Background: Class I and II diterpene synthases, although poorly understood, generate diverse products.
Results: Reported here is the structure of the bifunctional abietadiene synthase and supporting experimental/computational work.
Conclusion: Visualization of the class I and II active sites confirms known and implicates new determinants of product formation.
Significance: Residues, previously unrecognized, are assigned specific roles in substrate binding and catalysis.
Keywords: Enzyme Catalysis, Enzyme Mechanisms, Enzyme Structure, Plant Biochemistry, Terpenoids, Terpene Synthases
Abstract
Abietadiene synthase from Abies grandis (AgAS) is a model system for diterpene synthase activity, catalyzing class I (ionization-initiated) and class II (protonation-initiated) cyclization reactions. Reported here is the crystal structure of AgAS at 2.3 Å resolution and molecular dynamics simulations of that structure with and without active site ligands. AgAS has three domains (α, β, and γ). The class I active site is within the C-terminal α domain, and the class II active site is between the N-terminal γ and β domains. The domain organization resembles that of monofunctional diterpene synthases and is consistent with proposed evolutionary origins of terpene synthases. Molecular dynamics simulations were carried out to determine the effect of substrate binding on enzymatic structure. Although such studies of the class I active site do lead to an enclosed substrate-Mg2+ complex similar to that observed in crystal structures of related plant enzymes, it does not enforce a single substrate conformation consistent with the known product stereochemistry. Simulations of the class II active site were more informative, with observation of a well ordered external loop migration. This “loop-in” conformation not only limits solvent access but also greatly increases the number of conformational states accessible to the substrate while destabilizing the nonproductive substrate conformation present in the “loop-out” conformation. Moreover, these conformational changes at the class II active site drive the substrate toward the proposed transition state. Docked substrate complexes were further assessed with regard to the effects of site-directed mutations on class I and II activities.
Introduction
Terpenoids form the largest group of natural products, with some 50,000 known (1). Underlying the observed diversity are the manifold hydrocarbon skeletal backbones produced by the cyclization and/or rearrangement of acyclic precursors catalyzed by the relevant terpene synthases (2). They are first subdivided based on the number of five-carbon isoprenoid repeats; with the ten-carbon monoterpenoids generally derived from geranyl diphosphate, the fifteen-carbon sesquiterpenoids from farnesyl diphosphate, the twenty-carbon diterpenoids from geranylgeranyl diphosphate (GGPP),3 and the thirty-carbon triterpenoids from squalene (3).
As might be suspected from the composition of their precursors, the skeletal backbone structures of the lower (mono-, sesqui-, and di-) terpenes are generally produced by cyclization and/or rearrangement reactions initiated by ionization of the substrate allylic diphosphate ester bond, whereas triterpenes are cyclized via a protonation-initiated reaction (i.e. of a terminal C=C double bond or the oxido-ring of the derived oxido-squalene). The corresponding enzymes have been termed classes I and II, respectively (4). Notably, biosynthesis of the large superfamily of labdane-related diterpenoids (∼7,000 known) combines both types of reactions (5), in which GGPP is bicyclized by a class II diterpene cyclase (EC 5.5.1.x) prior to further cyclization and/or rearrangement catalyzed by a class I diterpene synthase (EC 4.2.3.x).
The labdane-related diterpenoids include many important plant natural products, such as the gibberellin phytohormones and others that act in defense (e.g. the ubiquitous conifer resin acids (6)). Indeed, it has been suggested that the diterpene synthases involved in gibberellin biosynthesis are the ancestral progenitors to the extensive family of lower (i.e. class I) terpene synthases found in plants (7). This is possible because, while catalyzing mechanistically distinct reactions, the corresponding class II ent-copalyl diphosphate synthase and subsequently acting class I ent-kaurene synthase are clearly homologous. Nevertheless, the vast majority of the plant lower terpene synthases are significantly smaller than these diterpene synthases (8).
Structural characterization of plant and microbial lower terpene synthases demonstrated that the class I reaction is catalyzed in a conserved α-helical bundle domain (9–18) that has been termed the α domain (19). This includes the placement of two highly conserved acidic motifs, DDXXD and (N/D)DXX(S/T)XXXE (hereafter called NTE), which coordinate the trio of Mg2+ required for catalysis (2, 20). However, the plant enzymes seem to invariably contain an additional N-terminal domain that appears to be relictual. In particular, this clearly is derived from the ancestral diterpene synthases, although these enzymes further contain an additional N-terminal large sequence element as well (8), with these domains termed β and γ, respectively (19).
The β domain in class II diterpene cyclases contains a DXDD motif required for such catalysis (21, 22). This invokes similarities to the squalene-hopene cyclase that also catalyzes such energetically difficult C=C protonation-initiated cyclization using an analogous motif. Furthermore, structural homology of this β domain (4), including modeled positioning (23) and the catalytic role (24) of the DXDD motifs, has been found between these seemingly unrelated but mechanistically similar enzymes. Conservation of the additional γ domain in class II diterpene cyclases has been used to argue that this also might be involved in class II catalysis (25), drawing further parallels to the triterpene cyclases, which are composed of a bidomain γβ structure wherein the active site is located between these domains (26, 27).
These observations have led to the hypothesis that the γβα tridomain structure of eukaryotic diterpene synthases originated from functional fusion of separate, smaller class II (γβ domain) and I (α domain) enzymes, specifically those found in bacteria (28). Consistent with this hypothesis is the presence of bifunctional diterpene synthases in fungi (29, 30), as well as gymnosperms and lower plants (31–34), which further have been suggested to closely resemble the ancestral plant terpene synthase (7, 35).
