Background: IRE1α regulation in ER stress response and protein homeostasis need to be fully studied.
Results: IRE1α binds the Cdc37-Hsp90 complex. Interfering this interaction enhanced basal IRE1α activity and impaired insulin synthesis in INS-1 cells.
Conclusion: Negative regulation of IRE1α activity by Hsp90/Cdc37 is important for insulin synthesis.
Significance: Hsp90/Cdc37-mediated regulation of IRE1α adds new mechanistic insight to ER stress signal and regulation.
Keywords: Endoplasmic Reticulum Stress, Heat Shock Protein, Insulin Synthesis, Protein Complexes, Signal Transduction, Cdc37, Hsp90, IRE1
Abstract
IRE1α is an endoplasmic reticulum (ER) localized signaling molecule critical for unfolded protein response. During ER stress, IRE1α activation is induced by oligomerization and autophosphorylation in its cytosolic domain, a process triggered by dissociation of an ER luminal chaperone, binding immunoglobulin-protein (BiP), from IRE1α. In addition, inhibition of a cytosolic chaperone protein Hsp90 also induces IRE1α oligomerization and activation in the absence of an ER stressor. Here, we report that the Hsp90 cochaperone Cdc37 directly interacts with IRE1α through a highly conserved cytosolic motif of IRE1α. Cdc37 knockdown or disruption of Cdc37 interaction with IRE1α significantly increased basal IRE1α activity. In INS-1 cells, Hsp90 inhibition and disruption of IRE1α-Cdc37 interaction both induced an ER stress response and impaired insulin synthesis and secretion. These data suggest that Cdc37-mediated direct interaction between Hsp90/Cdc37 and an IRE1α cytosolic motif is important to maintain basal IRE1α activity and contributes to normal protein homeostasis and unfolded protein response under physiological stimulation.
Introduction
The endoplasmic reticulum (ER)2 is a cellular organelle where secretory and transmembrane proteins are synthesized, folded, and modified into mature proteins. The folding capacity of the ER is tightly regulated to match the flux of protein synthesis through a highly conserved ER stress signaling pathway. An increase in ER load triggers a coordinated compensatory response, including inhibition of protein synthesis, increase in ER chaperone expression, and removal of unfolded proteins via proteasome-dependent degradation. This so-called unfolded-protein-response (UPR) is critical to maintain ER homeostasis and essential for cellular survival and function (1, 2). However, abnormal ER stress can also be proapoptotic (3, 4) and contributes to diseases ranging from diabetes to neurological disorders (5).
The full spectrum of UPR is mainly mediated by the coordinated activation and downstream signaling of three ER membrane proteins: IRE1, protein kinase RNA-like ER kinase (PERK), and ATF6 (6–8). Under basal condition, an ER luminal chaperone protein, BiP/GRP78, binds to these molecules from the ER lumen and suppresses their basal activity. During ER stress, misfolded proteins accumulate in the ER lumen and remove BiP from IRE1, PERK, and ATF-6, leading to their activation and downstream signaling. For IRE1, removal of BiP binding uncovers an IRE1 dimerization domain located in the N-terminal domain within the ER lumen that initiates IRE1 oligomerization. Oligomerized IRE1 triggers trans-autophosphorylation in its cytosolic domain (9) and activation of its endonuclease activity for non-conventional splicing of its downstream target mRNAs such as Xbp-1 (10–12). Spliced XBP-1 protein is a transcription factor necessary for UPR gene expression, including both ER chaperones and ER-associated degradation proteins (13). In addition to the dimerization domain within the ER lumen, recent biochemical and structural analysis has also revealed that IRE1 possesses an oligomerization motif in the cytosolic kinase domain that mediates IRE1 interaction and endonuclease activation (14). However, the mechanism that regulates the cytosolic domain-mediated oligomerization and autophosphorylation activity of IRE1α remains to be established.
Hsp90 is an abundant cytosolic protein that functions with cochaperone Cdc37 to selectively interact with client kinases and regulate their stability and activity. Inhibition of Hsp90 induces ER stress associated with increased expression BiP (15) and mitochondrial mediated apoptosis (16). Earlier studies have implied that heat shock protein Hsp90 regulates IRE1 protein stability and UPR through a cytosolic domain (14). However, the detailed molecular mechanism remains unexplored. In this report, we find that Hsp90 and Cdc37 are molecular components of the IRE1α signaling complex. Cdc37 interacts with IRE1α directly via a cytosolic motif located in the kinase/oligomerization domain of IRE1α. Cdc37 expression is both necessary and sufficient for IRE1α inactivation under basal conditions. Disrupting the Cdc37 interaction with IRE1α impairs the normal UPR during ER stress. In INS-1 cells, Hsp90 inhibition or interrupting Cdc37 interaction with IRE1 results in a significant impairment of glucose-induced insulin synthesis and secretion. Therefore, our study has revealed Cdc37 as a new molecular component of the IRE1 signaling complex. Cdc37-Hsp90 activity is necessary to maintain basal IRE1α inactivation through direct interaction in the cytosol. This mechanism also plays a role in the normal ER stress response and protein homeostasis important for glucose induced insulin synthesis/secretion in pancreatic β cells.
EXPERIMENTAL PROCEDURES
Cell Culture
INS-1 was grown in RPMI 1640 media supplemented with 10% FBS, 10 mm HEPES, 10 mm sodium pyruvate, 50 μm β-mercaptoethanol, and 100 units/ml penicillin:streptomycin solution. HeLa cells were cultured in DMEM with 10% FBS and 100 units/ml penicillin:streptomycin solution. Cells were treated with 1 μg/ml of geldanamycin (GA) for the indicated time. HeLa cells were infected with an adenovirus expressing IRE1α (Adv-IRE1α) and were incubated for an additional 24 h before the experiment. Cotransfection of HeLa cells with Adv-IRE1α and Adv-shRNA was also performed 24 h prior to the experiment.
