Background: Hypochlorite is strongly bactericidal and used as disinfectant; yet, a response regulator allowing adaptation to the inflicted stress is so far unknown.
Results: The transcription factor YjiE specifically confers hypochlorite resistance and is an atypical dodecameric regulator that undergoes DNA-induced dissociation to dimers and tetramers.
Conclusion: YjiE protects cells from hypochlorite-induced oxidative damage by triggering a specific stress response.
Significance: This is the first described hypochlorite-specific regulator.
Keywords: DNA Binding Protein, Oxidative Stress, Reactive Oxygen Species (ROS), Stress Response, Transcription Factors
Abstract
Hypochlorite is a powerful oxidant produced by neutrophils to kill invading microorganisms. Despite this important physiological role of HOCl in fighting bacterial infections, no hypochlorite-specific stress response has been identified yet. Here, we identified a hypochlorite-responsive transcription factor, YjiE, which is conserved in proteobacteria and eukaryotes. YjiE forms unusual dodecameric ring-like structures in vitro that undergo large DNA-induced conformational changes to form dimers and tetramers as shown by transmission electron microscopy and analytical ultracentrifugation. Such smaller oligomers are predominant in hypochlorite-stressed cells and are the active species as shown by fluorescence anisotropy and analytical ultracentrifugation. YjiE regulates a large number of genes upon hypochlorite stress. Among them are genes involved in cysteine, methionine biosynthesis, and sulfur metabolism (up-regulated) and genes involved in iron acquisition and homeostasis (down-regulated), thus supposedly replenishing oxidized metabolites and decreasing the hypochlorite-mediated amplification of intracellular reactive oxygen species. As a result, YjiE specifically confers hypochlorite resistance to E. coli cells. Thus, to our knowledge, YjiE is the first described hypochlorite-specific transcription factor.
Introduction
The reactive oxygen species (ROS)6 hypochlorite (HOCl) is produced by neutrophils in humans and higher eukaryotes upon bacterial infection and by mucosal barrier epithelia in the gut of Drosophila to inhibit bacterial colonization (1, 2). HOCl is also known as bleach and used worldwide as a powerful disinfectant in households, hospitals, water disinfection, and the food industry. In contrast to redox-cycling drugs and other ROS such as superoxide anion and hydrogen peroxide (H2O2), HOCl is extremely reactive and bactericidal. It reacts with iron forming hydroxyl radicals via the Fenton reaction making it cytotoxic (3, 4). HOCl targets and damages a variety of macromolecules, including DNA, lipids, and proteins. It is especially reactive toward the side chains of the amino acids methionine and cysteine, causing oxidation to methionine sulfoxide as well as formation of disulfides and oxidation to oxy acids (e.g. sulfenic, sulfinic, sulfonic acid), respectively (5). This eventually leads to genome-wide mutations, protein inactivation, and perturbations in membrane function (5, 6).
Despite this physiological importance of HOCl in killing bacteria, bacterial protection mechanisms against HOCl-derived damage have not received much investigative attention. Previous studies showed that HOCl causes the induction of the heat shock and SoxR responses (7, 8). Further studies revealed reasons for the bactericidal effect of HOCl as oxidative protein unfolding leading to irreversible aggregation (9) and proteome-wide oxidation of methionine residues (10). In contrast to the majority of cellular proteins that are inactivated by HOCl, the redox-regulated E. coli chaperone Hsp33 (11) is activated by oxidative unfolding and serves to protect cellular proteins from aggregation, thus increasing E. coli resistance to HOCl (9, 12).
Oxidative stress responses in prokaryotes and eukaryotes are typically driven by transcription factors specific to the drug or ROS that causes the oxidative stress (13). Examples for redox-cycling drugs and H2O2 are the well characterized oxidative stress transcription factors SoxR and OxyR, respectively (14, 15). In contrast, no HOCl-specific transcription factor protecting bacteria from HOCl damage has been identified until now. In this study, we identified a novel transcription factor, YjiE, which confers HOCl resistance to E. coli cells. Based on homology, YjiE belongs to the large family of LysR-type transcriptional regulators (LTTRs). LTTRs are ubiquitous and act as repressors or activators of transcription. They contain an N-terminal DNA-binding helix-turn-helix motif and a C-terminal co-inducer response domain (16). LTTRs typically form dimers and tetramers (e.g. OxyR (17), CysB (18), TsaR (19), nitrogen assimilation control protein (20)), but also octamers (CrgA (21)). Activation of LTTRs usually occurs through the binding of a small molecule co-inducer (18, 19) or covalent modifications (17). We show here that YjiE is the first HOCl-specific transcription factor that increases cellular viability by reducing the intracellular iron levels.
EXPERIMENTAL PROCEDURES
Bacterial Strains, Cloning, Cultivation Conditions, and Analysis of Cellular Viability
The generation and screen of the genomic library were performed as described (22). Details on the generation of bacterial strains, plasmids, and primer are given in the supplemental data. Bacteria were cultivated at 37 °C in LB medium supplemented with adequate antibiotics until an A600 of 0.45 to 0.5 was reached (equivalent to at least four doublings). Viability assays were performed as described (9, 22). The viability of cells (after 1:20 dilution into fresh LB) was tested at 37 °C in LB medium supplemented with the indicated ROS (4 mm HOCl, 4 mm H2O2, or 5 mm diamide). Samples were removed at various time points, serially diluted in LB, and spotted onto LB agar plates. Recovery of cells was analyzed in micro titer plates using a micro titer plate reader (BioTek). Cells (at 1:20 dilution) were mixed with various concentrations of HOCl, subsequently continuously shaken at 37 °C, and A600 values were recorded at 10-min intervals. (HOCl was not quenched.)
Gene Expression Analysis
The procedure for transcriptional profiles is described in the supplemental data.
Quantitative Real-time PCR (qRT-PCR)
3 ml of culture at an A600 of 0.45 were either treated with HOCl (37 °C, 10 min) or left untreated. Then, 2 ml of 5× LB medium was added to quench the remaining HOCl, cells were harvested (5000 rpm, 5 min), and the cell pellet was frozen in liquid nitrogen. RNA was isolated using the SV total RNA isolation system (Promega) according to the manufacturer's protocol. The concentration was determined at 260 nm using a NanoDrop spectrometer (PeqLab). qRT-PCR was performed in a one-step reaction using the Brilliant III Ultra-Fast SYBR® Green QRT-PCR Master Mix (Agilent Technologies) supplemented with 100 ng of isolated RNA as template and 150 nm primers (see supplemental Table S2). The cDNAs were synthesized at 50 °C for 10 min and subsequently denatured at 95 °C for 3 min. Then, the cDNAs were amplified through 40 cycles (denaturation, 95 °C and 20 s; annealing/extension, 60 °C and 30 s). Nonspecific products or primer-dimers were detected by increasing the incubation temperature from 25 to 95 °C after amplification. The amplification efficiency was validated by standard curves for each primer pair. Besides the no-template control and no RT control omitting the reverse transcriptase/RNase block provided in the master mix kit, the tufB gene was used as endogenous control to normalize the variation of RNA template in each sample.
Analysis of Intracellular Iron Levels
Cells were cultivated as described above. At an A600 of 0.45, cells were HOCl-stressed (2.75 mm HOCl, 10 min) or left untreated and harvested. Further sample treatment and EPR measurements of incorporated iron were exactly performed as described (23).
Reporter Gene Fusion
The construction of translational lacZ reporter gene fusions and β-galactosidase assays were performed as described in the supplemental data.