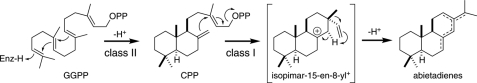
One such bifunctional enzyme is abietadiene synthase from the gymnosperm Abies grandis (AgAS), which is involved in grand fir tree resin acid biosynthesis (Fig. 1). AgAS was the first bifunctional diterpene synthase to be cloned (31) and has been suggested to closely resemble the ancestral (di)terpene synthase (7, 8). In addition, its enzymatic activities have been subjected to extensive mechanistic investigation, including labeling and reaction intermediate analog binding studies (21, 36–39), as well as mutational analysis of catalysis (20, 23, 40, 41), and identification of single residue switches for both product outcome (42) and biochemical regulation via susceptibility to Mg2+-dependent inhibition (43). Here we report the crystal structure of AgAS at 2.3 Å resolution and use it along with molecular dynamics simulations to correct and clarify previous hypotheses, via derived site-directed mutagenesis, regarding the structure-function relationships underlying both class I and II reactions.
FIGURE 1.
Class II and class I reactions catalyzed by AgAS. Isopimar-15-en-8-yl+ is a key intermediate of the class I reaction, with formation of a mixture of allylic double-bond isomers (38). Subsequent oxidation reactions transform the resulting abietadienes into resin acids (6).
EXPERIMENTAL PROCEDURES
Cloning, Expression, and Purification of AgAS
For crystallization the Δ84 pseudo-mature form of AgAS was recombinantly expressed using a previously described pSBET construct (38), typically in the C41 OverExpress strain of Escherichia coli (Lucigen) grown in NZY medium. For selenomethionine labeling, AgAS was expressed in B834(DE3) methionine auxotrophic E. coli (Novagen) grown in selenomethionine medium base plus nutrient mix (Molecular Dimensions) with the addition of l-selenomethionine (Fisher). AgAS was then purified much as previously described (38). Briefly, clarified bacterial lysates were initially fractionated over type II ceramic hydroxyapatite, and AgAS was purified over Mono Q and type II ceramic hydroxyapatite again using a BioLogic LP system (Bio-Rad), before a final polishing purification step over a Mono Q column on an ÄKTAfplc system (GE Healthcare). The resulting AgAS (>98% pure by SDS-PAGE) was then dialyzed against protein storage buffer (10 mm Bis-Tris, pH 6.8, 10% (v/v) glycerol, 150 mm KCl, 10 mm MgCl2, and 5 mm DTT), following which it could be stored at −80 °C for several months without significant loss of activity.
Crystallization of AgAS
Initial crystallization trials were carried out at the Hauptman-Woodward Institute. Promising conditions were replicated in our lab by the hanging drop vapor diffusion methods at both 4 and 18 °C. Typically, a 2-μl drop of AgAS in storage buffer was mixed with a 1.6-μl drop of precipitant solution (24% (v/v) PEG 8000, 0.1 m sodium citrate, pH 5.1, 0.1 m dibasic ammonium phosphate), as well as with 0.4 μl of additional 0.1 m l-proline or 0.1 m phenol from an additive screen (Hampton Research), and equilibrated against 1 ml of precipitant solution in the well. Plate-shaped crystals with maximal dimensions of 0.3 × 0.3 × 0.05 mm appeared within a week. Crystals were briefly transferred to a cryoprotectant solution (25% PEG 8000, 0.1 m sodium citrate, pH 5.1, 0.1 m dibasic ammonium phosphate, 10% (v/v) glycerol) and then flash-frozen. One selenomethionine labeled crystal (crystal I) was soaked in 5.5 mm tantalum bromide (Ta6Br122+; Jena Bioscience) prior to flash-freezing. Although this derivative was not successfully used for phasing, it appears to have altered the crystal lattice (from P21 to P21212), which facilitated localization of the selenium for phasing. This crystal, which diffracted to 2.5 Å resolution, belonged to space group P21212 with unit cell dimensions a = 100.14 Å, b = 190.175 Å, and c = 92.932 Å (two molecules in the asymmetric unit). The data also were collected for another selenomethionine-labeled crystal (crystal II), which diffracted to 2.3 Å, and belonged to space group P21 with unit cell parameters a = 92.84 Å, b = 189.09 Å, and c = 99.90 Å, β = 91.45° (four molecules in the asymmetric unit).
X-ray Diffraction Data Collection, Processing, Phasing, and Model Refinement
X-ray diffraction data were collected at the Advanced Photon Source, that for crystal I on Beamline 24-ID-D, and that for crystal II on Beamline 19-ID, using λ = 0.9792 Å, corresponding to the absorption peak of selenium. All of the diffraction data were indexed and merged using HKL-3000 (44). Selenium sites in crystal I (located by the Solve routine in Phenix (45)), led to the automatic generation of several peptide fragments by Autosol. A complete model (Rfree = ∼0.29, resolution of 2.8 Å) for crystal I resulted from cycles of model building (Xtalview) (46) and restrained refinement by Refmac (47) and CNS (48). The model from crystal I enabled a molecular replacement structure determination of crystal II using Phaser (49). CNS refinement of the structure from crystal II was refined against data to 2.3 Å resolution using standard restraints on stereochemistry and thermal parameters, and auxiliary restraints on donor-acceptor backbone distances in regions of regular secondary structure. Ramachandran plot statistics came from PROCHECK (50). Atomic coordinates and crystallographic structure factors are in the Protein Data Bank. The figures of protein structure were generated using PyMOL.
Site-directed Mutagenesis and Analysis
AgAS R356A, N451A, and V727T mutants were constructed via overlapping PCR of previously described AgAS pENTR (Invitrogen) constructs (51), either wild type (all three) or an existing D621A mutant (R356A and N451A only). The latter double mutants (AgAS:R356A/D621A and AgAS:N451A/D621A) enabled separate analysis of class II activity, as the D621A mutation selectively abrogates AgAS class I activity (21). The resulting pENTR mutant constructs were verified by complete gene sequencing and then transferred to pDEST expression vectors by directional recombination, specifically to pDEST17 for expression as His6-tagged fusion proteins. The product outcome mediated by the single mutants was characterized by co-expression with a GGPP synthase in a previously described modular metabolic engineering system (51). The R356A and N451A mutants also were expressed, purified, and kinetically characterized, as previously described, separately analyzing either class II (AgAS:R356A/D621A and AgAS:N451A/D621A) (43) or class I (AgAS:R356A and AgAS:N451A) (42) activity.