Western Blot Analysis
Proteins were homogenized in 1% Triton X-100 lysis buffer containing 50 mm Tris-HCl (pH 7.4), 150 mm NaCl, 1 mm EGTA, 1 mm EDTA, 1% Triton X-100, 1 mm β-glycerophosphate, 2.5 mm Na4P2O7, 20 mm NaF, 1 mm Na3VO4, 1 mm PMSF, and protease inhibitor mixture (Roche). Proteins were separated on 12% Bis/Tris PAGE (Invitrogen) and transferred onto nitrocellulose membranes. Proteins were detected by immunoblots using antibodies against IRE1α, XBP-1, Actin (Santa Cruz Biotechnology, Inc.), IRE1α (Ser-724) (Novus Biologicals), BiP, IRE1α, Cdc37 (Santa Cruz Biotechnology, Inc.), Cdc37, Hsp90α/β, insulin (Abcam), penta-His (Qiagen), Myc, p-Akt, and Akt (Cell Signaling Technology, Inc.).
Immunoprecipitation Western
Phospho-IRE1α was detected by first immunoprecipitating IRE1α from the total protein extract using anti-IRE1α (Santa Cruz Biotechnology, Inc.) with protein A-conjugated agarose beads (GE Healthcare) overnight at 4 °C. Precipitant was boiled in loading buffer containing 0.2% β-mercaptoethanol and separated on 4–12% Bis/Tris SDS-PAGE (Invitrogen) following the company's protocol.
Sucrose Gradient
Cells were lysed in 1% Triton X-100 lysis buffer, and 2 mg of total cellular protein lysates were loaded on a column of sucrose gradient ranging from 10–50% or 10–40% and centrifuged for 16 h at 132,000 × g at 4 °C.
Glucose-stimulated Insulin Secretion Assay
INS-1 cells were cultured as described earlier, and fresh media were added a day before the assay. Cells were starved in KRBH buffer (120 mm NaCl, 5 mm KCl, 1.2 mm KH2PO4, 5 mm NaHCO3, 2.5 mm CaCl2, 1.2 mm MgSO4, 0.2% fatty acid-free BSA, and 10 mm HEPES (pH 7.4)) for 2 h at 37 °C in the presence of 0.01% DMSO, 1 μg/ml GA, or 1 μg/ml tunicamycin (TM). Cells were then treated with KRBH + 16.7 mm glucose for 1 h at 37 °C. Media from each sample was snap-frozen and kept at −80 °C for an insulin secretion assay by ELISA (Crystal Chem, Inc.). Total protein lysates were prepared in 1% Triton X-100 lysis buffer, and RNA was extracted using TRIzol (Invitrogen) following the company's protocol.
Quantitative RT-PCR
RNA was isolated from each sample using TRIzol, and cDNA was synthesized using SuperScriptII reverse transcriptase (Invitrogen) following the company's protocol. Quantitative RT-PCR was done using iQ SYBR Green Supermix and MyiQ (Bio-Rad). All the reactions and analyses were done using iCycler (Bio-Rad). Primer sequences, melt curve, and efficiency of each PCR are shown in supplemental Fig. S6, A–C, respectively.
RESULTS
Hsp90 Regulates IRE1 Activity without Activating Full ER Stress Response
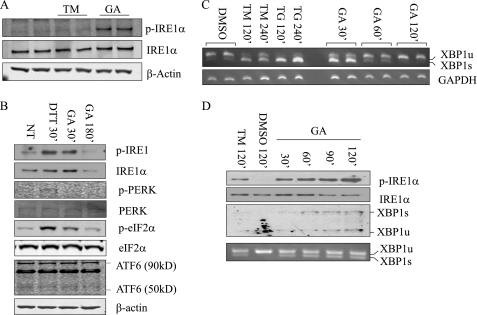
Hsp90 inhibition by GA is known to regulate IRE1 protein stability (14) and participates in the unfolded protein response regulation (14, 17). To determine the specificity of GA on IRE1, INS-1 (a rat pancreatic β cell line) was treated with GA, and its effect was compared with other ER stress inducers, including DTT, TM, and thapsigargin (TG). DTT is a strong reducing agent that quickly induces ER stress to activate UPR pathways, whereas TM and TG are slow-acting inducers by inhibiting protein glycosylation or sarcoplasmic/ER calcium ATPase (SERCA), respectively. We observed that GA treatment for 30 min resulted in a robust induction of IRE1α phosphorylation. In contrast, cells treated with TM failed to induce IRE1α phosphorylation during the same time frame (Fig. 1A). When the effect of GA was compared with DTT, both equally induced IRE1α phosphorylation, whereas the PERK pathway indicated by p-PERK and its downstream substrate p-eIF2α or ATF6 was not activated by GA to the same extent as DTT (Fig. 1B). Therefore, acute GA treatment specifically induced IRE1α phosphorylation without inducing the general ER stress pathway. Similar results were seen in INS-1 cells treated with another Hsp90 inhibitor, 17-allylamino-17-demethoxygeldanamycin (supplemental Fig. S1).
FIGURE 1.
Hsp90 inhibition induced IRE1α activation and degradation in INS-1 cells. A, INS-1 cells were treated with 1 μg/ml GA or 1 μm TM for 30 min and immunoblotted for p-IRE1α and total IRE1α after immunoprecipitation with IRE1α antibodies. β-Actin immunoblot analysis was performed on total extracts as a loading control. B, INS-1 cells were treated with 1 μg/ml GA for 30 or 180 min and with DTT for 30 min. Unfolded protein response proteins from total extracts were analyzed by immunoblotting as indicated. NT, non-treated. C, XBP-1 pro-RNA splicing was detected by RT-PCR from INS-1 cells treated with GA for 0.5, 1, or 2 h; 1 μm TM or 1 μm TG for 2 or 4 h; and DMSO for 2 h as indicated. D, INS-1 cells were cultured with fresh media containing 1 μg/ml GA every 30 min for 2 h, and immunoblot for p-IRE1α and XBP-1 protein as indicated. The bottom panel includes XBP-1 pro-RNA splicing detected by RT-PCR.
