasmic (Cyto) fractions were applied in each lane. D, Pd1222 and PdKO7 strains with or without an expression plasmid for ORF5 (pRK-ORF5) were grown in the minimum medium supplemented with n-butylamine. Subcellular localization of the α and β subunits of QHNDH was analyzed by Western blotting with an anti-QHNDH antibody. Total proteins of ∼5 μg from the periplasmic and cytoplasmic fractions were applied in each lane. E, subcellular localization of the γ subunit was analyzed by Western blotting with antibodies against a C-terminal region of the γ subunit (upper panel) and the leader peptide of the γ subunit (lower panel). Total proteins of ∼50 μg from the periplasmic and cytoplasmic fractions were applied in each lane.
Essentiality of ORF5
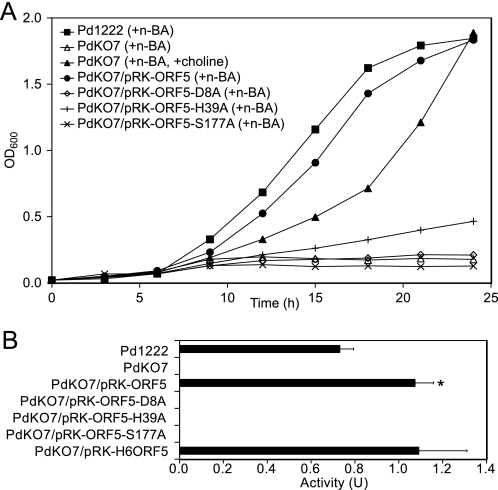
Pd1222 cells grew efficiently in minimum mineral medium containing n-butylamine as the sole carbon and energy source and showed a high level of QHNDH activity, whereas cells of a ΔORF5 mutant strain (PdKO7, Fig. 2A) neither grew nor showed QHNDH activity in the same medium (Fig. 3, A and B). Nevertheless, the QHNDH subunit polypeptides were produced even in PdKO7 cells grown in the medium containing n-butylamine and choline (Fig. 2, D and E). The latter compound was added to support the n-butylamine-independent growth of P. denitrificans (14) (Fig. 3A). These results indicate that the QHNDH subunit polypeptides are produced in PdKO7 but do not form active QHNDH. Therefore, ORF5 is essential for bacterial growth in medium containing n-butylamine as the sole carbon and energy source, which is oxidized by active QHNDH in the periplasm.
FIGURE 3.
Bacterial growth and QHNDH activity. Pd1222 (WT) and PdKO7 (mutant) cells of P. denitrificans with or without the indicated plasmids were grown in minimum mineral medium supplemented with n-butylamine (n-BA) or n-BA and choline. A, cell densities measured by the absorbance at 600 nm were plotted against cultivation time (h). B, QHNDH activity in the periplasmic fraction of cells cultivated for 24 h are shown as units/gram of wet cells. Each bar represents the mean ± S.E. from three independent experiments. *, p < 0.05 indicate a statistically significant difference compared with the activity in Pd1222 cells.
Subcellular Localization of QHNDH Subunits
Consistent with the presence of N-terminal-flanking signal peptides in both the α and the β subunits (4, 5), the two subunits were localized exclusively to the periplasmic fractions of Pd1222 and PdKO7 (Fig. 2D). By using an antibody against the C-terminal region of the γ subunit, the γ subunit polypeptide was also detected in the periplasmic fraction of Pd1222 (Fig. 2E). In contrast, this polypeptide was predominantly detected in the cytoplasmic fraction of PdKO7. Moreover, the cytoplasmic γ subunit of PdKO7 retained the N-terminal leader peptide, which was completely undetectable in the γ subunit of Pd1222 (Fig. 2E). The mass difference of ∼4 kDa is also consistent with the presence of the leader peptide in the cytoplasmic γ subunit of PdKO7. Similar cytoplasmic localization and retention of the leader peptide has been observed in the γ subunit of a ΔORF2 mutant strain, PdKO4 (8).