Molecular Dynamics Simulations and Structural Comparisons
Normal and accelerated molecular dynamics simulations (52, 53) employed NAMD (54), version 2.8, using the CHARMM27 force field (55). Topology and parameter files for the class II active site substrate GGPP and the class I active site substrate copalyl diphosphate (CPP) came from the ParamChem web server (56). The system was chain B of the AgAS crystal structure reported here, immersed in a rectangular TIP3P water box (57) with a 15 Å buffer. Sodium chloride to a concentration of 0.1 molar balanced electrostatic charge associated with the protein. Periodic boundary conditions were applied, and the Particle-mesh Ewald algorithm (58) was used for the calculation of long range electrostatic interactions. The time step was 2 fs with the SHAKE algorithm (59) applied. The cutoff for nonbonded van der Waals interactions was 12 Å. For accelerated molecular dynamics simulations, the threshold energy E and acceleration factor α were estimated as E = V + 4*N and α = 4*N/5, respectively, where V is the average dihedral potential from a normal molecular dynamics simulation, and N is the number of residues in protein. For every simulation, a 100-picosecond minimization step with gradient temperature increasing and a 100-picosecond equilibrium step with NPT condition preceded the productive 10-ns run. Results from molecular dynamics simulations are analyzed primarily with VMD (60) and carma (61). Root mean square deviation-based structural clustering was done according to Ref. 62 with a cutoff of 1 Å.
RESULTS
Crystal Structure of AgAS (Protein Data Bank Code 3S9V)
The initial 84 amino acid residues coded by the gene for AgAS represent a plastid-targeting segment not present in the mature protein. Hence, by the convention established in the literature, the first residue (methionine) is enumerated as 84 for the truncated recombinant protein used here in the structure determination.
Statistics of data collection and refinement of the AgAS structure are in Table 1. The four monomers of AgAS in the asymmetric unit are nearly identical, exhibiting root mean square deviations of 0.25 Å in pairwise superpositions of Cα atoms. Electron density for residues 84–109 is absent for each monomer, although Edman sequencing (performed by the Iowa State University Protein Facility) indicated a mixture of polypeptides beginning with residues 84 or 85. Electron density for residues 846–849 is absent in chains A and D, but present, albeit weakly, in chains B and C. All of the monomers have electron density in the class II active site cleft, which may represent a tripeptide (it is not a component of the crystallization buffer), but this is not included in the reported coordinates, nor in further analyses.
TABLE 1.
Statistics of data collection and refinement
| Crystal I | Crystal II | |
|---|---|---|
| Data collection | ||
| Wavelength (selenium peak) | 0.9793 | 0.9792 |
| Resolution range (Å) | 50.00–2.50 | 50.00–2.30 |
| No. of reflections (total/unique) | 271,627/61,432 | 474,752/148,465 |
| Completeness (%)a | 99.2 (98.4) | 97.7 (96.6) |
| I/σa | 25.1 (2.4) | 18.5 (3.3) |
| Rmergea,b | 0.104 (0.739) | 0.092 (0.360) |
| FOM for SADc | 0.46 | |
| Refinement | ||
| Rwork/Rfreed | 0.208/0.287 | 0.196/0.250 |
| Protein atomse | 24,815 | |
| Solvent atomse | 1346 | |
| Root mean square deviations | ||
| Bonds (Å) | 0.008 | 0.006 |
| Angles (°) | 1.19 | 1.10 |
| Average B factors (Å2) | ||
| Protein chains | 34.3 | |
| Solvent | 35.3 | |
| Ramachandran plot | ||
| Allowed (%) | 91.5 | |
| Additionally allowed (%) | 8.4 | |
| Generously allowed (%) | 0.1 | |
| Disallowed (%) | 0 | |
a Numbers in parentheses refer to the outer 0.04 Å shell of data.
b Rmerge = Σ|I − <I>|/ΣI, where I is the observed intensity and <I> is the average intensity calculated from replicate data.
c FOM, figure of merit; SAD, single wavelength anomalous dispersion.
d Rwork = Σ||Fo| − |Fc||/Σ|Fo| for reflections contained in the working set, where |Fo | and |Fc bar] are the observed and calculated structure factor amplitudes, respectively, and Rfree is calculated in the same way for reflections contained in the test set held aside during refinement.
e Per asymmetric unit.
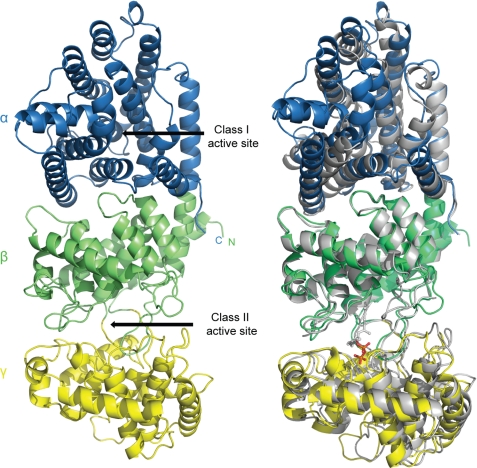
AgAS has three helical domains (termed γβα; Fig. 2), consistent with prediction (19), and resembling recently reported structures of the class I taxadiene synthase from Taxus brevifolia (TbTS) (63) and class II ent-copalyl diphosphate synthase from Arabidopsis thaliana (AtCPS) (64). AgAS differs from TbTS and AtCPS in having functional class I and class II active sites rather than a single active site. Putative active sites in AgAS are clefts that include relevant catalytic motifs and correspond to binding sites for substrate analogs in the TbTS and AtCPS structures. The absence of a channel connecting the two actives sites in AgAS is consistent with the diffusive transfer of the CPP intermediate (21).
FIGURE 2.
AgAS structure. Left panel, α (residues 559–868), β (residues 110–133, 350–558), and γ (residues 134–349) domains are blue, green, and yellow, respectively. Right panel, superposition of AgAS (colored as in the left panel) onto AtCPS (gray). The pyrophosphate moiety of the substrate analog bound to the class II active site of AtCPS is red.