Purification of YjiE and Preparation for Analysis
Expression was performed using BL21(DE3) cells. Cultures (1–6 liters) were inoculated and incubated overnight at 37 °C without shaking. Then, the cultures were shaken at 25 °C, isopropyl 1-thio-β-d-galactopyranoside was added after 45 min (0.1 mm final concentration), and cells were harvested after 4 h. Washed cells were resuspended in ice-cold buffer A (50 mm NaH2PO4, pH 7.5, 300 mm NaCl, 10 mm imidazole) containing Protease Inhibitor Mix HP (Serva), disrupted, and centrifuged (8 °C, 48,400 × g). All chromatography steps were performed at ambient temperature, and protein fractions were collected on ice. The lysate was applied onto a HisTrap FF column (GE Healthcare) and eluted with a linear gradient from buffer B (50 mm Tris, pH 7.5, 300 mm NaCl, 10 mm imidazole) to buffer C (buffer B with 750 mm imidazole). YjiE-containing fractions were diluted with buffer D (10 mm NaH2PO4, pH 7.5, 5 mm EDTA), immediately applied onto a Hi-Trap Q FF (GE Healthcare), and eluted with a linear gradient from buffer E (10 mm NaH2PO4, pH 7.5) to buffer F (buffer E with 600 mm NaCl). YjiE-His containing fractions were pooled and stored at −80 °C after addition of 5% (v/v) final concentration of glycerol. To remove the His tag, His-T-YjiE or His-T-YjiE-flash containing fractions after the Hi-Trap Q FF column were digested with thrombin (bovine, high activity (Calbiochem)) for 40 h at 4 °C. Thrombin was removed using a Superdex75 column (GE Healthcare) (buffer E with 400 mm NaCl). For storage at −80 °C, glycerol was added to a final concentration of 5% (v/v) (storage buffer B (9.5 mm NaH2PO4, pH 7.5, 380 mm NaCl, 5% glycerol)). Unless otherwise stated, YjiE was reduced with tris(2-carboxyethyl)phosphine (TCEP, 1 mm, 1 h, 37 °C), centrifuged (13,300 rpm, 4 °C, 20 min), and the protein concentration was determined at 280 nm using a NanoDrop spectrometer (PeqLab) prior to analysis. For purification of YjiE-HOCl (i.e. YjiE-His isolated from HOCl-treated cells), HOCl was added to cultures to a final concentration of 2 mm immediately before cells were harvested (10 min, 8 °C). Cells were then resuspended in buffer B containing Protease Inhibitor Mix HP, disrupted, and YjiE-HOCl purified and stored as described above for YjiE-His.
Sample Preparation for Analytical Ultracentrifugation
For the analysis of the oligomerization state of YjiE, the protein was analyzed at 15, 3, and 0.6 μm in storage buffer. YjiE-HOCl was analyzed at 12 μm in storage buffer. To analyze the dissociation of YjiE by DNA, 50 nm YjiE-flash* (see below) was incubated with 50 nm DNA (158 bp) in 10 mm NaH2PO4, pH 7.5, 375 mm NaCl. After 20 to 30 min at 25 °C, sedimentation velocity ultracentrifugation was carried out. To analyze binding of YjiE to different DNA, the protein was reduced with TCEP (1 mm, 2 h, 37 °C), and TCEP was removed using a NAP-5 column (elution in 2.5 mm NaH2PO4, pH 7.5, 100 mm NaCl). DNA-binding samples were prepared containing 30–50 nm DNA (yjiE, metN, cydA), different amounts of protein, and a final buffer concentration of 1 mm NaH2PO4, pH 7.5, 30 mm NaCl.
Analytical Ultracentrifugation (aUC)
aUC experiments were carried out using a ProteomeLab XL-A analytical ultracentrifuge (Beckman Coulter) equipped with a fluorescence detection system (Aviv) (24, 25). For sedimentation velocity experiments, samples were spun at 35,000 rpm or 42,000 rpm (monitoring fluorescence) at 20 °C. Scans were collected continuously during sedimentation recording absorbance at 235 and 280 nm for unlabeled protein or monitoring fluorescence at an excitation of 488 nm and emission cut off at 505 nm when YjiE-flash* or Alexa Fluor 488-labeled DNA should be detected.
Sedimentation velocity data were analyzed using the dC/dT method (25) and/or c(s) method provided with the SedFit software. For two-dimensional spectrum analysis (26), sedimentation profiles were analyzed at a grid resolution of 62,500 using 25 grid repetitions. Confidence levels for statistics were derived from two-dimensional spectrum analysis data refinement using genetic algorithm followed by 50 Monte Carlo simulations. Buffer density and viscosity were determined using a falling ball viscosimeter type C (Haake), and all aUC data were calculated based on the experimentally determined buffer parameters. The partial specific volume (vbar) of 0.737 ml/g for YjiE was calculated based on the amino acid sequence. Sedimentation equilibrium experiments were performed at 8,000 rpm at 4 °C with 15 μm YjiE-His. Absorbance scans at 280 nm and interference scans were recorded after the sample was at equilibrium. These data were analyzed by a single species fit. The calculation of the YjiE-DNA-complex stoichiometry is described in the supplemental data.
Sucrose Gradient Centrifugation
Preparation and analysis of sucrose gradients are described in the supplemental data.
Transmission Electron Microscopy and Image Processing
YjiE samples were adsorbed for 2 min onto carbon-coated grids that were glow discharged in air before the application of 5 μl of protein solution. YjiE alone and HOCl-treated YjiE were analyzed at 0.1 mg/ml and 0.05 mg/ml, respectively, in 10 mm NaH2PO4, pH 7.4, 200 mm NaCl. HOCl-treated YjiE (0.5 mg/ml (14 μm)) was incubated with a 10-fold molar ratio of HOCl (25 °C, 30 min) before HOCl was quenched by the addition of methionine. For DNA-binding experiments, TCEP-reduced (1 mm, 1 h, 37 °C) YjiE was diluted to a final concentration of 50 mm NaCl and incubated with an equimolar ratio of 315-bp yjiE DNA (final YjiE concentration, 0.018 mg/ml (504 nm)). Excess protein solution was blotted off, and the samples were stained negatively with uranyl acetate (5 μl 2% (w/v), 30 s). Electron micrographs were recorded at a nominal magnification of 50,000 and at defocus values ∼500 to 700 nm using a JEOL JEM 100CX electron microscope operated at 100 kV. Image preparation was performed as described (27). Centered images (1,500 for YjiE+DNA; 9,918 for YjiE alone; 8,320 for HOCl-treated YjiE) were analyzed by multivariate statistical analysis and multireference-alignments within the Imagic suite (28) to obtain the final class averages. Quantification, three-dimensional reconstruction, and the hydrodynamic simulations are described in the supplemental data.
FlAsH Labeling of YjiE in Vitro and in Vivo
AD2 cells were induced with 0.1% arabinose (30 °C, 4 h), which yielded YjiE levels ∼100 times higher than wild-type levels (final level ∼0.1% of the total cellular protein, as inferred from quantitative Western blot analysis using YjiE-specific antibodies) and was required to obtain a significant fluorescence signal. For preparation of the lysate, cell pellets were resuspended in 20 mm NaH2PO4, pH 7.5, 115 mm NaCl. Cells were disrupted using a bead mill (4-mm glass beads, frequency 30 s−1, 6 min). After centrifugation, the protein concentration in the soluble fraction was determined by Bradford assay.
Purified YjiE-flash and yjiE-flash expressing cell lysates were incubated with 1 mm TCEP and 0.4 μm FlAsH-EDT2 (Invitrogen) for 90 min (29) prior to analysis by aUC (see above) or sucrose gradient centrifugation (see supplemental data). Lysate (50–75 μg) was used for aUC monitoring fluorescence in a total volume of 450 μl. In vivo labeling was performed as described before (30) with minor modifications as follows using AD2 cells at an A600 of 0.5. Lysozyme-treated, washed cells were resuspended in LB medium supplemented with ampicillin, and 0.4 μm FlAsH-EDT2 and 2 μm ethanedithiol were added (37 °C, 30 min). Then, 0.1% arabinose was added to induce expression of yjiE-flash at 30 °C for 4 h, and cells were harvested as described above. For the in vivo HOCl treatment, cells were treated with 2 mm HOCl (5 min), and the reaction was quenched with LB medium directly before cell disruption. Lysate (120 μg) was used for aUC in a total volume of 300–450 μl.