ER stressors such as TM and TG lead to sustained IRE1 activation, as indicated by increased levels of Xbp-1 pro-RNA splicing for up to 4 h post-treatment (Fig. 1C). In contrast, Hsp90 inhibition resulted in only a transient induction of IRE1 activity. After 120 min post-GA-treatment, total IRE1, phospho-IRE1α, and Xbp-1 pro-RNA splicing were all decreased (Fig. 1, B and C). This is in an agreement with a previous report that long-term Hsp90 inhibition causes IRE1 protein degradation (14). Unexpectedly, however, the IRE1 induction was maintained up to 120 min when the culture media was replaced every 30 min with fresh media containing 1 μg/ml GA. In contrast, total IRE1 protein was reduced after 90 min of GA treatment compared with the earlier time points by the same treatment (Fig. 1D). These data suggest that Hsp90 regulates IRE1 through two different mechanisms: one regulates IRE1 protein stability, and another regulates IRE1 activity through a so far uncharacterized mechanism independent of general ER stress induction.
IRE1α Interacts with Cdc37 in an Hsp90-dependent Manner
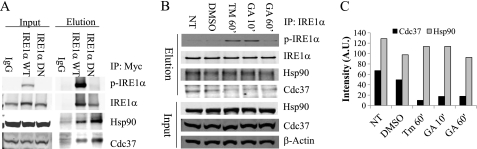
To identify the IRE1α binding proteins, we employed LC-MS/MS analysis of the IRE1α immunocomplex isolated from INS-1 cells expressing FLAG-tagged IRE1α protein and identified Hsp90 and its cochaperone Cdc37 as part of the protein complex. Using coimmunoprecipitation assay in HeLa cells expressing cMyc-tagged IRE1α (WT) or IRE1α kinase dead mutant (KD), we found that both wild-type IRE1α and the KD mutant bind to the endogenous Hsp90 and Cdc37 chaperone complex (Fig. 2A). However, when INS-1 cells were treated with TM or GA for 10 or 60 min, the amount of Cdc37 interacting with the IRE1α was decreased with no marked change in the interaction between IRE1α and Hsp90. Therefore, Cdc37 appears to be released from the IRE1α complex upon ER stress induction or Hsp90 inhibition (Fig. 2, B and C). This observation suggests that IRE1 interaction with Hsp90 is not sensitive to Hsp90 inhibition. However, Cdc37 interaction with IRE1 is Hsp90 activity-dependent.
FIGURE 2.
IRE1α interacts with the Hsp90-Cdc37 chaperone complex. A, the IRE1α complex was isolated from INS-1 cells overexpressing IRE1α WT or the IRE1α KD mutant as indicated using IgG or anti-Myc antibody followed by immunoblot analysis for p-IRE1α, total IRE1α, Hsp90 (both α/β), and Cdc37 as indicated. B, INS-1 cells were treated with DMSO, 1 μm TM, or 1 μg/ml GA for 10 or 60 min. Following immunoprecipitation (IP) using anti-IRE1α antibody, an immunoblot analysis for p-IRE1α, IRE1α, Hsp90, and Cdc37 was performed as indicated. C, the changes in the amount of Hsp90 and Cdc37 interacting with the IRE1α are quantified and plotted. A.U., arbitrary unit; NT, non-treated.
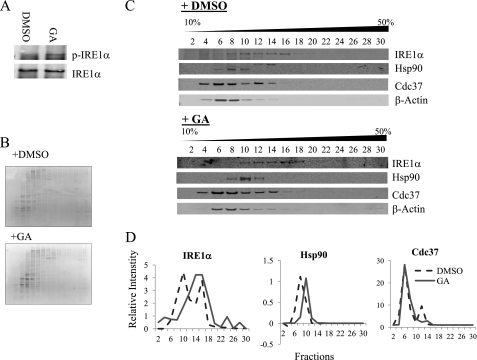
To further investigate the interaction between endogenous IRE1α and Hsp90-Cdc37 chaperones, INS-1 cells were treated with DMSO or GA for 15 min, and the total lysates were fractionated on a sucrose gradient. Increase in phospho-IRE1α is detected in GA-treated samples as expected (Fig. 3A). Coomassie staining of SDS-PAGE (Fig. 3B) or immunoblotting on β-actin (C) from the fractionated samples demonstrated no apparent differences in the overall protein distribution profile between DMSO- and GA-treated samples. In protein extracts prepared from DMSO-treated INS-1 cells, IRE1α displayed a broad spectrum of distribution from fraction 8 to 16, indicating different forms of IRE1 complexes in intact cells. Both Hsp90 and Cdc37 comigrated with IRE1α in lower molecular weight fractions (8 to 12). Upon GA treatment, IRE1α signals were shifted to higher molecular weight fractions, consistent with IRE1α oligomerization (Fig. 3, C and D). Similarly, distribution of Hsp90 was also observed to shift to higher molecular weight fractions. This is consistent with the notion that Hsp90 remained associated with IRE1 upon Hsp90 inhibition (as shown in Fig. 2), as well as previously reported by Marcu et al. (14). In contrast, Cdc37 showed no change in sucrose gradient distribution after GA treatment. These data again support the earlier observation that Cdc37 interacts with IRE1 in an Hsp90 activity-dependent manner.
FIGURE 3.