For the most part, the γ-, β-, and α-domains include residues from the first, middle, and last thirds of the amino acid sequence, respectively; however, the N-terminal segment is part of all three domains. Lys86 and Arg87, for instance, are important for class I, but not class II, activity (40), indicating that some of the first 26 residues (electron density absent in the AgAS structure) are part of the α-domain. Moreover, the N-terminal segment also makes up the first helix of the β-domain, before the γ-domain begins. Hence, similar to the triterpene cyclases (26, 27), as well as TbTS and AtCPS (63, 64), the AgAS γ-domain falls between the first and second helices of the β-domain.
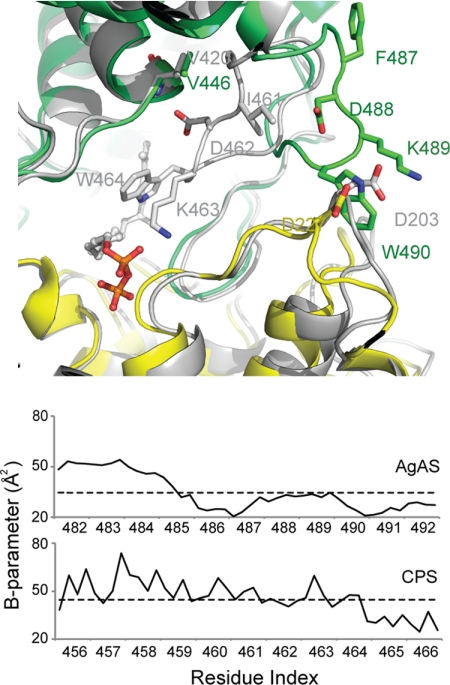
The AgAS class II active site exhibits a conformation distinct from other class II systems, specifically loop 482–492 is at the exterior of the protein (hereafter the loop-out conformation), whereas in ligated complexes of AtCPS (64) the corresponding loop penetrates deeply into the active site crevice (the loop-in conformer) (Fig. 3). Despite significantly different environments for conserved residues, the respective loop in the AgAS and AtCPS structures are well ordered, with thermal parameters at or below average values for their respective structures. Residues 402–405, corresponding to the class II DXDD catalytic motif, also are well ordered and in strong electron density.
FIGURE 3.
Comparison of AgAS and AtCPS class II active sites. Top panel, the class II active sites define the in conformation of loop 456–466 of AtCPS (gray) and the out conformation of loop 482–492 of AgAS (colored as in Fig. 2). Bottom panel, B-parameters of loop 482–492 in AgAS and the corresponding loop 456–466 in AtCPS are near or below the average value (dotted line) for each structure.
The class I active site of AgAS has no bound ligand. Residues 846–849, which have weak or absent electron density in each of the four AgAS molecules in the asymmetric unit, correspond to a loop that covers ligated active sites of other class I structures. Residues belonging to the class I catalytic motifs (DDXXD, residues 621–625, and NTE, residues 765–773) are ordered and well placed in electron density.
Mutagenesis of AgAS
The identification of the class II active site of AgAS rests largely on similarities in sequence and structure of related systems (26, 63–66), e.g. the location of the class II DXDD catalytic motif. In class II diterpene and triterpene cyclases, the “middle” aspartic acid of this motif acts as the general acid (24, 65). In the C=C protonating squalene-hopene cyclase, this aspartic acid is activated by a basic residue, conserved as histidine or arginine (26, 65, 66). In the mechanistically similar class II diterpene cyclases, this position is not conserved as a basic residue, rather than as an asparagine, leading to the suggestion that another basic residue might serve to activate the general acid, which in AgAS by sequence and structural similarity is Asp404 (43). However, in the AgAS crystal structure, it is the conserved asparagine (Asn451) that forms a hydrogen bond with Asp404, and an analogous interaction also has been observed in AtCPS (64). To determine the relevance of this interaction to catalytic activity, Asn451 was mutated to alanine, and the resulting AgAS:N451A mutant was kinetically characterized. Consistent with a role in the class II reaction, this mutation reduces class II activity ∼100-fold, with essentially no effect on class I activity, indicating that there is no loss of overall structural integrity (Table 2). Although there also appears to be some effect on substrate binding as well as catalytic activity, this is complicated by the weakened substrate inhibition observed with AgAS:N451A relative to the wild-type enzyme.
TABLE 2.
Kinetic analysis
| Class II activitya |
Class I activity |
|||
|---|---|---|---|---|
| Km | kcat | Km | kcat | |
| μm | s−1 | μm | s−1 | |
| WT | 0.5 ± 0.1 | 1.1 ± 0.1 | 2.0 ± 0.1 | 2.6 ± 0.1 |
| R356A | 6 ± 3 | 1.1 × 10−3 | 0.7 ± 0.1 | 1.1 ± 0.6 |
| N451A | 6 ± 4 | 0.016 | 1.4 ± 0.3 | 1.9 ± 0.1 |
a Determined in D621A background to abrogate class I activity.
Although we had originally speculated that Arg356, in agreement with the conservation pattern in squalene-hopene cyclase, activates the catalytic aspartic acid in class II diterpene cyclases (43), Arg356 is not part of the AgAS class II active site, protruding outward from the protein opposite the active site cleft. Nonetheless, Arg356 forms hydrogen bonds with residues that cross over from the γ- to β-domain and could conceivably influence the size of the active site crevice. Our original hypothesis that the basic residues at this position would be directly involved in class II catalysis was largely based on the >1,000-fold reduction in activity observed with mutation of the corresponding histidine in AtCPS to alanine (43). To verify a similar effect in AgAS, alanine was substituted for Arg356, which also led to a ∼1,000-fold decrease in class II catalytic activity, with a relatively small loss of class I catalytic activity again indicative of retention of overall structural integrity (Table 2). Consistent with the hypothesis that GGPP substrate inhibition is synergistic with inhibitory Mg2+ binding (67) and a role for Arg356 in this (43), AgAS:R356A exhibited no substrate inhibition.