Rescue of Cell Growth and QHNDH Activity
When PdKO7 was transformed with a plasmid carrying ORF5 (pRK-ORF5, supplemental Fig. S2), both cell growth and QHNDH activity were significantly recovered in the medium containing n-butylamine as the sole carbon and energy source (Fig. 3, A and B). Noteworthy is that this recovery of QHNDH activity exceeded the activity in Pd1222 by about 50% (Fig. 2B) and appeared to correlate with the intracellular amount of ORF5 (∼50% more in PdKO7/pRK-ORF5 than in Pd1222, Fig. 2B). However, the recovered activity was not increased further, even with N-His6-ORF5 (Fig. 3B), which was produced abundantly (Fig. 2B). Presumably, the increase in QHNDH activity was limited by the amount of QHNDH subunits. The amounts of the β subunit were similar in Pd1222, PdKO7/pRK-ORF5, and PdKO7/pRK-H6ORF5 (Fig. 2B). Furthermore, the γ subunit of PdKO7/pRK-ORF5 was translocated to the periplasm and completely lost the N-terminal leader peptide (Fig. 2E). These results unequivocally show that ORF5 is necessary for the processing of the γ subunit leader peptide, which eventually leads to the periplasmic formation of active QHNDH, essential for n-butylamine-dependent bacterial growth.
Mutagenesis of the Catalytic Triad in ORF5
To examine whether ORF5 functions as a subtilisin-like protease, the putative catalytic triad (Asp-8/His-39/Ser-177) was mutated individually to Ala. When PdKO7 was transformed with expression plasmids (pRK-ORF5-D8A, pRK-ORF5-H39A, or pRK-ORF5-S177A) constructed for the mutant proteins (supplemental Fig. S2), the cells neither grew well nor exhibited QHNDH activity (Fig. 3, A and B). Furthermore, Western blot analysis revealed that the γ subunit was mainly present in the cytoplasmic fractions of PdKO7 transformed with these plasmids (supplemental Fig. S3D) and also retained the N-terminal leader peptide (supplemental Fig. S3E). The results strongly suggest that ORF5 is a subtilisin-like protease possessing the Asp/His/Ser catalytic triad and directly cleaves the N-terminal leader peptide. In addition, the γ subunit polypeptides detected slightly in the periplasmic fractions of PdKO7 transformed with the mutant plasmids (supplemental Fig. S3B) still contained the N-terminal leader peptide (supplemental Fig. S3C). This observation suggests that removal of the N-terminal leader peptide itself is not a prerequisite for the periplasmic translocation of the γ subunit and that the leader peptide does not function as a signal peptide, unlike those of the α and β subunits (4, 5). Alternatively, the leader peptide may function as a signal peptide (i.e. by directing translocon-dependent translocation into the periplasm) but lacks a signal peptide cleavage site, and the cleavage is instead carried out by ORF5. Although the mechanism of the periplasmic translocation of the γ subunit remains to be clarified, removal of the N-terminal leader peptide is probably necessary for efficient translocation of the γ subunit.
Mass Spectrometric Analysis of the γ Subunit
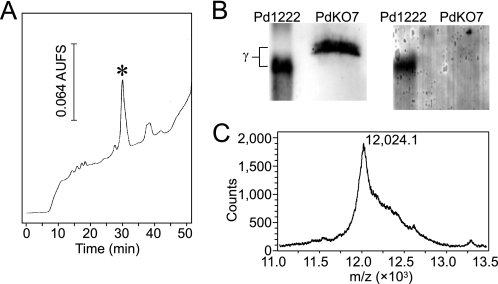
To elaborate the posttranslational modification of the γ subunit in PdKO7, the γ subunit polypeptide was purified by repeated HPLC with a C18 reversed-phase column (Fig. 4A). We reported previously that, for purification of the γ subunit from PdKO4, treatment of the peptide with 4-vinylpyridine is necessary before HPLC to block the free SH groups (8), which hamper peptide isolation by uncontrollable disulfide bridge formation. In contrast, the γ subunit polypeptide of PdKO7 was eluted as a single peak from the reversed-phase column without 4-vinylpyridine treatment, suggesting that it contains few or no free SH groups. Furthermore, the purified γ subunit was not stained at all by the redox-cycling method (15) for detection of redox-active quinone groups (Fig. 4B), indicating that the CTQ-precursor Trp residue (Trp43) was not oxidized to tryptophylquinone.
FIGURE 4.
Isolation and MALDI-TOF mass spectrometric analysis of the γ subunit produced in PdKO7. A, an HPLC profile for purification of the γ subunit produced in the cytoplasmic fraction of PdKO7. The elution was performed with a 40-min linear gradient of 35–50% CH3CN in 0.1% TFA. Fractions containing the asterisked peak were collected. B, the purified γ subunit polypeptide was analyzed by Western blotting with an antibody against the C-terminal region of the γ subunit (left panel) or was stained for a redox-active quinone group (right panel). The purified WT enzyme (2 μg) was used as the positive control. C, a mass spectrum of the γ subunit purified from PdKO7.