Dynamic interaction between IRE1α and Cdc37. A, INS-1 cells were treated with DMSO or 1 μg/ml GA for 15 min, and cell extracts were fractionated in a 10 to 50% sucrose gradient. B, images of Coomassie-stained gels of protein fractions from DMSO and GA-treated INS-1 cell extracts. C, immunoblot analysis of protein fractions for IRE1α, Hsp90, Cdc37, and β-actin as indicated. D, bands were quantified and plotted to show the difference in migration pattern.
Cdc37 Regulates IRE1α Activity without Affecting IRE1α Stability
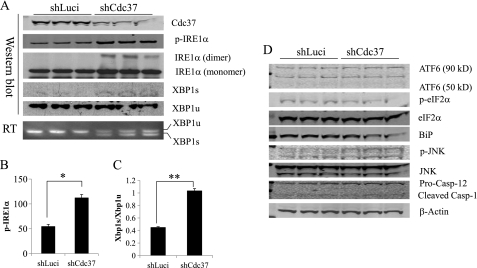
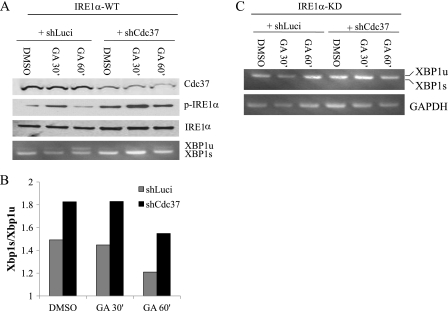
To investigate the functional significance of Cdc37-Hsp90 interaction with IRE1, shRNA was used to down-regulate endogenous Cdc37 expression in HeLa cells where the Hsp90 inhibition-induced IRE1 activation was also observed (supplemental Fig. S2). As shown in Fig. 4, A–C, IRE1α activities, as indicated by phospho-IRE1α, IRE1α dimerization, and Xbp-1 pro-RNA splicing at both protein and RNA levels, were all significantly increased in Cdc37 knockdown cells compared with controls (shLuci), whereas there was no change in ATF6 or PERK (indicated by p-eIF2α) activities (Fig. 4D). Moreover, GA-induced IRE1 activation was further enhanced in Cdc37 knockdown cells (Fig. 5, A and B), suggesting that Cdc37 is a negative regulator of IRE1α. The specificity of IRE1α-mediated XBP1 splicing was demonstrated in Fig. 5C, where the IRE1α kinase dead mutant (KD) failed to trigger XBP1 splicing under any treatment. Chronic inactivation of Cdc37 did not result in a loss of total IRE1α protein expression or a change in its sucrose gradient distribution profile (supplemental Fig. S3), implying that Cdc37 is not involved in Hsp90-mediated regulation of IRE1α protein stability or oligomerization. Overexpression of Cdc37 in INS-1 resulted in a modest but significant reduction of phospho-IRE1α at the basal level (supplemental Fig. S4). When the cells were treated with DTT or GA, there was a trend toward a decrease in IRE1α activation, but it did not reach statistical significance. Thus, although necessary, base-line expression of Cdc37 does not appear to be sufficient to modulate IRE1 activation. This may be explained by the notion that Hsp90/Cdc37 are very abundant proteins relative to other ER regulatory components (18).
FIGURE 4.
Cdc37 is necessary for negative regulation of basal IRE1α activity. A, shRNA-mediated knockdown of Cdc37 in HeLa cells expressing IRE1αWT with shLuci as a negative control. Immunoblots on p-IRE1α, total-IRE1α, XBP-1 spliced (s), and unspliced (u) forms were performed as indicated. B, quantitative measurement of phospho-IRE1α levels in A. *, p < 0.01 shCdc37 versus shLuci samples. C, XBP-1 pro-RNA splicing detected by RT-PCR. D, total lysates were immunoblotted for ATF6 and eIF1α as an indicator of PERK activities, JNK, caspase-12, and BiP as indicated.
FIGURE 5.
Cdc37 in IRE1α regulation induced by Hsp90 inhibition. A, response to Hsp90 inhibition in control (shLuci) and Cdc37 knockdown (shCdc37) HeLa cells expressing IRE1α-WT was measured by immunoblots for p-IRE1α and IRE1α as indicated or by XBP-1 pro-RNA splicing by RT-PCR (bottom panel). B, response to Hsp90 inhibition in control (shLuci) and Cdc37 knockdown (shCdc37) HeLa cells expressing the IRE1αKD mutant was measured by XBP-1 pro-RNA splicing.