Generation of DNA Fragments for Binding Studies
DNA fragments comprising the upstream region of yjiE, metN, or cydA were generated by PCR using the following primers: −263-bp yjiE (forward) and +52-bp yjiE (reverse) (315-bp DNA), or −106-bp yjiE (forward) metN or cydA and +52-bp yjiE, metN, or cydA (reverse) (158-bp DNA), respectively, and purified. Positions are given with respect to the translational start sites. To obtain single fluorescently labeled DNA fragments, the reverse primer was 5′-labeled with Alexa Fluor 488.
Fluorescence Anisotropy Measurements (FA)
FA was analyzed using a Jasco FP-6500 fluorescence spectrometer at 37 °C (excitation/emission at 488 nm/515 nm). YjiE was reduced as indicated (1 h, 37 °C, 10 mm TCEP, or 3 h, 37 °C, 10 mm DTT) prior to addition to DNA. Analysis was performed in the presence of reductants. YjiE-His was always analyzed in its non-reduced state (in vitro HOCl-treated, from unstressed cells, and from HOCl-treated cells). For in vitro oxidation, YjiE-His (0.5 mg/ml) was incubated with a 10-fold molar excess of HOCl (70 μm; 30 min, 25 °C) before HOCl was quenched by the addition of methionine (final concentration, 1 mm). Single Alexa Fluor 488-labeled 158-bp DNA at 10 nm (yjiE, metN, or cydA) was preincubated in 10 mm NaH2PO4, pH 7.5, 5% glycerol in a stirred cuvette for 5 min. Then, the respective YjiE variant was added in 100 or 200 nm steps to the DNA, and anisotropy was monitored for 60 s (data pitch, 2 s). The recorded values (20–50 s) were averaged.
RESULTS
Identification of Putative Transcription Factor That Confers Specific Resistance to HOCl Stress
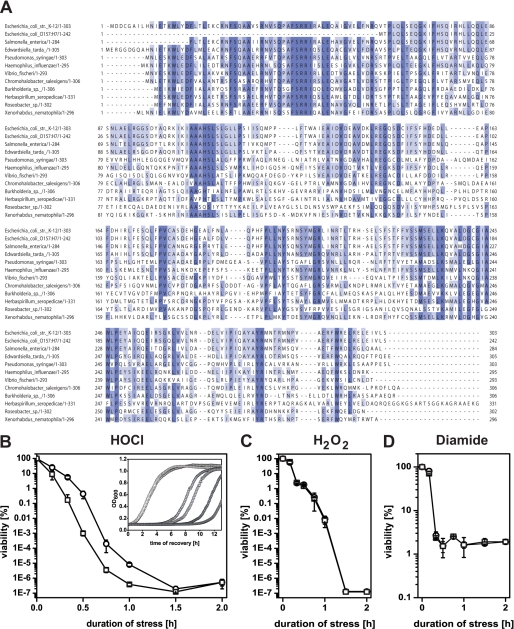
In an attempt to identify a potential cellular response system to HOCl, we screened a genomic expression library and selected clones that survived exposure to 6 mm HOCl in liquid LB medium (22). We identified one clone with a 150-fold increased number of surviving cells (5 mm HOCl) that contained the complete gene yjiE and parts of iadA and iraD, suggesting that yjiE is responsible for the observed HOCl resistance. Based on homology, yjiE encodes a putative LTTR and is conserved in α-, β-, and γ-proteobacteria and in eukaryotes (Xenopus tropicalis). Multiple sequence alignment showed that YjiE contains many conserved amino acids in the N-terminal DNA-binding domain and the C-terminal co-inducer response domain (Fig. 1A). To confirm that yjiE confers the HOCl-resistant phenotype of the originally selected library clone, we deleted yjiE in E. coli C600 (KMG214) and compared its survival after exposure to HOCl to the parental strain C600. Deletion of yjiE resulted in an up to 350-fold reduced viability upon HOCl stress and delayed recovery, resulting in an increased lag phase in the growth curve (Fig. 1B and inset). YjiE appears to act specifically during HOCl stress and does not confer resistance to other ROS such as H2O2 and diamide (Fig. 1, C and D). We conclude that YjiE improves cell viability and growth upon HOCl stress.
FIGURE 1.
A, sequence alignment of YjiE from 10 representative species from α-, β-, and γ-proteobacteria and eukaryotes using ClustalW. Conserved amino acids are highlighted in blue, and the degree of conservation is proportional to the darkness of the color. B–D, growth of C600 (WT, circles) and KMG214 cells (yjiE−, squares) at different stresses. Cells were grown at 37 °C, supplemented with the indicated stressor, and either viability or recovery analyzed. Viability at 4 mm HOCl (B), 4 mm H2O2 (C), and 5 mm diamide (D), respectively. Shown is the mean ± error from two to six independent experiments. Inset, recovery of cells at 0 mm (open symbols), 2.8 mm (light gray symbols), and 3 mm HOCl (dark gray symbols). Shown is the result of one representative experiment.
What is the function of YjiE upon HOCl stress? To obtain insight into the potential regulatory role of YjiE and to assess how YjiE protects cells from HOCl stress, we performed gene expression analyses of yjiE+ and yjiE− cells (C600, KMG214) in response to HOCl treatment. Under the conditions chosen the viability of cells was 100% for all tested HOCl concentrations. Considering that most LTTRs autoregulate their expression (16), we compared RNA levels of YjiE in unstressed and HOCl stressed cells. No significant change in RNA levels was observed, and this was confirmed by qRT-PCR (data not shown). Likewise, translational fusions of the putative yjiE promoter region with lacZ yielded similar β-galactosidase activity in unstressed and HOCl-stressed yjiE+ and yjiE− cells (supplemental Fig. 2). This suggests that yjiE transcription is not altered by HOCl stress and that YjiE does not up-regulate its own expression, unlike other LTTRs (16).
Only 11 genes had slightly different RNA levels (2.1–3.5-fold up/down) under non-stress conditions in yjiE− cells compared with yjiE+ cells. Upon HOCl treatment, however, >300 genes were altered at the transcript level by at least 2-fold in response to HOCl in yjiE+ or yjiE− cells. Some of these genes showed stronger regulation in one of both strains, and some were exclusively regulated in yjiE+ cells but remained unaffected by HOCl in yjiE− cells. These genes are supposedly regulated by YjiE (supplemental Tables S3 and S4). Up-regulated genes can mainly functionally be classified into genes involved in sulfur, methionine, and cysteine metabolism (metB, metK, metN, cysH, cysK, cysN, cysPUW, sbp, sufA), biosynthesis of other amino acid, and genes with unknown function. Down-regulated genes can mainly be classified into genes involved in iron acquisition and homeostasis (entC, entH, fecABCDE, fecR, fepCD, ryhB, tonB, yncE; and with a much lower amplitude entEBA, ybiX (1.5 to 2-fold change)) as well as transporter (sugar, peptides, amino acids) and RNA-related genes.