A single-residue switch that substantially alters product outcome was first identified in ent-kaurene synthases, which react with enantiomeric (ent-)CPP (68). In short, replacement of an ethyl group with a hydroxyl (a threonine for isoleucine substitution) redirects ent-kaurene synthases to form pimaradienes (68). A similar effect has been observed in AgAS, where substitution of Ala723 with serine also leads to pimaradiene production. However, this position is four residues N-terminal to that corresponding to the Ile → Thr product switching position in ent-kaurene synthases (42), which is a valine in AgAS. The Val727 side chain also protrudes into the class I active site, so to investigate the effect on product outcome of hydroxyl group introduction at this position, we substituted in threonine. This AgAS:V727T mutant enzyme produces the same mixture of abietadiene double-bond isomers as wild-type AgAS, with no significant increase in pimaradiene content.
Molecular Dynamics Simulations: Class II Active Site
Given the large change in conformation of loop 482–492 in our AgAS crystal structure (loop-out) relative to that of the corresponding loop in the substrate analog containing AtCPS crystal structure (loop-in), we were interested in the role of this loop. In particular, we wanted to discover whether the loop-in conformer is accessible in the absence of substrate and what the effect of the loop-in conformer is on the class II active site. This was examined by selected molecular dynamics simulations of our AgAS structure.
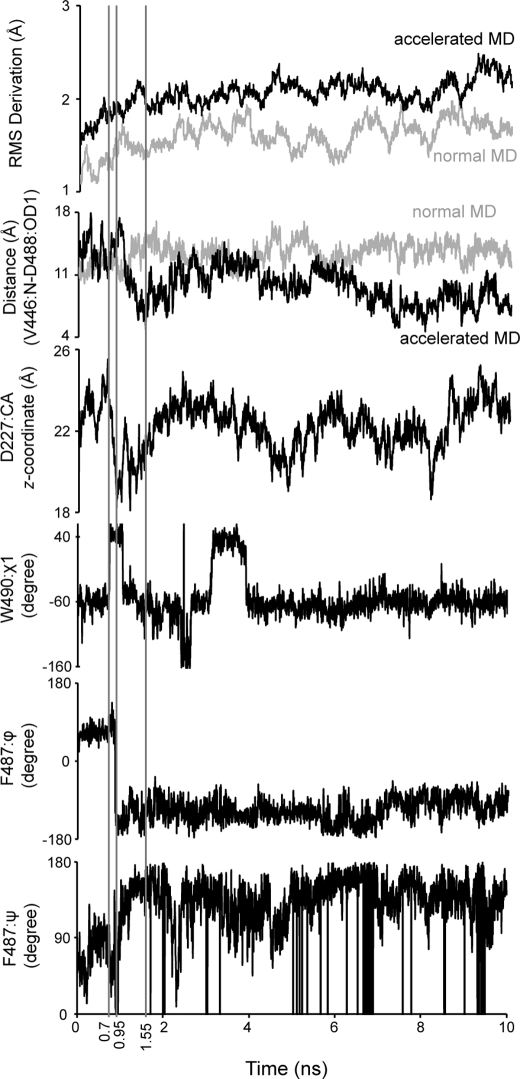
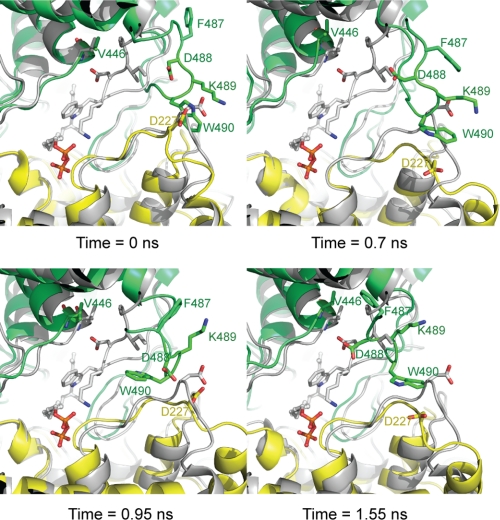
Loop 482–492 remains in its starting (loop-out) conformation in normal molecular dynamics simulations. Asp227 (γ-domain) is in contact with Trp490 (β-domain) and blocks loop 482–492 from entering the active-site crevice (Fig. 3). In accelerated simulations, however, loop 482–492 transiently enters the active-site bringing Asp488 in proximity to Val446 and the N terminus of helix 446–457 (Figs. 4 and 5). Note that the lone hydrogen bond between this loop and the rest of the protein in the AtCPS loop-in structure is between the side chain of Asp462 and the backbone amide of Val420, residues that correspond to Asp488 and Val446 of AgAS, respectively. Phi and psi angles of Phe487 undergo abrupt and significant change during the simulation, indicating points of articulation. In addition, movement of Asp227 correlates strongly with that of loop 482–492. Hence, Asp227 appears to act as a gate in providing loop 482–492 access to the active site.
FIGURE 4.
Selected parameters of the molecular dynamics simulation of ligand-free AgAS as a function of time. The parameters illustrated from top to bottom are the root mean square (RMS) deviation of all atoms, distance between the backbone amide nitrogen of Val446 and atom OD1 of Asp488, variation in the z-coordinate of the Cα atom of Asp227, variation in the χ1 angle of Trp490, variation of ϕ-angle of Phe487, and variation in ψ-angle of Phe487.
FIGURE 5.
Movement of loop 482–492. Snapshots of loop 481–492 (colored as in Fig. 2) at the indicated times during the simulation. The AtCPS structure (gray) defines the loop-in conformation.
The binding locus for the pyrophosphate moiety of the substrate (as suggested by the bound analog in the AtCPS structure) is near residues 223–225, suggesting a substrate-induced displacement of Asp227 that enables movement of loop 482–492 into the active site. Alternatively, as the accelerated simulation reveals movement toward the loop-in conformation in the absence of substrate, the two conformational states of loop 482–492 may exist at equilibrium. The loop-in conformer, however, puts Lys489 proximal to the pyrophosphate binding locus. Hence, substrate binding could shift the equilibrium population in favor of the loop-in conformer.