By MALDI-TOF mass spectrometric analysis, the γ subunit polypeptide purified from the cytoplasmic fraction of PdKO7 showed a mass at m/z 12,021.5 ± 2.7 (an averaged value of [M+H], n = 3, Fig. 4C), which is much larger than the mass of the fully processed γ subunit of Pd1222 (m/z 8,857.57, calculated value of [M+H]+ for amino acid residues Asn-2–Ser-82 including CTQ and the three intrapeptidyl thioether cross-links) and very close to that of the cytoplasmic γ subunit of PdKO4 (m/z 12,007.52, calculated value of [M+H]+ for Met (–28)–Ser-82, including two Met sulfoxide modifications), which does not undergo posttranslational modifications (formation of CTQ and the three intrapeptidyl thioether cross-links) but retains the N-terminal leader peptide (8). When the spontaneous oxidation of Met residues to sulfoxide or sulfone is uncontrolled during peptide isolation, the mass difference (Δm/z = ∼14) between the γ subunit polypeptides of PdKO4 and PdKO7 is too small to assess the extent of intrapeptidyl thioether cross-link formation (Δm/z = −2 per thioether cross-link formation) as well as the oxidation state of three Met residues (Met-1, Met-30, and Met-65). Therefore, to chemically analyze the presence of the free SH groups (i.e. Cys residues not forming intrapeptidyl thioether cross-links), we had previously modified the SH groups with 4-vinylpyridine. No pyridylethyl group was incorporated into the fully processed γ subunit of the WT strain, whereas four groups were incorporated into the cytoplasmic γ subunit of PdKO4, yielding a total mass increase of m/z 420.4 (Δm/z = 105.1 per pyridylethylation) (8). After the treatment with 4-vinylpyridine, the γ subunit polypeptide of PdKO7 had an averaged mass at m/z 12,169.4 ± 10.2 (n = 3). The mass increase (Δm/z = 147.9) resulting from pyridylethylation strongly suggests that the γ subunit polypeptide of PdKO7 contains only one reactive SH group with two or three Met residues further oxidized to sulfones. Considering the structural features of the four Cys residues in the γ subunit polypeptide, the free SH group is most likely ascribable to Cys-37, which is converted into CTQ. The other three (Cys-7, Cys-27, and Cys-41) are involved in the formation of thioether cross-links with the methylene carbon atoms of Asp or Glu (Fig. 1B). Collectively, we conclude that the γ subunit of PdKO7 already contains the three Cys-to-Asp/Glu thioether cross-links and still retains the N-terminal leader peptide but does not show CTQ formation. Therefore, the formation of the Cys-to-Asp/Glu thioether cross-links precedes the cleavage of the leader peptide and formation of CTQ.
Cleavage of Synthetic Peptides
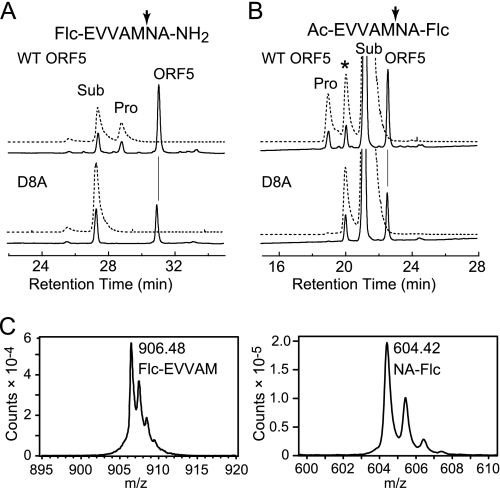
The proteolytic activity of N-His6-ORF5 expressed in E. coli and purified by Ni affinity chromatography was assayed with N- or C-Flc-labeled synthetic heptapeptides, which corresponded to a part of the N-terminal leader peptide of the γ subunit. Although the proteolytic activity of ORF5 was extremely low with the synthetic peptides, cleaved fluorescent products from both the substrates could be detected by HPLC after a 20-h reaction (Fig. 5, A and B). A catalytic triad mutant (D8A) of ORF5 exhibited no activity in both the substrates. Furthermore, MALDI-TOF mass spectrometric analysis clearly indicated the presence of cleaved fluorescent products from both the substrates (Flc-EVVAM, [M+H]+ = m/z 906.323; NA-Flc, [M+H]+ = m/z 604.204; Fig. 5C). The cleavage sites in these synthetic peptides were identical to each other (after the fifth Met) and agreed with the cleavage site in the N-terminal leader peptide of the γ subunit (Fig. 1B). Determination of the amount of product in the reaction mixture by using a series of enzyme-to-substrate ratios (1:5–40) for various incubation times revealed that the proteolytic activity of ORF5 proceeded without turnover in the N-Flc substrate or with only a few turnovers in the C-Flc substrate (supplemental Fig. S4). The N-Flc substrate was cleaved faster than the C-Flc substrate, suggesting that the presence of the bulky fluorophore close to the cleavage site was unfavorable for the reaction by ORF5.