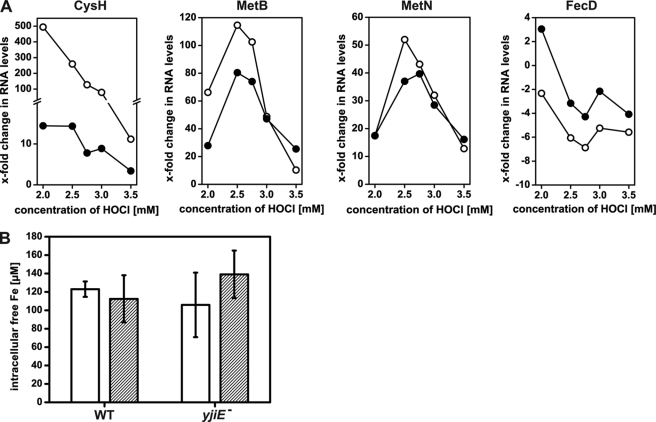
To confirm the regulatory effect of YjiE during HOCl stress, we monitored the levels of selected RNAs (CysH, MetB, MetN, FecD) by qRT-PCR. All of these genes showed an YjiE-dependent transcription over a HOCl concentration range from 2–3.5 mm with an optimum at 2.5–2.75 mm HOCl (Fig. 2A). (Differences in absolute values in the gene expression analysis and the qRT-PCR analysis are most likely due to a different experimental setup.) It should be noted that the viability of cells was 100% for all HOCl concentrations tested. Thus, the qRT-PCR data support the gene expression analysis.
FIGURE 2.
A, qRT-PCR analysis of the RNA levels of CysH, MetB, MetN, and FecD at the indicated concentration of HOCl. WT (open symbols) and yjiE− cells (closed symbols) were grown at 37 °C and supplemented with HOCl. After 10 min, 5× concentrated LB was added to quench HOCl, cells were harvested, RNA was isolated, and the RNA levels were analyzed by qRT-PCR using specific primer pairs. Shown is the result of one representative experiment. Please note that CysH, MetB, and MetN are up-regulated, whereas FecD is down-regulated. B, analysis of intracellular free iron levels in WT and yjiE− cells. Cells were cultivated in LB medium at 37 °C without stress (white bars) or subjected to stress (2.75 mm HOCl, 10 min; striped bars) and prepared for EPR analysis. Shown is the mean ± error from two to four independent experiments.
The up-regulation of cysteine and methionine biosynthesis may reflect the requirement to replenish methionine, which is quickly oxidized upon HOCl stress (5) and thus possibly depleted. The potential role of genes with unknown functions is yet to be analyzed. Genes that were down-regulated are involved mainly in iron homeostasis. They are regulated by Fur, a transcriptional regulator and repressor that is activated by iron (31). Dukan and Touati (4) showed that fur mutants, which have higher intracellular iron levels because they lack down-regulation of iron acquisition, are more sensitive to HOCl stress. This suggests that E. coli cells with higher intracellular iron levels suffer from increased ROS generation via the Fenton reaction (4). Thus, the primary role of YjiE may be to down-regulate iron acquisition genes to prevent further synthesis of iron uptake proteins.
To test whether YjiE influences iron levels, we analyzed intracellular free iron levels in yjiE+ and yjiE− cells in the absence or presence of HOCl stress. Cells were treated with 2.75 mm HOCl at which the levels of FecD were most strongly affected as shown by qRT-PCR (see Fig. 2A). The unincorporated free iron levels were similar in untreated yjiE+ and yjiE− cells as expected because YjiE is inactive in the absence of HOCl. Unincorporated free iron levels were also similar in HOCl-stressed yjiE+ and yjiE− cells (Fig. 2B). Such similar levels in HOCl-stressed and unstressed cells suggest that HOCl does not particularly damage iron-containing proteins. Significant iron release from proteins may, however, occur depending on the applied HOCl concentration as shown in vitro for lactoperoxidase (32). Therefore, a direct effect of YjiE on intracellular unincorporated iron levels cannot be observed because HOCl stress does not increase iron levels. Thus, the role of YjiE-regulated genes upon HOCl stress is apparently multifaceted, and they may contribute collectively to cell survival.
YjiE Forms Unusually Large Oligomers
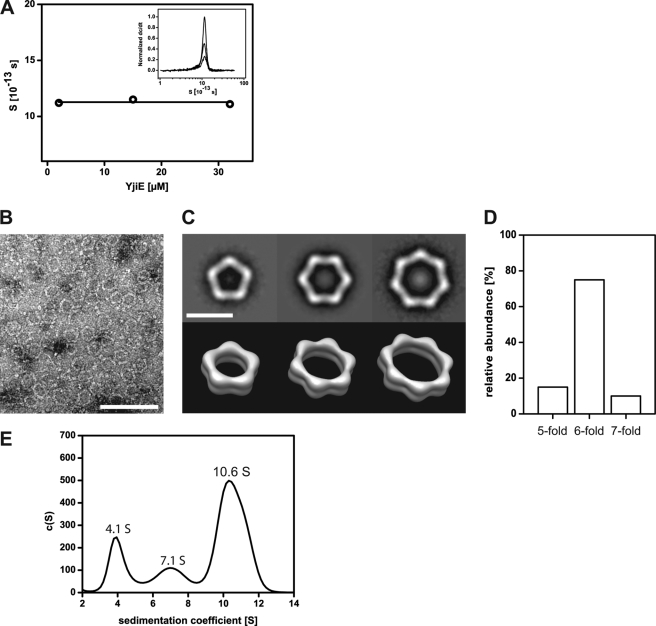
Many LTTRs form dimers or tetramers (16). We thus analyzed the oligomerization state of purified YjiE by aUC. In sedimentation equilibrium experiments, we determined a molecular mass of ∼420 kDa for C-terminally His-tagged (YjiE-His) and untagged YjiE, suggesting that it forms a dodecamer under equilibrium conditions (YjiE monomer, 35 kDa). In sedimentation velocity experiments, YjiE sedimented with an s value of 11.1 S ± 0.3 S independent of the protein concentration within the tested range of 0.6–15 μm (Fig. 3A). In agreement with the sedimentation equilibrium experiments, two-dimensional spectrum analysis yielded a molecular mass of 395 ± 10 kDa for the 11.1 S particles and a frictional ratio of f/f0 = 1.66, suggesting that the YjiE oligomer adopts an elongated shape in solution (data not shown). (Note that an f/f0 value of 1 is expected for a spherical particle.)
FIGURE 3.
A, concentration-dependent analysis of the sedimentation coefficient of YjiE. The indicated concentrations of YjiE in storage buffer were analyzed by aUC. Inset, sedimentation coefficient distribution of 15, 3, and 0.6 μm YjiE using the dC/dT method. Sedimentation was performed at 35,000 rpm. B, negative stain transmission electron microscopy image of YjiE (0.1 mg/ml) negatively stained with uranyl acetate. Scale bar, 100 nm. C, typical class averages of the pentagonal, hexagonal, and heptagonal YjiE oligomers (upper panel) and corresponding surface representations of the simulated three-dimensional structures (lower panel). Scale bar, 20 nm. D, distribution of oligomers shown in C. E, YjiE-flash that was FlAsH-labeled in lysates was analyzed by aUC, and fluorescence was monitored. The 4.1 S, 7.1 S, and 10.6 S YjiE particles likely correspond to dimers, tetramers, and dodecamers.
To obtain further structural information about YjiE, we subjected the samples to negative stain transmission electron microscopy. YjiE adsorbed onto carbon-coated grids and stained with uranyl acetate mainly formed ring-like pentagonal, hexagonal, and heptagonal structures with hexagons being the predominant assembly form (Fig. 3, B–D). The hexagons have a diameter of 24 nm with a segment length and width of ∼11 and 3 nm, respectively. The height of all oligomers is ∼6 nm, as indicated by three-dimensional reconstructions (Fig. 3C, lower panel). According to hydrodynamic simulations, the s values of the corresponding three-dimensional volumes are between 10.4 and 11.9 S and fit well the observed mean s of ∼11 S (supplemental Fig. 1A).