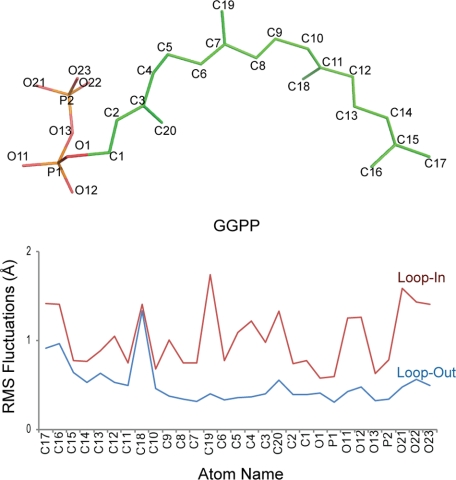
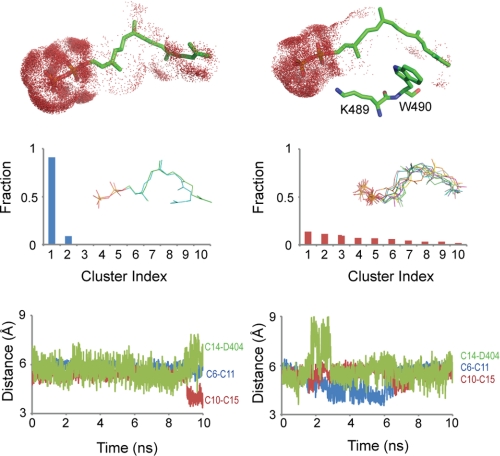
Normal simulations of the AgAS enzyme-substrate complex employ the substrate (GGPP) with loop 482–492 in its out- and in-conformations. (The loop-in conformation of AgAS was modeled after the Cα coordinates of the corresponding loop of the AtCPS structure and the starting conformation of GGPP after the coordinates of the substrate analog bound to the class II active site of the same structure). Atoms of the substrate exhibit low root mean square fluctuations in the loop-out conformer and high fluctuations in the loop-in conformer (Fig. 6). The isoprenoid moiety of the substrate, however, is hydrated to a greater extent in the loop-out relative to the loop-in conformation (Fig. 7). Cluster analysis reveals one dominant conformational type for the substrate in the loop-out simulation but many conformational types of equal weight in the loop-in simulation. Moreover, distances between the atoms that participate in covalent bond formation or proton transfer during the catalyzed reaction approach those expected for the occurrence of such chemistry among the loop-in conformers but are largely absent among loop-out conformers (Fig. 7). Given that the starting coordinates for the GGPP substrate were derived from the analog bound in the AtCPS crystal structure, which is not appropriately positioned for cyclization, this was a striking result that we further investigated.
FIGURE 6.
Atom names and atom fluctuation in GGPP during simulations of loop-in and loop-out conformations of AgAS. Top panel, carbon atom numbering of GGPP is provided. Bottom panel, root mean square fluctuations of atoms of GGPP in the loop-in simulation (red) are greater than the loop-out simulation (blue). Note that these are grouped by proximity, rather than numbering.
FIGURE 7.
Hydration and conformational variations of GGPP during simulations. Results from the loop-out simulation are in the left panels, and those from the loop-in simulation are in the right panels. Top panels, the distribution of water molecules (red dots) within 3 Å of GGPP show higher levels of hydration of the terminal isoprenoid unit in the loop-out relative to the loop-in conformation. The positions of Lys489 and Trp490 relative to GGPP are shown for the loop-in conformer. Middle panels, GGPP conformers during the loop-out simulation fall in two clusters, whereas during the loop-in simulation conformers distribute equally over multiple clusters. Bottom panels, distances of C6–C11, C10–C15, and C14–Asp404:OD1 are plotted against simulation time for loop-out and loop-in conformers. The atom pairs that define the distances in the bottom panel either form covalent bonds (C6–C11 and C10–C15) or participate in proton transfer (C14–Asp404). The loop-in simulation fosters a higher frequency of atom contacts consistent with the transition state, although at no point during the simulation were all three distances simultaneously less than 4 Å.
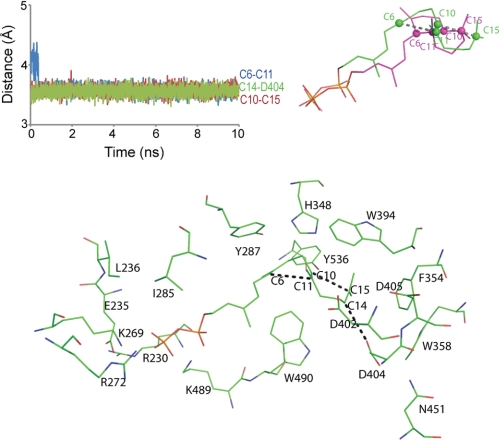
As a means of equilibrating the active site about the putative transition state, force restraints were applied to pairs of atoms that undergo covalent bond formation or proton transfer (Fig. 8). The docked transition state complex reveals contacts between planar isoprenoid groups and the side chains of Tyr287 and Phe354, with the side chains of Tyr287 and/or Tyr536 in position to accept a proton from atom C19 after, or in concert with the cyclization reaction. Lys269, Lys489, and Arg272 in the model interact with the pyrophosphate moiety.
FIGURE 8.
Model of the productive complex for AgAS-GGPP from a restrained molecular dynamics simulation. Plots of distances C6–C11, C10–C15, and C14–Asp404:OD1 are plotted against time (top left panel). The final model from the simulation is aligned with the product of the reaction (top right panel). Atoms C6, C10, C11, and C15 are represented by spheres. Shown is the protein in the vicinity of GGPP in its productive conformation (bottom panel).