FIGURE 5.
HPLC assays of the proteolytic activity of ORF5. Purified ORF5 (WT, 20 μg; D8A, 7.6 μg) was incubated with 20.2 μm N-Flc (A) or 204 μm C-Flc peptide (B) at 30 °C for 15 h. A 20-μl aliquot of the reaction mixture was analyzed by HPLC as described under supplemental experimental procedures. The absorbance at 220 nm (solid line) and fluorescence at 520 nm (dotted line) upon excitation at 445 nm were monitored. The cleavage sites by ORF5 are indicated with the arrows in the synthetic peptide sequences shown at the top of each panel. The peaks indicate the cleaved product (Pro), substrate (Sub), and the protein (ORF5). The peak with the asterisk is a contaminant in the synthetic peptide. C, MALDI-TOF mass spectra of the reaction product from the N-Flc (left panel) or C-Flc (right panel) peptide shown with the assigned chemical structures.
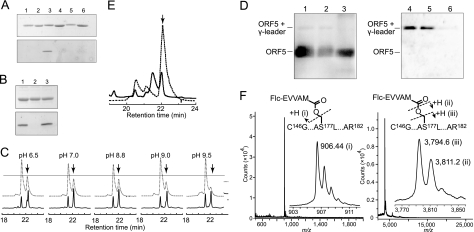
More intriguingly, we found that the cleaved fluorescent product from the N-Flc substrate bound to ORF5 even after SDS treatment (95 °C for 8 min) and PAGE (Fig. 6A). The fluorescent band was detected neither in the reaction of the inactive D8A mutant with the N-Flc substrate nor in the reaction of WT ORF5 with the C-Flc substrate, indicating that the fluorescent band bound to ORF5 was not derived from an uncleaved substrate but from the cleaved N-Flc substrate. The binding of the cleaved peptide to ORF5 was stable at weak acidic-to-neutral pH but was considerably labile at alkaline pH (Fig. 6B). Quick HPLC separation of the reaction mixture also revealed that the cleaved fluorescent product from the N-Flc substrate coeluted with ORF5 and that the fluorescent peak height decreased gradually after pretreatment of the reaction mixture at increasing pH (Fig. 6C). These observations strongly suggest that the cleaved N-terminal leader peptide binds to ORF5, most likely through a covalent bond such as an acyl ester linkage, which is stable at acidic-to-neutral pH but labile at alkaline pH. Alkaline-labile binding was also observed between the leader peptide of the endogenous γ subunit and N-His6-ORF5, abundantly expressed in PdKO7/pRK-H6ORF5 (Fig. 6D).
FIGURE 6.