YjiE in Lysates Forms Small and Large Oligomeric Species
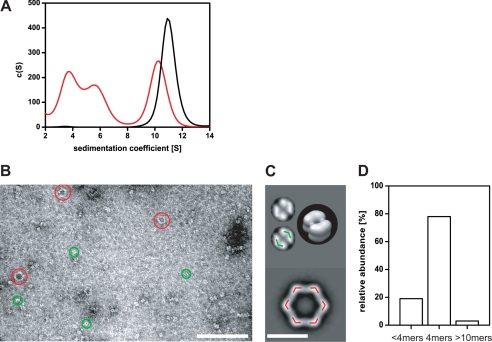
To test whether YjiE also forms atypical large complexes in vivo, we fused a CCGPCC tag (flash tag), which is known to specifically react with the biarsenical fluoresceine analog FlAsH forming strongly fluorescent adducts (29), to the C terminus of YjiE (yielding YjiE-flash). YjiE-flash showed a wild-type-like oligomerization and DNA binding (see below) and conferred wild-type-like HOCl resistance to ΔyjiE cells (data not shown). We then labeled YjiE-flash with the FlAsH dye in vitro, in vivo, and in lysates to form fluorescent YjiE-flash (YjiE-flash*) and analyzed the lysates by sucrose gradient centrifugation (supplemental Fig. 1C). YjiE-flash* was detected in two fractions, in which it was distributed equally, and co-migrated with proteins of molecular weights between 140–380 kDa. A smaller fraction migrated at ∼70 kDa, presumably dimers (supplemental Fig. 1C, fractions 16/18). We then analyzed YjiE-flash* by aUC monitoring fluorescence. Purified YjiE-flash* showed a wild-type-like s value of 10.9 S (see Fig. 5A, black line). To detect YjiE-flash* fluorescence in cell lysates, yjiE-flash had to be expressed to ∼0.1% of the total cellular protein (∼100-fold greater than wild-type levels). Labeling was YjiE-specific as a lysate of FlAsH-labeled cells that did not express yjiE-flash showed no significant fluorescence (supplemental Fig. 1B, gray line). YjiE-flash* sedimented predominantly with 10.6 S (Fig. 3E and supplemental Fig. 1B, black line), demonstrating that YjiE dodecamers form in cell lysates. As in sucrose gradient centrifugation, smaller YjiE particles (4.1 and 7.1 S) were detected in addition to the 11 S particles. Two-dimensional spectrum analysis and hydrodynamic estimations from the oligomeric three-dimensional structures (see supplemental Fig. 1A) estimate that the 4.1 S particles correspond to dimers, whereas the 7.1 S particles are likely to be tetramers (Fig. 3E). The ratio of small to large oligomers depended on the cellular concentration of YjiE. Smaller oligomers (i.e. dimers, tetramers) comprised the most abundant species in wild-type E. coli cells, whereas large oligomers (i.e. dodecamers) were detected predominantly when yjiE was overexpressed as used for purification purposes (as inferred from analytical gel filtration of cell lysates; data not shown). These results indicate that the YjiE oligomers are in an equilibrium governed by the cellular concentration of YjiE.
FIGURE 5.
A, 50 nm yjiE DNA and 50 nm YjiE-flash* (at 400 mm NaCl) were analyzed by aUC monitoring fluorescence. Dodecameric YjiE-flash* alone (black line) sedimented with ∼11 S. YjiE-flash* dissociated into smaller oligomers with 3.7 S and 5.5 S in the presence of DNA (red line), most likely representing dimers and tetramers. B, electron microscopic image of YjiE (0.018 mg/ml, 50 mm NaCl) that was incubated with an equimolar ratio of 315 bp yjiE DNA and stained with uranyl acetate. Please note large oligomers (red) and smaller oligomers (green). Scale bar, 100 nm. C, upper panel: typical class average (top view) of YjiE tetramers shown in B. The green overlay indicates the positions of YjiE dimers of an YjiE tetramer. Lower panel, red overlay indicating the positions of YjiE dimers on the top view class average of an YjiE dodecamer. Inset, surface representation of the simulated tetrahedral three-dimensional structure. Scale bar, 10 nm. D, distribution of oligomers shown in B.
YjiE Binds to DNA
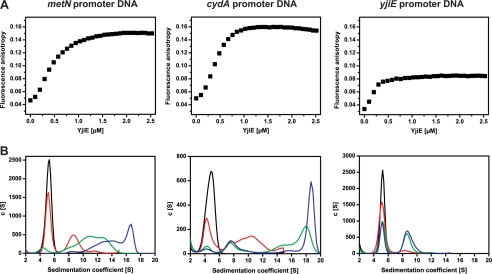
DNA binding is a prerequisite for the activity of YjiE as an LTTR and occurs independent of activation (16). To test for DNA binding, we mixed purified YjiE with fluorescent labeled DNA (Alexa Fluor 488 DNA) and subsequently analyzed the samples by FA and aUC monitoring fluorescence. As double-stranded DNA substrates, we chose the promoter regions of two of the identified target genes, cydA and metN (158 bp in length, comprising the 106-bp upstream region of the respective gene) and the yjiE promoter region in our analysis. First, we titrated YjiE to Alexa Fluor 488-labeled DNA and monitored changes in FA. Changes in FA were observed with all three DNA substrates and followed a sigmoidal binding curve with half-maximal amplitudes at ∼0.3 μm YjiE (Fig. 4A). The change in FA could be reverted by addition of excess unlabeled YjiE promoter DNA, demonstrating that binding of YjiE to DNA is dynamic and reversible (supplemental Fig. 1D). To corroborate further evidence for the interaction between YjiE and the double-stranded DNA substrates, we subjected the samples to aUC and monitored the sedimentation of Alexa Fluor 488 DNA in the absence and presence of YjiE. Alexa Fluor 488 DNA sedimented with 5.1 S. Because we monitored DNA-fluorescence, any shift in the s value reports on the interaction of YjiE with the DNA. Upon titration of YjiE to DNA, the amount of free DNA decreased, and we observed a shift toward larger s values, demonstrating the formation of DNA-YjiE complexes. YjiE interacted with all three DNA substrates; however, the YjiE-yjiE DNA complex (8.7 S) was significantly smaller compared with complexes between YjiE and cydA and metN DNAs (∼16–20 S) (Fig. 4, A and B). This is in agreement with the yjiE DNA yielding the smallest FA amplitude. The 8.7 S complex (Fig. 4B) fits to an YjiE-dimer bound to DNA (according to two-dimensional spectrum analysis and simulations). 8 S complexes were also populated in metN and cydA samples at intermediate YjiE concentrations, whereas the larger ∼16–20 S complexes were populated at high YjiE concentrations, suggesting that target DNA might be bound by multiple YjiE molecules (presumably dimers). To exclude aggregate formation, we subjected these samples to SDS-PAGE analysis and found that YjiE was soluble after fractionation by centrifugation (see Fig. 6A, inset, lanes 1 and 2). We conclude that YjiE exhibits the ability to distinguish between target DNA and nonspecific DNA (i.e. yjiE DNA) and forms distinct DNA-YjiE complexes.
FIGURE 4.
A, binding of reduced YjiE to Alexa Fluor 488 DNA was monitored by FA. TCEP-reduced YjiE at the indicated concentrations was added stepwise to 10 nm of the indicated Alexa Fluor 488 DNA in 100 nm steps. The change in FA upon titration of YjiE was measured using a Jasco FP-6500 fluorescence spectrometer. B, binding of YjiE to Alexa Fluor 488 DNA was analyzed by aUC, and fluorescence was monitored. Reduced YjiE was added to the indicated DNA at a ratio of 10:1 (red), 40:1 (green), and 80:1 (blue) for metN and cydA DNA and 10:1 (red), 24:1 (green), and 48:1 (blue) for yjiE DNA (at 30 mm NaCl).