Molecular Dynamics Simulations: Class I Active Site
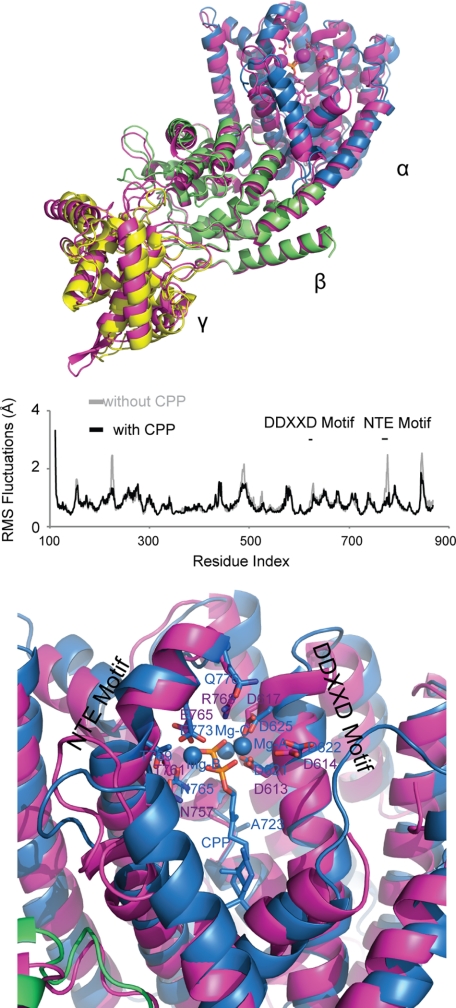
The TbTS complex with a substrate analog (FGG) (63) was the basis for modeling the CPP substrate in the class I active site of AgAS. A normal molecular dynamics simulation of ligand-free and CPP-ligated AgAS reveals substantial reductions in root mean square fluctuations of several regions throughout the entire AgAS structure with ligand bound (Fig. 9). The NTE motif, which is in contact with the pyrophosphate-Mg2+ moiety of CPP, exhibits less movement than in the absence of CPP. Notable is the reduction in movement of loop 482–492 and residues neighboring and including Asp227. Further analysis, however, reveals no evidence for correlated (or anti-correlated) motions between structural elements of the class I active site and loop 482–492. Hence, the enhanced stability of loop 482–492 may result by chance alone (the absence of a relatively rare and low frequency movement of loop 482–492 in the simulation of the ligated structure).
FIGURE 9.
Class I active site of AgAS. Top and middle panels, the structure of AgAS (colored as in Fig. 2) is aligned with that of taxadiene synthase (magenta, top panel). Reductions in the root mean square (RMS) fluctuations for the AgAS-CPP complex (black) relative to apo-AgAS (gray) appear in the α-domain (near CPP) as well as the γ- and β-domains (middle panel). Bottom panel, the final AgAS-CCP complex from molecular dynamics (colored as in Fig. 2) is superimposed over taxadiene synthase (magenta).
The simulation of the AgAS-CPP complex did not result in significant conformational changes in the protein, consistent with crystallographic studies of other ligand-free and ligated class I terpene synthases from plants. Ligated and ligand-free forms of bornyl diphosphate synthase (Protein Data Bank codes 1N1B and 1N1Z), isoprene synthase (Protein Data Bank codes 3N0F and 3N0G),and 5-epi-aristolochene synthase (Protein Data Bank codes 5EAS and 5EAT) superimpose with overall root mean square deviations of 0.6 Å or less based on at least 525 corresponding Cα atoms. Atoms of CPP had root mean square fluctuations ranging from 0.5–0.9 Å, similar in magnitude to those of GGPP in the loop-out conformation of the class II active site. Atom C16 of CPP exhibits the greatest fluctuation because of the “flipping” of the isoprenoid unit proximal to the pyrophosphate group of CPP. Flipping of this isoprenoid unit infers a mixture of isopimar-15-en-8-yl+ intermediates (Fig. 1) that is not supported by the single dominant stereochemistry of the reaction product. Evidently, the CPP molecule during simulation has too much conformational freedom in the AgAS class I active site. That active site, however, is incomplete in two respects: N-terminal residues (Lys86 and Arg87) critical for catalysis are absent, and loop 846–849, which covers ligated active sites in other class I terpene synthases, is not in place. Given the effects of loop 482–492 on the conformational behavior of GGPP in the class II active site, the missing elements of the AgAS class I active site may be responsible in part for defining a single productive conformer of CPP.
DISCUSSION
AgAS has served as a model diterpene synthase for over a decade, yet despite extensive previous mechanistic and mutational analysis, the determination of its structure reported here has further elucidated its activity. Although no substrate analogs are present in this structure, it has been noted that such analogs in the known co-crystal structures often are not in a catalytically relevant conformation (2, 69), including those in the only two other reported diterpene synthase crystal structures (63, 64). Thus, the AgAS structure reported here was followed up by both site-directed mutagenesis and molecular dynamics simulations to probe the inferred structure-function relationships underlying the class I and class II cyclization reactions catalyzed by this bifunctional enzyme.
The wild-type class I active site favors proton and methyl migration from the initial isopimar-15-en-8-yl+ intermediate, resulting primarily in the formation of abietadienes (Fig. 1). The pyrophosphate co-product, presumably still bound to the active site, should favor such carbocation migration through electrostatic attraction (69). We have previously hypothesized that substitution of Ala723 with serine stabilizes the isopimar-15-en-8-yl+ intermediate, allowing deprotonation and the formation of isopimara-7,15-diene as the major product (42). The AgAS structure reported here is consistent with this hypothesis. Moreover, mutation of Val727 to threonine, to put a nearby hydroxyl group in the active site, has no effect on product distribution. Hence, the single-residue switch behavior of position 723 is localized and specific, although difficult to understand without complete knowledge of a fully assembled class I active site.