Analyses of the cleaved product bound to ORF5. ORF5 (0.2 mg/ml) was incubated with the N-Flc and C-Flc substrates (20.2 and 204 μm, respectively) at 30 °C for 15 h. A 5-μl aliquot of the reaction mixture containing ∼1 μg of protein was subjected to SDS-PAGE (15%) and analyzed by Coomassie Brilliant Blue staining (upper panel) and fluorescence scan (lower panel). A, lane 1, ORF5 alone; lane 2, D8A (mutant) alone; lane 3, ORF5 reacted with the N-Flc peptide; lane 4, D8A reacted with the N-Flc peptide; lane 5, ORF5 reacted with the C-Flc peptide; and lane 6, D8A reacted with the C-Flc peptide. B, the reaction mixture of ORF5 and the N-Flc substrate (30 °C for 15 h) was treated at pH 3 (lane 1) or pH 11.9 (lane 2) for 30 min and subjected to SDS-PAGE (lane 3, no treatment). C, the reaction mixture of ORF5 and the N-Flc substrate (30 °C for 15 h) was incubated with 200 mm buffer with the indicated pH values for 30 min in the presence of 5 M urea and analyzed by HPLC (30-min linear gradient of 0–60% CH3CN in 0.1% TFA). The absorbance at 220 nm (solid line) and fluorescence at 520 nm (dotted line) upon excitation at 445 nm were monitored. The arrows indicate the peaks of the fluorescent product bound to ORF5. D, ORF5 purified from PdKO7/pRK-H6ORF5 was treated at pH 3 (lanes 1 and 4), 7 (lanes 2 and 5), or 11.9 (lanes 3 and 6) for 30 min at 30 °C; subjected to SDS-PAGE; and analyzed by Western blotting with antibodies against ORF5 (lanes 1–3) and the γ subunit leader peptide (lanes 4–6). E, the fluorescent product bound to ORF5 was digested with trypsin and separated by HPLC. The arrow indicates the peak collected for mass spectrometric analysis. F, MALDI-TOF mass spectra of the collected peak (arrow in E) are shown for low (upper panel) and high (lower panel) molecular mass ranges. The insets are enlarged spectra with m/z values of the detected peaks.
To analyze the chemical structure, the cleaved fluorescent product bound to ORF5 was digested with trypsin and purified by HPLC (Fig. 6E). MALDI-TOF mass spectrometric analysis of the isolated fluorescent peptide showed the presence of 2 fragments with m/z ∼900 and ∼3800 (Fig. 6F), instead of a single fragment expected for the cleaved N-terminal leader peptide covalently bound to a tryptic peptide of ORF5. The fragment with m/z ∼900 was assigned to the cleaved N-Flc peptide (Flc-EVVAM, [M+H]+ = m/z 906.323). The fragment with m/z ∼3800 was found to consist of two fragments with m/z 3794.6 and 3811.2. The minor fragment with m/z 3811.2 could be assigned to a tryptic fragment of ORF5 (C146G···AS177L···AR182, including carboxyamidomethylated Cys-146 and Cys-163, [M+H]+ = m/z 3,810.72) containing catalytic nucleophile Ser-177. On the other hand, the major fragment with m/z 3794.6 could most likely be assigned to the same tryptic fragment that suffered MALDI-induced cleavage at the β-hydroxyl oxygen atom of Ser-177. The β-hydroxyl group of a Ser residue in the free state is generally uncleavable by MALDI. However, if it is involved in an ester linkage, it may be cleaved by MALDI in an analogous manner to that observed in the cleavage of peptide N-Cα bonds, induced by hydrogen transfer to the adjacent carbonyl oxygen from the matrix (16, 17).
To further demonstrate the formation of acyl-enzyme intermediate in the ORF5 reaction, we examined the effect of neutral hydroxylamine, which has been classically known to accelerate the product release in the serine protease reaction by acting as a nucleophile on the acyl-enzyme intermediate (18). As shown in supplemental Fig. S5, addition of hydroxylamine indeed decreased the fluorescence peak derived from the covalent complex of ORF5 and N-Flc substrate in a concentration-dependent manner in the quick HPLC analyses of the reaction mixture. This result also supports the formation of an acyl-enzyme intermediate in the ORF5 reaction with the leader peptide of γ subunit, although MALDI-TOF mass spectrometric analysis failed to detect the product hydroxamate in the reaction mixture (data not shown).
Altogether, these results strongly suggest that the cleaved N-Flc peptide remained covalently bound to ORF5 as an unstable acyl-enzyme intermediate attached to the active-site Ser residue. An attempt to identify the intact acyl-enzyme intermediate by a milder ionization method (electrospray ionization MS spectrometry) was unsuccessful, yielding again a mixture of cleaved N-Flc peptide and whole ORF5 protein (supplemental Fig. S6).
DISCUSSION
By an extensive sequence similarity search of QHNDH homologs in the up-to-date protein database compiled by the National Center for Biotechnology Information, we have identified QHNDH homologs in at least 17 bacterial species (supplemental Fig. S7) belonging to the Proteobacteria with a periplasmic compartment. Among them, three species (Aromatoleum aromaticum EbN1, Oceanospirillum sp., and Thauera sp.) have two sets of QHNDH homolog genes encoding at different genomic loci in the same or opposite orientation. The four genes (ORF1-ORF4) encoding the QHNDH α subunit, [Fe-S] cluster/AdoMet-binding protein (8), γ subunit, and β subunit constitute a gene cassette strictly conserved in this arrangement, with ORF2 always preceding ORF3.