FIGURE 6.
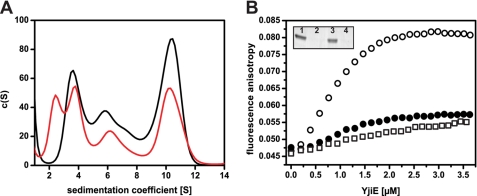
A, YjiE in HOCl-stressed cells forms a higher proportion of small oligomers compared with YjiE in non-stressed cells. yjiE-flash expressing cells were in vivo FlAsH-labeled, and cultures were split and either left untreated (black line) or HOCl-stressed (2 mm, 5 min; red line) immediately before cell lysis. Then, lysates were analyzed by aUC monitoring fluorescence. B, YjiE-His purified from HOCl-stressed cells (open circles) shows a higher DNA-binding capacity than YjiE-His purified from non-stressed cells (filled circles) or in vitro HOCl-treated YjiE-His (HOCl:YjiE 10:1; squares). Binding of YjiE to Alexa Fluor 488 metN DNA was monitored by FA. YjiE protein was not reduced prior to analysis and was added in 200 nm steps to 10 nm DNA. The change in FA was measured using a Jasco FP-6500 fluorescence spectrometer. Inset, samples after FA analysis were spun, and the supernatant (1 and 3) and the pellet (2 and 4) were analyzed by SDS-PAGE. Samples correspond to YjiE from non-stressed cells (1 and 2) and YjiE from HOCl-stressed cells (3 and 4).
YjiE Dissociates in Presence of DNA
In vitro, free YjiE sediments with ∼11 S, but the DNA-YjiE complexes at a given DNA:YjiE ratio sedimented with smaller s values. We also detected smaller oligomers in vivo (see above), suggesting that the dodecamer dissociates into smaller particles and that this process might be DNA-driven. To visualize changes in YjiE oligomerization, we monitored the sedimentation of YjiE-flash* in the absence and presence of unlabeled DNA. YjiE-flash* (50 nm) sedimented with ∼11 S in the absence of DNA. In the presence of equimolar concentration of DNA, 3.7 and 5.5 S particles occurred in addition to the 11 S particles (Fig. 5A), indicating dodecamer dissociation into dimers and tetramers in the presence of DNA substrates.
The presence of considerably smaller oligomers was confirmed by analyzing samples containing YjiE and DNA by negative stain transmission electron microscopy (Fig. 5, B–D). Smaller oligomers (i.e. ∼80%) assembled into small, ring-like structures, assumedly tetramers, with mean diameters of ∼11 nm (Fig. 5, B–D). An overlay of the tetramer with the dodecameric structure elements shows that the basic building block of the dodecamer is likely to be a dimer (see colored substructures in Fig. 5C; see also supplemental Fig. 1A).
YjiE Forms Predominantly Smaller Oligomers in HOCl-stressed E. coli Cells
Given that YjiE dissociates in the presence of DNA and to test whether and how YjiE is activated in vivo, the oligomerization state of YjiE was analyzed in lysates prepared from HOCl-stressed E. coli cells. To this end, E. coli cells were labeled with FlAsH prior to the induction of yjiE-flash expression (i.e. in vivo labeling). (Please note that yjiE-flash was overexpressed, yielding ∼100 times the wild-type levels.) Shortly before cell lysis, cells were either treated with HOCl or left untreated, and then the lysate prepared and analyzed by aUC. This ensured low and very similar YjiE levels under both conditions. YjiE-flash* showed 3.8 S, 5.9 S, and 10.5 S particles in the absence of HOCl, which correspond to dimers, tetramers, and dodecamers (Fig. 6A, black line). Similar species were observed after HOCl stress, but their distribution was shifted toward the smaller oligomers (Fig. 6A, red line). An additional species was observed with 2.6 S, likely corresponding to a monomer (see supplemental Fig. 1A). These small oligomers may represent the physiologically relevant species, whereas the dodecamer may be a storage form. Considering that YjiE-flash levels were ∼100 times the wild-type levels, and the oligomer distribution depends on the cellular YjiE concentration, it seems reasonable that the effect of HOCl on YjiE would be even stronger pronounced in wild-type cells.
YjiE Is Not Directly Activated upon HOCl Treatment in Vitro
LTTRs are typically regulated by binding of and/or modification by co-inducer molecules. To this end, we tested whether the activation of the protective function of YjiE to E. coli cells is by direct interaction of HOCl with YjiE. First, purified YjiE was incubated with a 5–10-fold molar excess of HOCl, and its secondary and quaternary structure was examined for conformational changes. A 10-fold molar excess of HOCl was found sufficient to activate the redox-regulated chaperone Hsp33 and to cause major unfolding of the chaperone GrpE (8). In the case of YjiE, four of the five cysteines of YjiE were oxidized as opposed to one to two oxidized cysteines in untreated YjiE, which is due to air oxidation (as inferred from Ellman's assay). However, neither the secondary structure and thermal stability nor the dodecameric ring-like conformation of YjiE was altered significantly as shown by circular dichroism, aUC, and negative stain transmission electron microscopy (supplemental Fig. 3, A–D). Also, the ability of HOCl-treated YjiE to interact with DNA was similar to the untreated protein (Fig. 6B, squares and filled circles). (Note that samples were not reduced prior to analysis.) indicating that neither binding of HOCl to YjiE nor oxidation of cysteine residues per se are sufficient to activate YjiE in vitro. Similarly, purified YjiE that was treated with reaction products of HOCl with iron (to generate hydroxyl radicals) and ammoniumchloride or Tris (to generate chloramines) showed a low DNA-binding activity (data not shown).
Next, we overexpressed and purified YjiE from HOCl-stressed cells (YjiE-HOCl). Similar to YjiE purified from unstressed cells, YjiE-HOCl formed a dodecamer in solution (data not shown) and exhibits a secondary structure that is very similar to untreated YjiE, indicating that no major structural and conformational changes occurred (data not shown). In sharp contrast to purified YjiE that was HOCl-treated in vitro, YjiE-HOCl showed a high change in anisotropy that was significantly larger than that of untreated YjiE (Fig. 6B, filled circles). (Note that samples were not reduced prior to analysis.) This indicates formation of large DNA-YjiE complexes and is suggestive for YjiE activation in HOCl-stressed E. coli cells. It is noteworthy to add that neither YjiE nor YjiE-HOCl showed any aggregation in the time course of the experiments, thus indicating that the detected changes in FA did probably not arise from aggregate formation (Fig. 6B, inset).
DISCUSSION
ROS damage macromolecules and cells, therefore making it pivotal for cells to employ response systems that are specifically activated by the ROS inflicting stress. Here, we identified E. coli YjiE as the first transcription factor specifically protecting cells from HOCl-derived killing. YjiE was described recently as cell density-dependent motility repressor, QseD (33). However, in this work, we did not observe an effect of YjiE on the motility of various E. coli strains (data not shown). YjiE is specific for HOCl and not activated upon H2O2 or diamide stress. This makes YjiE a very specific defense molecule for strong oxidative stress as HOCl is much more reactive toward amino acids than those ROS and causes oxidative protein unfolding (9, 34). In contrast to the specific activation of YjiE, a recent transcriptome study in Bacillus subtilis identified the regulator OhrR to be inactivated by HOCl. In this case, HOCl causes S-bacillithiolation of OhrR thus inactivating its repressor function and concomitantly up-regulating ohrA, which encodes a peroxiredoxin (35). Considering that OhrR is also inactivated by organic hydroperoxides leading to S-cysteinylation (36), this suggests that OhrR does not specifically respond to HOCl per se but is responsive to reactive species causing oxidation of cysteines to sulfenic acid that can form mixed disulfides.