Cdc37 Regulates IRE1α Activity through Direct Interaction with the IRE1 Kinase Domain Dimer Interface
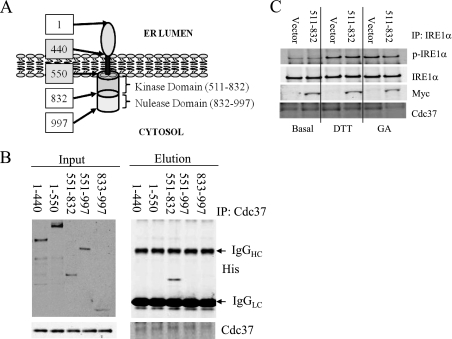
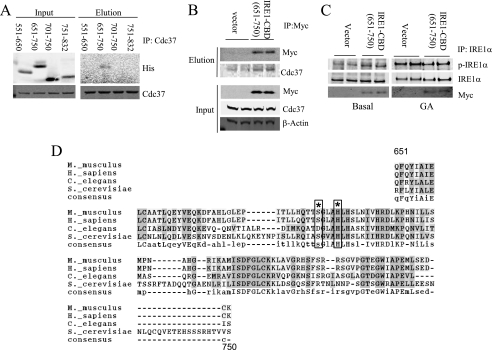
To determine the molecular basis of Cdc37/IRE1α interaction, we mapped the Cdc37 binding domain on IRE1α by a coimmunoprecipitation assay using several truncation mutants of IRE1α as shown in Fig. 6A. The full-length IRE1α containing the N terminus transmembrane domain could not be purified because of its insolubility. However, a specific interaction was detected between Cdc37 and the IRE1α aa 551–832 fragment containing the kinase domain (Fig. 6, B and C). When expressed in INS-1 cells, this IRE1α fragment did not reduce the endogenous IRE1α interaction with Cdc37 under basal conditions but reduced Cdc37 binding after 15 min of DTT or GA treatment (Fig. 6D). The aa 551–832 region was further divided into smaller fragments containing different functional motifs, including aa 551–650 for the ATP binding pocket, aa 651–750 for both the catalytic loop and activation loop, and aa 701–750 for the activation loop. As shown in Fig. 7A, the Cdc37 interaction was only detected with the fragment aa 651–750, which we postulate to contain a Cdc37 binding domain (CBD) (supplemental Fig. S5). This interaction was further validated using a coimmunoprecipitation assay in INS-1 cells expressing Myc-IRE1α-CBD (Fig. 7B). Consistent with the observation made in the Cdc37 knockdown cells, the expression of IRE1α-CBD fragment increased the basal phosphorylation level of the endogenous IRE1α (Fig. 7C). However, overexpression of IRE1α-CBD attenuated GA-induced IRE1α autophosphorylation, indicating Cdc37-interaction with IRE1α can also impact on the full activity. Earlier structural studies have revealed that the IRE1α aa 551–650 fragment contains several conserved residues that are directly involved in an IRE1 kinase domain dimer interface interaction critical for the IRE1 autophosphorylation (Fig. 7D, Ref. 12). Therefore, our data suggest that Cdc37 interacts with IRE1α via its highly conserved kinase domain in the cytosol. This interaction may modulate IRE1 autophosphorylation by regulating the kinase domain dimer interface interaction.
FIGURE 6.
Cdc37 interacts with kinase domain of IRE1α. A, schematic illustration of domains of human IRE1α with the amino acid number indicated in boxes. B, coimmunoprecipitation (IP) assay using His-tagged recombinant IRE1α deletion mutant proteins (as indicated by amino acid numbers) and Cdc37 recombinant protein as described under “Experimental Procedures.” C, IRE1α truncated protein 551–832) or vector control were expressed in INS-1 cells and treated with 1 μm DTT or 1 μg/ml GA for 15 min as indicated. Immunoprecipitation on total lysates was performed with anti-IRE1α followed by immunoblotting for p-IRE1α, IRE1α, Myc, and Cdc37 as indicated.
FIGURE 7.
Cdc37 interacts with the IRE1 dimer interface region and regulates IRE1 activity. A, IRE1α aa 551–832 was further truncated into smaller fragments, and a binding assay was performed by coimmunoprecipitation (IP) as in Fig. 6. B, Myc-IRE1α aa 651–750 was expressed in INS-1 cells. Coimmunoprecipitation was performed with anti-Myc followed by immunoblotting with anti-Myc and anti-Cdc37 as indicated. C, the same samples were also examined for IRE1α activation by immunoblotting for p-IRE1α, total IRE1α, and Myc as indicated. D, sequence alignment of conserved Cdc37 binding domain in IRE1 showing amino acid residues involved in the IRE1 dimer interface interaction (asterisk) and boxes (12).
Role of Cdc37-Hsp90-mediated IRE1 Regulation in Insulin Production and Secretion
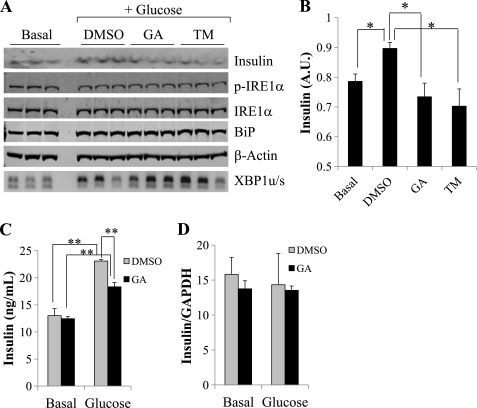
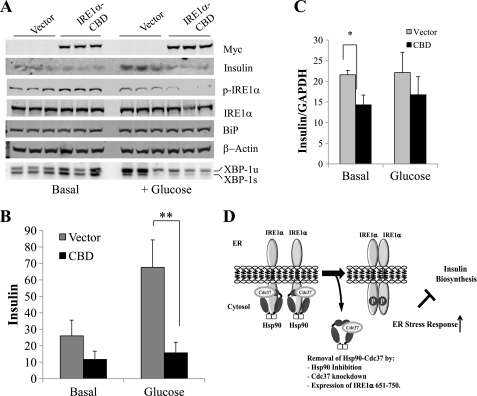
Insulin synthesis in pancreatic β-cells requires a balanced ER stress response to maintain ER flux (19, 20). An increase in IRE1α activity is associated with insulin biosynthesis, whereas sustained IRE1α activity or Adv-mediated expression of XBP1s decreased insulin expression in β-cells (21, 22). To determine the effect of Hsp90 inhibition on insulin expression and secretion, INS-1 cells were starved for 2 h and treated with or without 16.7 mm glucose for 1 h in the presence of 0.01% DMSO or 1 μg/ml GA. As shown in Fig. 8A, GA treatment induced a robust increase in Xbp-1 pro-RNA splicing, whereas there was no detectable difference in IRE1α phosphorylation levels at the same time point. The intracellular insulin level was significantly reduced in GA-treated cells compared with the DMSO-treated control (Fig. 8B). A similar effect was also observed in TM-treated cells. Glucose-induced insulin secretion was measured by ELISA from conditioned culture media and found to be reduced in GA-treated cells compared with DMSO controls (Fig. 8C). In contrast, there were no differences in the insulin mRNA levels between DMSO- and GA-treated cells at both basal conditions and in response to glucose stimulation (Fig. 8D). The quality of RT-PCR according to MIQE guidelines is shown in supplemental Fig. S6. Therefore, Hsp90 inhibition is sufficient to decrease insulin production and secretion similar to what is observed under ER stress induction (21, 22). To further demonstrate the role of Cdc37-mediated IRE1α regulation in this process, the IRE1α-CBD was expressed in INS-1 cells followed by glucose stimulation as described earlier (Fig. 9A). Western blot analyses showed a significant decrease in the insulin protein level at both basal conditions and following glucose stimulation in the IRE1α-CBD expressing INS-1 cells compared with controls. In addition, RT-PCR also showed a significant decrease in insulin mRNA expression at the basal level when IRE1α-CBD was overexpressed (Fig. 9B). Therefore, disrupting the Cdc37-Hsp90-IRE1α complex by either Hsp90 inhibition or IRE1α-CBD peptide leads to impaired insulin synthesis and secretion. All these data suggest that Cdc37-Hsp90-mediated IRE1α regulation is important for protein homeostasis (Fig. 9C).