As described here, ORF5 encodes a subtilisin-like serine protease indispensable for QHNDH biogenesis and n-butylamine-dependent bacterial growth. Although the genetic loci of ORF5 homologs relative to the ORF1-ORF4 gene cassette are different among bacterial species, they are fully conserved and nearby ORF1-ORF4 in all bacteria except Pseudomonas entomophila L48, carrying an ORF5 homolog at a distant locus (supplemental Fig. S7). Therefore, similar to the [Fe-S] cluster/AdoMet-binding protein, encoded by ORF2 and necessary for the formation of the three intrapeptidyl thioether cross-links in the γ subunit (8), ORF5 presumably plays a common essential role in these bacteria.
ORF5 was formed in the cytoplasmic fraction of Pd1222 cells grown in n-butylamine-containing medium (Fig. 2C), suggesting that it is an intracellular processing enzyme specifically acting on the γ subunit precursor. Considering that the γ subunit accumulated in PdKO4 retains the N-terminal leader peptide, ORF5 presumably recognizes the γ subunit precursor after it has undergone intrapeptidyl thioether cross-link formation by the [Fe-S] cluster/AdoMet-binding protein and before it is translocated into the periplasm. In the crystal structure of the mature QHNDH complex (5, 6), the γ subunit contains only two short α-helices and consists mostly of featureless coils. Therefore, the complete formation of the three intrapeptidyl thioether cross-links (Fig. 1B) is probably crucial for the folded structure of the γ subunit to which ORF5 binds. In addition, the leader peptide, which is absent in the γ subunit of the mature QHNDH complex, was predicted to have a high α-helix-forming tendency by a program for secondary structure prediction (19). Collectively, it is concluded that ORF5 is not a signal peptidase that usually recognizes peptide chains in an extended conformation but a protease specifically acting on the folded γ subunit precursor in the cytoplasm. Furthermore, the periplasmic translocation of the cross-linked γ subunit appears to be independent of the presence of the leader peptide (supplemental Fig. S3). Although the mechanism of the periplasmic translocation remains to be studied, we have recently found that an ABC transporter-like protein encoded adjacent to ORF5 (supplemental Fig. S7) is also essential for QHNDH biogenesis and may participate in the γ subunit translocation.
A particularly interesting observation in this study was that purified N-His6-ORF5 had extremely low proteolytic activity for synthetic peptides surrogating for the N-terminal leader peptide of the γ subunit, with the cleavage reaction proceeding without turnover or with only a few turnovers (supplemental Fig. S4). The low activity may be due to the fact that the cleaved N-terminal peptide remains bound to ORF5, most likely as an unstable acyl-enzyme intermediate attached to the active-site Ser residue (Fig. 6F). Alkaline-labile binding of the cleaved peptide with ORF5 was also observed for the leader peptide of the endogenous γ subunit (Fig. 6D). In this regard, the behavior of the γ subunit precursor as a substrate of ORF5 is similar to that of the serpins (serine proteinase inhibitors) employing a suicide substrate-like inhibitory mechanism, during which an acyl-enzyme intermediate is formed by a covalent bond (20, 21). Therefore, in γ subunit processing, ORF5 functions as a cis-acting enzyme nearly in a disposable manner. Considering that ORF5 would be cotranslated with the QHNDH subunits following n-butylamine induction, its single-turnover cleavage of the N-terminal leader peptide should be enough for the entire process of QHNDH biogenesis, even if it is rapidly degraded together with the covalently bound leader peptide that has finished its role in thioether cross-link formation within the γ subunit.
Accessory genes such as ORF2 and ORF5 in the n-butylamine-utilizing operon are reportedly required for the biosynthesis of the quinone cofactor pyrroloquinoline quinone, in which six genes, designated pqqA-pqqF, are involved (22). The Glu and Tyr residues in the consensus Glu-X-X-X-Tyr motif contained in the center of PqqA, the 23-residue peptide precursor of pyrroloquinoline quinone (23), are coupled covalently by radical AdoMet protein PqqE (24) followed by cleavage of the flanking peptide regions by Zn-dependent metalloprotease PqqF to form an early pyrroloquinoline quinone precursor (25). Therefore, ORF2 and ORF5, essential for QHNDH biogenesis, may correspond to PqqE and PqqF, respectively. However, whether this similarity is merely fortuitous or implies an evolutionary relationship between the biosynthetic mechanisms of the quinone cofactor and quinoprotein is unknown. Finally, ORF5 may represent a rare bacterial version of processing enzymes essential for the irreversible activation of otherwise latent enzymes or proteins that are identifiable frequently in higher eukaryotic organisms.