YjiE appears to be activated in HOCl-stressed cells but not in vitro by HOCl directly. This may suggest that activation of YjiE is caused by some reaction product of HOCl with a cellular component or by subtle conformational changes that are sufficient for increased DNA binding but could not be detected by CD spectroscopy. Also, YjiE seems to be exclusively regulated at the level of activity. As the activation of already existing YjiE molecules is much faster than the synthesis of new YjiE, YjiE should be able to regulate the transcription of specific genes before cells are irreversibly damaged by HOCl. This fits well to the kinetics of action of HOCl, which damages cells within seconds (37) and activates the molecular chaperone Hsp33 quickly (9).
Most LTTRs are believed to form tetramers from a dimer of dimers (16). YjiE is an oligomerically versatile molecule. We observed dimers-tetramers-dodecamers with large oligomers dissociating to smaller ones in the presence of DNA. Binding of YjiE to DNA occurs very quickly (see supplemental Fig. 1D) with the DNA binding competent small oligomers being potentially quickly removed from the small oligomer-dodecamer equilibrium, thus leading to dissociation. This has not been observed for a transcription factor before and may be unique for YjiE. The advantage may be that once the cellular YjiE level exceeds a certain threshold, YjiE molecules form a dodecameric storage form that can dissociate and become activated quickly on demand. Yet, the main building block of YjiE is likely a dimer, similar to other LTTRs. Dimers may associate to tetramers that apparently represent the DNA-binding state in the case of YjiE, with their ratio being determined by the cellular YjiE level or the availability of target genes. Based on the observation that YjiE showed similar affinity but forms different complexes with nonspecific or target DNA, it appears that the regulation of transcriptional activity of target genes may predominantly involve acquisition of the right amount of YjiE molecules into the functional complex. However, the exact mechanism of YjiE activation needs yet to be unraveled.
Similar to the other E. coli oxidative stress transcription factors OxyR and SoxR, YjiE is specifically activated by a ROS. Regulation of many genes is also similar to OxyR and SoxR because the inflicted damage is multifaceted, yet it is unusual for LTTRs that typically regulate only few genes (16). For SoxR and OxyR, the corresponding gene products decrease the extent of intracellular oxidative stress (e.g. catalase, superoxide dismutase) and repair damaged macromolecules (15, 38). YjiE differs from OxyR and SoxR as it does not regulate genes that are apparently involved in the degradation of ROS. Whether the genes with unknown function play a central role in the protection of E. coli from HOCl stress needs to be analyzed. The main focus of the response orchestrated by YjiE appears to generally decrease the intracellular load of ROS by replenishing methionine and cysteine and down-regulating Fur-regulated genes involved in iron acquisition. OxyR and SoxR activate fur expression upon H2O2 or superoxide (i.e. redox-cycling drugs (14)) treatment by ∼10-fold, leading to a 2-fold increase in Fur protein (39). Although Fur-RNA levels were only slightly increased upon HOCl stress (as inferred from gene expression analysis and qRT-PCR; data not shown), this indicates that the control of iron homeostasis in E. coli is an integral part of the antioxidant defense response. In the case of HOCl stress, some but not all Fur-controlled genes are affected, suggesting some kind of fine-tuning of iron homeostasis that is facilitated by YjiE. Whether YjiE regulates genes directly or maybe in cooperation with another regulator is unclear as of now and needs to be analyzed.
In vitro and in vivo, HOCl can undergo Fenton reaction in the presence of iron generating cytotoxic hydroxyl radicals that cause cell damage (3, 4). One could speculate that HOCl-induced oxidative protein unfolding (9) damages iron-containing proteins, thereby increasing free iron levels and requiring immediate regulation of iron homeostasis genes. However, such increased iron levels after HOCl stress are not evident from our analysis, in contrast to the increase observed upon H2O2 stress (40). Even if HOCl stress increased iron levels, YjiE potentially does not directly affect intracellular free iron levels, considering that it acts as a repressor of iron acquisition genes. Repression is a slow-acting process; even though transcription and protein synthesis are repressed, cellular protein levels will remain similar until the respective protein is degraded or diluted out by cell growth.
To summarize, we identified the first, to our knowledge, bacterial transcription factor that specifically protects E. coli cells from HOCl stress but not peroxide or diamide stress. YjiE is present in proteobacteria and eukaryotes, suggesting that YjiE-mediated protection from HOCl stress arose early in evolution and is maintained throughout the kingdoms.
Supplementary Material
Acknowledgments
We thank Bernd Bukau, Barry Wanner, and Regine Hengge for strains/plasmids and Ursula Jakob, Johannes Buchner, Ryan Frisch, Robert Bender, Stefan Gleiter, Hauke Lilie, Jirka Peschek, and Sevil Weinkauf for discussions. James Imlay is gratefully acknowledged for discussions and comments on the manuscript, and Yuanyuan Liu is acknowledged for technical assistance. EPR analysis was performed at the Illinois EPR Research Center. Calculations were performed on the UltraScan LIMS cluster (Bioinformatics Core Facility, University of Texas Health Science Center, San Antonio), the Lonestar cluster (Texas Advanced Computing Center), and the National Supercomputer HLRB-II at the Leibnitz-Rechenzentrum Munich, Germany.
This work was supported by project pr28ci (to Johannes Buchner and T. M. F.), the Elitenetzwerk Bayern (to K. M. G., T. M. F., and J. G.), the Boehringer Ingelheim Fonds (to Y. L.), and the Emmy-Noether program of the Deutsche Forschungsgemeinschaft (to J. W.).

This article contains supplemental “Experimental Procedures,” Tables S1–S4, Figs. 1–3, and additional references.
- ROS
- reactive oxygen species
- aUC
- analytical ultracentrifugation
- FA
- fluorescence anisotropy
- LTTR
- LysR-type transcriptional regulator
- HOCl
- sodium hypochlorite
- qRT-PCR
- quantitative real-time PCR
- TCEP
- tris(2-carboxyethyl)phosphine.