FIGURE 8.
Inhibition of Hsp90 activity on glucose-stimulated insulin secretion. INS-1 cells were starved (basal) and stimulated with 16.7 mm glucose (glucose) for 1 h as described under “Experimental Procedures.” A, intracellular insulin protein expression as well as p-IRE1α, total IRE1α, BiP, and β-actin were determined by immunoblot analysis using total cell extracts as indicated. XBP-1 pro-RNA splicing was detected by RT-PCR (bottom panel) with insulin signal quantified in B. *, p < 0.05. A.U., arbitrary unit. C, the conditioned media were used to determine secreted insulin by ELISA as described under “Experimental Procedures” from INS-1 cells treated with DMSO or GA at basal levels or after 1 h of glucose stimulation. D, insulin mRNA expression was measured from the same treatment samples. **, p < 0.05.
FIGURE 9.
Cdc37-IRE1 interaction in glucose-stimulated insulin secretion. A, immunoblot analyses for Myc, insulin protein, p-IRE1α, total IRE1α, and BiP in cell extracts prepared from INS-1 cells expressing either vector or Myc-IRE1α-CBD (651–750) at basal levels or after 1-h treatment with 16.7 mm glucose. XBP-1 pro-RNA splicing (unspliced versus spliced) was determined by RT-PCR as indicated. B, quantitative levels of insulin protein in INS-1 cells shown in A. **, p < 0.05. C, the relative level of insulin mRNA was measured by RT-PCR at both basal levels and glucose stimulation from the same samples. *, p < 0.05. D, illustration of the working hypothesis for Cdc37/Hsp90-mediated regulation of IRE1 activity in the ER stress response and protein homeostasis.
DISCUSSION
IRE1α is known to be a client protein of Hsp90 (14), but the molecular basis and the functional significance of Hsp90-mediated regulation of IRE1 have not yet been established. In this report, we demonstrate that Cdc37, a kinase specific cochaperone of Hsp90, interacts directly with the kinase domain of IRE1α in an Hsp90 activity-dependent manner. This interaction may block IRE1α kinase autophosphorylation by interfering with the IRE1 dimerization via its kinase domain dimer interface and has a critical role in suppressing IRE1α activity under basal condition as well as its induction under ER stress. Disrupting this interaction by either Hsp90 inhibition, Cdc37 knockdown, or overexpression of the IRE1α-CBD peptide led to elevated basal activity of IRE1α. At the functional level, Hsp90 inhibition or IRE1α-CBD expression attenuated glucose-stimulated insulin production and secretion. Therefore, Cdc37 interaction with IRE1α and Cdc37-Hsp90-mediated regulation of IRE1α activity are critical to normal ER stress response and the dynamic regulation of protein homeostasis in cells.
It has been reported previously that IRE1α is a client protein of Hsp90 because of its ability to regulate IRE1α protein stability (14). In this report, we show that Hsp90 inhibition induces IRE1α autophosphorylation and oligomerization, which is also associated with the exit of Cdc37 from the IRE1α complex. Cdc37 knockdown resulted in an increase in IRE1α phosphorylation and oligomerization and an induction of XBP-1s. However, Cdc37 knockdown or disruption of Cdc37-IRE1α interaction had no effect on IRE1α protein stability. These data lead us to conclude that Hsp90 regulates IRE1α oligomerization and autophosphorylation in a Cdc37-dependent manner, whereas IRE1α protein stability is regulated independently of Cdc37, perhaps involving other yet to be characterized cochaperones. On the basis of our binding assays, Cdc37 directly interacts with the IRE1α kinase domain, which overlaps with dimer interface residues as defined by a previous structural analysis. Overexpression of the Cdc37 binding domain increased the basal IRE1α phosphorylation, whereas attenuated Hsp90 inhibition or ER stress induced IRE1α induction. Therefore, our study revealed a so far uncharacterized Cdc37-dependent cytosolic regulatory motif for IRE1α oligomerization and autophosphorylation. We can postulate that the dissociation of BiP from the ER luminal domain of IRE1α and dissociation of Cdc37 from the kinase domain in the cytosolic compartment represent a coordinated two-prompt regulatory process for IREα activation during the ER stress response. The exact molecular mechanism involved in the dynamic regulation of Cdc37-IRE1α interaction is unknown but may involve Cdc37-Hsp90-mediated conformational changes of IRE1α. It is known that conformation of the IRE1 cytosolic domain determines the amplitude as well as the specificity of its RNase activity. The specific contribution of Cdc37-Hsp90 chaperone complex activity toward IRE1 conformational change needs to be further established.