Acknowledgments
We thank the staff of the Comprehensive Analysis Center, Institute of Scientific and Industrial Research, Osaka University for technical assistance in MALDI-TOF mass analyses.
This work was supported by Japan Society for the Promotion of Science Grants in Aid for Scientific Research Category B 18370043 (to K. T.) and 18350085 (to T. O.) and Category C 23570135 (to T. N.), by a Kaneko/Narita research grant from the Protein Research Foundation (to T. N.), and by a research fund from the Japan Foundation of Applied Enzymology (to K. T.).

This article contains supplemental Figs. S1–S7, Table S1, and “Experimental Procedures.”
- QHNDH
- quinohemoprotein amine dehydrogenase
- CTQ
- cysteine tryptophylquinone
- AdoMet
- S-adenosylmethionine.
REFERENCES
- 1. Takagi K., Torimura M., Kawaguchi K., Kano K., Ikeda T. (1999) Biochemical and electrochemical characterization of quinohemoprotein amine dehydrogenase from Paracoccus denitrificans. Biochemistry 38, 6935–6942 [DOI] [PubMed] [Google Scholar]
- 2. Takagi K., Yamamoto K., Kano K., Ikeda T. (2001) New pathway of amine oxidation respiratory chain of Paracoccus denitrificans IFO 12442. Eur. J. Biochem. 268, 470–476 [DOI] [PubMed] [Google Scholar]
- 3. Adachi O., Kubota T., Hacisalihoglu A., Toyama H., Shinagawa E., Duine J. A., Matsushita K. (1998) Biosci. Biotechnol. Biochem. 62, 469–478 [DOI] [PubMed] [Google Scholar]
- 4. Vandenberghe I., Kim J. K., Devreese B., Hacisalihoglu A., Iwabuki H., Okajima T., Kuroda S., Adachi O., Jongejan J. A., Duine J. A., Tanizawa K., Van Beeumen J. (2001) The covalent structure of the small subunit from Pseudomonas putida amine dehydrogenase reveals the presence of three novel types of internal cross-linkages, all involving cysteine in a thioether bond. J. Biol. Chem. 276, 42923–42931 [DOI] [PubMed] [Google Scholar]
- 5. Datta S., Mori Y., Takagi K., Kawaguchi K., Chen Z. W., Okajima T., Kuroda S., Ikeda T., Kano K., Tanizawa K., Mathews F. S. (2001) Structure of a quinohemoprotein amine dehydrogenase with an uncommon redox cofactor and highly unusual crosslinking. Proc. Natl. Acad. Sci. U.S.A. 98, 14268–14273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Satoh A., Kim J. K., Miyahara I., Devreese B., Vandenberghe I., Hacisalihoglu A., Okajima T., Kuroda S., Adachi O., Duine J. A., Van Beeumen J., Tanizawa K., Hirotsu K. (2002) Crystal structure of quinohemoprotein amine dehydrogenase from Pseudomonas putida. Identification of a novel quinone cofactor encaged by multiple thioether cross-bridges. J. Biol. Chem. 277, 2830–2834 [DOI] [PubMed] [Google Scholar]
- 7. Sofia H. J., Chen G., Hetzler B. G., Reyes-Spindola J. F., Miller N. E. (2001) Radical SAM, a novel protein superfamily linking unresolved steps in familiar biosynthetic pathways with radical mechanisms. Functional characterization using new analysis and information visualization methods. Nucleic Acids Res. 29, 1097–1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ono K., Okajima T., Tani M., Kuroda S., Sun D., Davidson V. L., Tanizawa K. (2006) Involvement of a putative [Fe-S] cluster-binding protein in the biogenesis of quinohemoprotein amine dehydrogenase. J. Biol. Chem. 281, 13672–13684 [DOI] [PubMed] [Google Scholar]
- 9. Van Spanning R. J., Wansell C. W., Reijnders W. N., Harms N., Ras J., Oltmann L. F., Stouthamer A. H. (1991) A method for introduction of unmarked mutations in the genome of Paracoccus denitrificans. Construction of strains with multiple mutations in the genes encoding periplasmic cytochromes c550, c551i, and c553i. J. Bacteriol. 173, 6962–6970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Simon R., Priefer U., Pühler A. (1983) in Molecular Genetics of the Bacteria-Plant Interaction (Pühler A., ed) pp. 98–106, Springer Verlag, Heidelberg, Germany [Google Scholar]