REFERENCES
- 1. Ha E. M., Oh C. T., Bae Y. S., Lee W. J. (2005) A direct role for dual oxidase in Drosophila gut immunity. Science 310, 847–850 [DOI] [PubMed] [Google Scholar]
- 2. Winterbourn C. C., Hampton M. B., Livesey J. H., Kettle A. J. (2006) Modeling the reactions of superoxide and myeloperoxidase in the neutrophil phagosome: Implications for microbial killing. J. Biol. Chem. 281, 39860–39869 [DOI] [PubMed] [Google Scholar]
- 3. Candeias L. P., Stratford M. R., Wardman P. (1994) Formation of hydroxyl radicals on reaction of hypochlorous acid with ferrocyanide, a model iron(II) complex. Free Radic. Res. 20, 241–249 [DOI] [PubMed] [Google Scholar]
- 4. Dukan S., Touati D. (1996) Hypochlorous acid stress in Escherichia coli: Resistance, DNA damage, and comparison with hydrogen peroxide stress. J. Bacteriol. 178, 6145–6150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Davies M. J. (2005) The oxidative environment and protein damage. Biochim. Biophys. Acta 1703, 93–109 [DOI] [PubMed] [Google Scholar]
- 6. Imlay J. A. (2003) Pathways of oxidative damage. Annu. Rev. Microbiol. 57, 395–418 [DOI] [PubMed] [Google Scholar]
- 7. Dukan S., Dadon S., Smulski D. R., Belkin S. (1996) Hypochlorous acid activates the heat shock and soxRS systems of Escherichia coli. Appl. Environ. Microbiol. 62, 4003–4008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang S., Deng K., Zaremba S., Deng X., Lin C., Wang Q., Tortorello M. L., Zhang W. (2009) Transcriptomic response of Escherichia coli O157:H7 to oxidative stress. Appl. Environ. Microbiol. 75, 6110–6123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Winter J., Ilbert M., Graf P. C., Ozcelik D., Jakob U. (2008) Bleach activates a redox-regulated chaperone by oxidative protein unfolding. Cell 135, 691–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rosen H., Klebanoff S. J., Wang Y., Brot N., Heinecke J. W., Fu X. (2009) Methionine oxidation contributes to bacterial killing by the myeloperoxidase system of neutrophils. Proc. Natl. Acad. Sci. U.S.A. 106, 18686–18691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jakob U., Muse W., Eser M., Bardwell J. C. (1999) Chaperone activity with a redox switch. Cell 96, 341–352 [DOI] [PubMed] [Google Scholar]
- 12. Winter J., Linke K., Jatzek A., Jakob U. (2005) Severe oxidative stress causes inactivation of DnaK and activation of the redox-regulated chaperone Hsp33. Mol. Cell 17, 381–392 [DOI] [PubMed] [Google Scholar]
- 13. Mongkolsuk S., Helmann J. D. (2002) Regulation of inducible peroxide stress responses. Mol. Microbiol. 45, 9–15 [DOI] [PubMed] [Google Scholar]
- 14. Gu M., Imlay J. A. (2011) The SoxRS response of Escherichia coli is directly activated by redox-cycling drugs rather than by superoxide. Mol. Microbiol. 79, 1136–1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zheng M., Wang X., Templeton L. J., Smulski D. R., LaRossa R. A., Storz G. (2001) DNA microarray-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide. J. Bacteriol. 183, 4562–4570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Maddocks S. E., Oyston P. C. (2008) Structure and function of the LysR-type transcriptional regulator (LTTR) family proteins. Microbiology 154, 3609–3623 [DOI] [PubMed] [Google Scholar]
- 17. Choi H., Kim S., Mukhopadhyay P., Cho S., Woo J., Storz G., Ryu S. (2001) Structural basis of the redox switch in the OxyR transcription factor. Cell 105, 103–113 [DOI] [PubMed] [Google Scholar]
- 18. Lochowska A., Iwanicka-Nowicka R., Plochocka D., Hryniewicz M. M. (2001) Functional dissection of the LysR-type CysB transcriptional regulator. Regions important for DNA binding, inducer response, oligomerization, and positive control. J. Biol. Chem. 276, 2098–2107 [DOI] [PubMed] [Google Scholar]
- 19. Monferrer D., Tralau T., Kertesz M. A., Dix I., Sola M., Uson I. (2010) Structural studies on the full-length LysR-type regulator TsaR from Comamonas testosteroni T-2 reveal a novel open conformation of the tetrameric LTTR fold. Mol. Microbiol. 75, 1199–1214 [DOI] [PubMed] [Google Scholar]
- 20. Bender R. A. (2010) A NAC for regulating metabolism: The nitrogen assimilation control protein (NAC) from Klebsiella pneumoniae. J. Bacteriol. 192, 4801–4811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sainsbury S., Lane L. A., Ren J., Gilbert R. J., Saunders N. J., Robinson C. V., Stuart D. I., Owens R. J. (2009) The structure of CrgA from Neisseria meningitidis reveals a new octameric assembly state for LysR transcriptional regulators. Nucleic Acids Res. 37, 4545–4558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Barth E., Gora K. V., Gebendorfer K. M., Settele F., Jakob U., Winter J. (2009) Interplay of cellular cAMP levels, σS activity, and oxidative stress resistance in Escherichia coli. Microbiology 155, 1680–1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liu Y., Bauer S. C., Imlay J. A. (2011) The YaaA protein of the Escherichia coli OxyR regulon lessens hydrogen peroxide toxicity by diminishing the amount of intracellular unincorporated iron. J. Bacteriol. 193, 2186–2196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chen J., Feige M. J., Franzmann T. M., Bepperling A., Buchner J. (2010) Regions outside the α-crystallin domain of the small heat shock protein Hsp26 are required for its dimerization. J. Mol. Biol. 398, 122–131 [DOI] [PubMed] [Google Scholar]
- 25. MacGregor I. K., Anderson A. L., Laue T. M. (2004) Fluorescence detection for the XLI analytical ultracentrifuge. Biophys. Chem. 108, 165–185 [DOI] [PubMed] [Google Scholar]
- 26. Brookes E., Cao W., Demeler B. (2010) A two-dimensional spectrum analysis for sedimentation velocity experiments of mixtures with heterogeneity in molecular weight and shape. Eur. Biophys. J. 39, 405–414 [DOI] [PubMed] [Google Scholar]
- 27. Peschek J., Braun N., Franzmann T. M., Georgalis Y., Haslbeck M., Weinkauf S., Buchner J. (2009) The eye lens chaperone α-crystallin forms defined globular assemblies. Proc. Natl. Acad. Sci. U.S.A. 106, 13272–13277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. van Heel M., Harauz G., Orlova E. V., Schmidt R., Schatz M. (1996) A new generation of the IMAGIC image processing system. J. Struct. Biol. 116, 17–24 [DOI] [PubMed] [Google Scholar]
- 29. Griffin B. A., Adams S. R., Tsien R. Y. (1998) Specific covalent labeling of recombinant protein molecules inside live cells. Science 281, 269–272 [DOI] [PubMed] [Google Scholar]
- 30. Ignatova Z., Gierasch L. M. (2008) Chapter 3: A fluorescent window into protein folding and aggregation in cells. Methods Cell Biol. 89, 59–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. McHugh J. P., Rodríguez-Quinoñes F., Abdul-Tehrani H., Svistunenko D. A., Poole R. K., Cooper C. E., Andrews S. C. (2003) Global iron-dependent gene regulation in Escherichia coli. A new mechanism for iron homeostasis. J. Biol. Chem. 278, 29478–29486 [DOI] [PubMed] [Google Scholar]
- 32. Souza C. E., Maitra D., Saed G. M., Diamond M. P., Moura A. A., Pennathur S., Abu-Soud H. M. (2011) Hypochlorous acid-induced heme degradation from lactoperoxidase as a novel mechanism of free iron release and tissue injury in inflammatory diseases. PLoS One 6, e27641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Habdas B. J., Smart J., Kaper J. B., Sperandio V. (2010) The LysR-type transcriptional regulator QseD alters type three secretion in enterohemorrhagic Escherichia coli and motility in K-12 E. coli. J. Bacteriol. 192, 3699–3712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pattison D. I., Davies M. J. (2001) Absolute rate constants for the reaction of hypochlorous acid with protein side chains and peptide bonds. Chem. Res. Toxicol. 14, 1453–1464 [DOI] [PubMed] [Google Scholar]
- 35. Chi B. K., Gronau K., Mader U., Hessling B., Becher D., Antelmann H. (2011) Mol. Cell Proteomics 10, M111.009506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lee J. W., Soonsanga S., Helmann J. D. (2007) A complex thiolate switch regulates the Bacillus subtilis organic peroxide sensor OhrR. Proc. Natl. Acad. Sci. U.S.A. 104, 8743–8748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Barrette W. C., Jr., Albrich J. M., Hurst J. K. (1987) Hypochlorous acid-promoted loss of metabolic energy in Escherichia coli. Infect. Immun. 55, 2518–2525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Blanchard J. L., Wholey W. Y., Conlon E. M., Pomposiello P. J. (2007) Rapid changes in gene expression dynamics in response to superoxide reveal SoxRS-dependent and independent transcriptional networks. PLoS One. 2, e1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zheng M., Doan B., Schneider T. D., Storz G. (1999) OxyR and SoxRS regulation of fur. J. Bacteriol. 181, 4639–4643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jang S., Imlay J. A. (2007) Micromolar intracellular hydrogen peroxide disrupts metabolism by damaging iron-sulfur enzymes. J. Biol. Chem. 282, 929–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.