Although others have observed the induction of both the ATF6 and PERK pathways in response to Hsp90 inhibition, we only observed a slight increase in PERK activity, as indicated by the eIF2α phosphorylation level. This is consistent with a previous observation showing a very weak activation of PERK in response to GA (14). In our study, Cdc37 knockdown has a specific effect on IRE1α activity. Moreover, despite the increase in XBP-1s, we did not observe any increase in BiP expression after GA treatment or Cdc37 knockdown. Therefore, Cdc37 has a highly selective impact on IRE1α activity. All three UPR sensor proteins, PERK, ATF6, and IRE1, are under tight regulation. Although these proteins all interact with BiP in the luminal domain, the kinetics and the duration of the activation for each pathway are different (23). There is also cross-talk and cooperation among different pathways to enable concerted UPR (2,24–28). Once an ER stress is resolved, the IRE1 activity is the first to be terminated, followed by ATF6 and PERK (23). Some of the mechanisms implicated in the IRE1 inactivation include deactivation by phosphatase, such as Dcr2 in S. cerevisiae (29), or by cleaving its own mRNA(30). Interestingly, Lin et al. (23) found that prolongation of IRE1 activity under constant ER stress also enhanced cell survival. Thus, sustained or early termination of IRE1 activity can lead to cell apoptosis, highlighting the importance of tight regulation of IRE1 activity in ER stress signaling. Our report has identified yet another step in the IRE1α regulatory scheme involving the cytosolic interaction with Cdc37. Its specific impact on IRE1α versus other ER stress pathways, such as PERK and ATF6, may contribute to the differential activation profiles among the different ER stress signaling pathways and helps to determine the functional outcome of an UPR.
Pancreatic β-cells secrete insulin in response to hyperglycemia along with an induction in insulin gene expression (31). IRE1α is shown to be activated, and its activity is important to mediate compensatory UPR during insulin induction (21) (19). IRE1α knockdown resulted in a decrease in insulin expression at the basal level, whereas prolonged activation of IRE1α also leads to impaired insulin biosynthesis (21), suggesting that IRE1 activity needs to be tightly regulated for normal insulin production. We show here that Hsp90 inhibition and specific disruption of the Cdc37-IRE1α interaction all lead to defects in insulin secretion and expression. Therefore, a coordinated regulation of IRE1α from the ER lumen and cytosol is critical to maintain normal ER function. However, our current study is limited to in vitro analysis using INS-1 cells, and the physiological significance of this mechanism in glucose regulation or other ER stress related function in intact animals remains to be elucidated.
Supplementary Material
Acknowledgments
We thank Dr. Gang Lu for Adv-IRE1α WT and Adv-IRE1α KD and Mr. William Foster for the YFP-IRE1α construct. We also thank Dr. Beth Rose for assistance with proofreading.
This study was supported, in whole or in part, by National Institutes of Health Pre-doctoral Individual Fellowship Award F31AG032163 (to A. O.) and Grants HL070079 and 088640 (to Y. W.). This work was also supported by an American Heart Association Established Investigator award (to Y. W.).

This article contains supplemental Figs. S1–S5.
- ER
- endoplasmic reticulum
- UPR
- unfolded protein response
- Adv
- adenovirus
- TM
- tunicamycin
- GA
- geldanamycin
- TG
- thapsigargin
- DMSO
- dimethyl sulfoxide
- aa
- amino acids
- CBD
- Cdc37 binding domain
- BiP
- binding immunoglobulin protein
- shLuci
- shLuciferase.
REFERENCES
- 1. Patil C., Walter P. (2001) Intracellular signaling from the endoplasmic reticulum to the nucleus. The unfolded protein response in yeast and mammals. Curr. Opin. Cell Biol. 13, 349–355 [DOI] [PubMed] [Google Scholar]
- 2. Ma Y., Hendershot L. M. (2001) The unfolding tale of the unfolded protein response. Cell 107, 827–830 [DOI] [PubMed] [Google Scholar]
- 3. Urano F., Wang X., Bertolotti A., Zhang Y., Chung P., Harding H. P., Ron D. (2000) Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science 287, 664–666 [DOI] [PubMed] [Google Scholar]
- 4. Szegezdi E., Logue S. E., Gorman A. M., Samali A. (2006) Mediators of endoplasmic reticulum stress-induced apoptosis. EMBO Rep. 7, 880–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yoshida H. (2007) ER stress and diseases. FEBS J. 274, 630–658 [DOI] [PubMed] [Google Scholar]
- 6. Shen J., Chen X., Hendershot L., Prywes R. (2002) ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of Golgi localization signals. Dev. Cell 3, 99–111 [DOI] [PubMed] [Google Scholar]
- 7. Sommer T., Jarosch E. (2002) BiP binding keeps ATF6 at bay. Dev. Cell 3, 1–2 [DOI] [PubMed] [Google Scholar]
- 8. Bertolotti A., Zhang Y., Hendershot L. M., Harding H. P., Ron D. (2000) Dynamic interaction of BiP and ER stress transducers in the unfolded protein response. Nat. Cell Biol. 2, 326–332 [DOI] [PubMed] [Google Scholar]
- 9. Welihinda A. A., Kaufman R. J. (1996) The unfolded protein response pathway in Saccharomyces cerevisiae. Oligomerization and trans-phosphorylation of Ire1p (Ern1p) are required for kinase activation. J. Biol. Chem. 271, 18181–18187 [DOI] [PubMed] [Google Scholar]