- 11. Hirel P. H., Schmitter M. J., Dessen P., Fayat G., Blanquet S. (1989) Extent of N-terminal methionine excision from Escherichia coli proteins is governed by the side-chain length of the penultimate amino acid. Proc. Natl. Acad. Sci. U.S.A. 86, 8247–8251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tobias J. W., Shrader T. E., Rocap G., Varshavsky A. (1991) The N-end rule in bacteria. Science 254, 1374–1377 [DOI] [PubMed] [Google Scholar]
- 13. Keen N. T., Tamaki S., Kobayashi D., Trollinger D. (1988) Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene 70, 191–197 [DOI] [PubMed] [Google Scholar]
- 14. Page M. D., Ferguson S. J. (1994) Differential reduction in soluble and membrane-bound c-type cytochrome contents in a Paracoccus denitrificans mutant partially deficient in 5-aminolevulinate synthase activity. J. Bacteriol. 176, 5919–5928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Paz M. A., Flückiger R., Boak A., Kagan H. M., Gallop P. M. (1991) Specific detection of quinoproteins by redox-cycling staining. J. Biol. Chem. 266, 689–692 [PubMed] [Google Scholar]
- 16. Takayama M. (2001) J. Am. Soc. Mass Spectrom. 12, 420–427 [DOI] [PubMed] [Google Scholar]
- 17. Köcher T., Engström A., Zubarev R. A. (2005) Fragmentation of peptides in MALDI in-source decay mediated by hydrogen radicals. Anal. Chem. 77, 172–177 [DOI] [PubMed] [Google Scholar]
- 18. Nishino T., Kozarich J. W., Strominger J. L. (1977) Kinetic evidence for an acyl-enzyme intermediate in D-alanine carboxypeptidases of Bacillus subtilis and Bacillus stearothermophilus. J. Biol. Chem. 252, 2934–2939 [PubMed] [Google Scholar]
- 19. Guermeur Y., Geourjon C., Gallinari P., Deléage G. (1999) Improved performance in protein secondary structure prediction by inhomogeneous score combination. Bioinformatics 15, 413–421 [DOI] [PubMed] [Google Scholar]
- 20. Silverman G. A., Bird P. I., Carrell R. W., Church F. C., Coughlin P. B., Gettins P. G., Irving J. A., Lomas D. A., Luke C. J., Moyer R. W., Pemberton P. A., Remold-O'Donnell E., Salvesen G. S., Travis J., Whisstock J. C. (2001) The serpins are an expanding superfamily of structurally similar but functionally diverse proteins. Evolution, mechanism of inhibition, novel functions, and a revised nomenclature. J. Biol. Chem. 276, 33293–33296 [DOI] [PubMed] [Google Scholar]
- 21. Ye S., Goldsmith E. J. (2001) Serpins and other covalent protease inhibitors. Curr. Opin. Struct. Biol. 11, 740–745 [DOI] [PubMed] [Google Scholar]
- 22. Meulenberg J. J., Sellink E., Riegman N. H., Postma P. W. (1992) Nucleotide sequence and structure of the Klebsiella pneumoniae PQQ operon. Mol. Gen. Genet. 232, 284–294 [DOI] [PubMed] [Google Scholar]
- 23. Puehringer S., Metlitzky M., Schwarzenbacher R. (2008) The pyrroloquinoline quinone biosynthesis pathway revisited. A structural approach. BMC Biochem. 9, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wecksler S. R., Stoll S., Tran H., Magnusson O. T., Wu S. P., King D., Britt R. D., Klinman J. P. (2009) Pyrroloquinoline quinone biogenesis. Demonstration that PqqE from Klebsiella pneumoniae is a radical S-adenosyl-L-methionine enzyme. Biochemistry 48, 10151–10161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Velterop J. S., Sellink E., Meulenberg J. J., David S., Bulder I., Postma P. W. (1995) Synthesis of pyrroloquinoline quinone in vivo and in vitro and detection of an intermediate in the biosynthetic pathway. J. Bacteriol. 177, 5088–5098 [DOI] [PMC free article] [PubMed] [Google Scholar]