- 10. Lee K., Tirasophon W., Shen X., Michalak M., Prywes R., Okada T., Yoshida H., Mori K., Kaufman R. J. (2002) IRE1-mediated unconventional mRNA splicing and S2P-mediated ATF6 cleavage merge to regulate XBP1 in signaling the unfolded protein response. Genes Dev. 16, 452–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kaufman R. J. (1999) Stress signaling from the lumen of the endoplasmic reticulum. Coordination of gene transcriptional and translational controls. Genes Dev. 13, 1211–1233 [DOI] [PubMed] [Google Scholar]
- 12. Lee K. P., Dey M., Neculai D., Cao C., Dever T. E., Sicheri F. (2008) Structure of the dual enzyme Ire1 reveals the basis for catalysis and regulation in nonconventional RNA splicing. Cell 132, 89–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yoshida H., Matsui T., Hosokawa N., Kaufman R. J., Nagata K., Mori K. (2003) A time-dependent phase shift in the mammalian unfolded protein response. Dev. Cell 4, 265–271 [DOI] [PubMed] [Google Scholar]
- 14. Marcu M. G., Doyle M., Bertolotti A., Ron D., Hendershot L., Neckers L. (2002) Heat shock protein 90 modulates the unfolded protein response by stabilizing IRE1α. Mol. Cell Biol. 22, 8506–8513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lai M. T., Huang K. L., Chang W. M., Lai Y. K. (2003) Geldanamycin induction of grp78 requires activation of reactive oxygen species via ER stress responsive elements in 9L rat brain tumour cells. Cell Signal 15, 585–595 [DOI] [PubMed] [Google Scholar]
- 16. Taiyab A., Sreedhar A. S., Rao C. M. (2009) Hsp90 inhibitors, GA and 17AAG, lead to ER stress-induced apoptosis in rat histiocytoma. Biochem. Pharmacol. 78, 142–152 [DOI] [PubMed] [Google Scholar]
- 17. Davenport E. L., Moore H. E., Dunlop A. S., Sharp S. Y., Workman P., Morgan G. J., Davies F. E. (2007) Heat shock protein inhibition is associated with activation of the unfolded protein response pathway in myeloma plasma cells. Blood 110, 2641–2649 [DOI] [PubMed] [Google Scholar]
- 18. Csermely P., Schnaider T., Soti C., Prohászka Z., Nardai G. (1998) The 90-kDa molecular chaperone family. Structure, function, and clinical applications. A comprehensive review. Pharmacol. Ther. 79, 129–168 [DOI] [PubMed] [Google Scholar]
- 19. Hartley T., Siva M., Lai E., Teodoro T., Zhang L., Volchuk A. (2010) Endoplasmic reticulum stress response in an INS-1 pancreatic β-cell line with inducible expression of a folding-deficient proinsulin. BMC Cell Biol. 11, 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fonseca S. G., Urano F., Burcin M., Gromada J. (2010) Stress hypERactivation in the β-cell. Islets 2, 1–9 [DOI] [PubMed] [Google Scholar]
- 21. Lipson K. L., Fonseca S. G., Ishigaki S., Nguyen L. X., Foss E., Bortell R., Rossini A. A., Urano F. (2006) Regulation of insulin biosynthesis in pancreatic beta cells by an endoplasmic reticulum-resident protein kinase IRE1. Cell Metab. 4, 245–254 [DOI] [PubMed] [Google Scholar]
- 22. Allagnat F., Christulia F., Ortis F., Pirot P., Lortz S., Lenzen S., Eizirik D. L., Cardozo A. K. (2010) Sustained production of spliced X-box binding protein 1 (XBP1) induces pancreatic cell dysfunction and apoptosis. Diabetologia 53, 1120–1130 [DOI] [PubMed] [Google Scholar]
- 23. Lin J. H., Li H., Yasumura D., Cohen H. R., Zhang C., Panning B., Shokat K. M., Lavail M. M., Walter P. (2007) IRE1 signaling affects cell fate during the unfolded protein response. Science 318, 944–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Niwa M., Sidrauski C., Kaufman R. J., Walter P. (1999) A role for presenilin-1 in nuclear accumulation of Ire1 fragments and induction of the mammalian unfolded protein response. Cell 99, 691–702 [DOI] [PubMed] [Google Scholar]
- 25. Ma Y., Hendershot L. M. (2004) Herp is dually regulated by both the endoplasmic reticulum stress-specific branch of the unfolded protein response and a branch that is shared with other cellular stress pathways. J. Biol. Chem. 279, 13792–13799 [DOI] [PubMed] [Google Scholar]
- 26. Harding H. P., Zhang Y., Bertolotti A., Zeng H., Ron D. (2000) Perk is essential for translational regulation and cell survival during the unfolded protein response. Mol. Cell 5, 897–904 [DOI] [PubMed] [Google Scholar]
- 27. Yoshida H., Matsui T., Yamamoto A., Okada T., Mori K. (2001) XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 107, 881–891 [DOI] [PubMed] [Google Scholar]
- 28. Okada T., Yoshida H., Akazawa R., Negishi M., Mori K. (2002) Distinct roles of activating transcription factor 6 (ATF6) and double-stranded RNA-activated protein kinase-like endoplasmic reticulum kinase (PERK) in transcription during the mammalian unfolded protein response. Biochem. J. 366, 585–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Guo J., Polymenis M. (2006) Dcr2 targets Ire1 and down-regulates the unfolded protein response in Saccharomyces cerevisiae. EMBO Rep. 7, 1124–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tirasophon W., Lee K., Callaghan B., Welihinda A., Kaufman R. J. (2000) The endoribonuclease activity of mammalian IRE1 autoregulates its mRNA and is required for the unfolded protein response. Genes Dev. 14, 2725–2736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fonseca V., Davidson J., Home P., Snyder J., Jellinger P., Dyhr Toft A., Barnett A. (2010) Starting insulin therapy with basal insulin analog or premix insulin analog in T2DM. A pooled analysis of treat-to-target trials. Curr. Med. Res. Opin. 26, 1621–1628 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.