The AgAS structure forces a revision in the proposed arrangement of catalytic residues in the class II active site (43). Specifically, a conserved asparagine (Asn451 in AgAS) rather than the previously proposed Arg356 activates the catalytic aspartic acid, as not only suggested by the AgAS and AtCPS structures but further demonstrated here by site-directed mutagenesis. On the other hand, our results reaffirm the importance of Arg356 in class II catalysis (Table 2). Inspection of the crystal structure reveals a stacking interaction between the guanidine group of Arg356 and the aromatic ring of Tyr382. The side chain of Arg356 is buried, as is the corresponding histidine in AtCPS. The mutation of Arg356 to alanine likely creates a destabilizing void in the protein, the consequence of which is conformational change to minimize the volume of the void. One half-turn away from Arg356 on the same α-helix is a highly conserved aromatic residue, Phe354 in AgAS, which by simulation stacks against the terminal isoprenoid unit of the substrate (Fig. 8). The relationship of Phe354 to GGPP is consistent with carbocation stabilization via the aromatic π quadrupoles, despite such stabilization not being built into the molecular dynamics simulation. In addition, such π quadrupole interaction with the π orbitals of the sp2 hybridized atom C15 of GGPP may bias the double-bonded atom C14 toward accepting a proton from Asp404 to initiate the reaction.
Although Arg356 in AgAS and the corresponding His331 in AtCPS fulfill similar structural roles (planar group stacking with tyrosine), these residues have distinctly different and interchangeable effects on Mg2+-dependent inhibition of class II activity. AtCPS is susceptible to Mg2+-dependent inhibition, whereas AgAS is not (67). Interchanging residue types also swap susceptibility to Mg2+-dependent inhibition, with little to no change in catalytic activity (43). Indeed, this position is conserved as a histidine in all plant class II diterpene cyclases is involved in gibberellin phytohormone biosynthesis, such as AtCPS, but is an arginine in all enzymes dedicated to specialized or secondary metabolism, such as AgAS. The mechanism by which arginine at position 356 abrogates Mg2+-dependent inhibition is unclear; however, the electrostatic charge on a buried arginine (as opposed to a neutral histidine) can have long range effects through electrostatic interactions.
Although further work is necessary to fully understand the significance of Arg356, the current effort here has revealed a significant role for loop 482–492 in class II activity. First, Lys489 and Trp490 are conserved residues, and as is evident from Fig. 8, these residues make substantial contacts with GGPP. Moreover, the loop-in conformer plays a counterintuitive role by increasing the conformational flexibility of GGPP, even as it reduces the volume of the active site crevice. In the loop-out conformation, the substrate adopts a favored conformation, which evidently is much lower in energy than alternative substrate conformations. The loop-in conformation raises the energy of this conformer so that it is on par with alternative conformations. These alternative conformers bring pairs of atoms into contact consistent with the proposed mechanism of cyclization. In principle, given sufficiently long simulation times, we are more likely to observe the productive substrate conformation in the loop-in conformer.
Trp358 does not interact with GGPP in the class II complex that arises from constrained simulation. Nevertheless, this is an absolutely conserved residue, and the AgAS:W358A mutant enzyme has been shown to exhibit 1000-fold less class II activity and decreased affinity for the 14,15-dihydro-15-azageranylgeranyl-S-thiolodiphosphate analog that approximates the initial carbocation of the proposed reaction mechanism (23). Such tryptophan to alanine substitution will leave the mutant enzyme with a substantial internal void and the concomitant loss of energy-favorable nonbonded contacts. Conformational change to reduce the volume of the void is likely, including perturbation of the adjacent catalytic pairing of Asp404 and Asn451.
As noted for TbTS and AtCPS (63, 64), the AgAS structure is consistent with the previously advanced hypothesis that class II diterpene cyclases evolved from the mechanistically similar triterpene cyclases (19), because they all share structural homology (i.e. across the γβ domains). The diterpene synthase structures also are consistent with the previous suggestion that plant diterpene synthases originated from fusion of bacterial class II and I diterpene synthases (28). This ancestral bifunctional diterpene synthase was presumably involved in gibberellin biosynthesis and underwent gene duplication and subfunctionalization to give rise to the separate class II and class I enzymes that carry out such biosynthesis in higher plants (35).
More speculatively, given the ancestral role for diterpene synthases in the extensive plant sterpene synthase family (7), the overall arrangement of the γβα domain structure (Fig. 2) provides a rationale for the observed βα domain structure of most plant class I terpene synthases (70). In particular, only the β domain makes extensive contacts with the α domain, such that loss of the γ domain might have relatively little effect in monofunctional class I terpene synthases. Consistent with such speculation, the γ domain appears to have been independently lost multiple times in plant terpene synthase evolution (71). By contrast, the βα domain interface appears to be important, because it has been shown for AgAS that class I activity for separately expressed α domain requires the presence of the γβ domains (40).
In addition, the presence of the N terminus in the α domain and its passage through the β domain provides a rationale for the observed conservation pattern of the majority of βα domain plant terpene synthases, wherein loss of the γ domain occurred with retention of the corresponding N-terminal sequence, leading to the original “internal” nomenclature for the sequence element corresponding to the γ domain (7). Moreover, the presence of a pair of basic residues at the N terminus important for class I catalysis in AgAS (40) draws some interesting parallels to a similar N-terminal motif found in many βα domain terpene synthases (15, 70) and also is consistent with retention of the N-terminal sequence in γ-domain loss. Thus, coupled to the extensive previous studies of this model diterpene synthase, the AgAS structure not only provides insights into the class II and class I diterpene cyclization reactions catalyzed by this bifunctional enzyme, but also into the evolution of plant terpene synthases more generally.
Acknowledgment
We thank Prof. Gaya Amarasinghe (Iowa State University) for collecting diffraction data on crystal I.
This work was supported, in whole or in part, by National Institutes of Health Grants GM076324 (to R. J. P.) and NS010546 (to R. B. H.). This work was also supported in part by Department of Enery Grants W-31–109-ENG-38 and DE-AC02–06CH11357 and by the National Science Foundation.
The atomic coordinates and structure factors (code 3S9V) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- GGPP
- (E,E,E)-geranylgeranyl diphosphate
- AgAS
- abietadiene synthase from A. grandis
- AtCPS
- ent-CPP synthase from Arabidopsis thaliana
- CPP
- copalyl diphosphate
- TbTS
- taxadiene synthase from Taxus brevifolia.
REFERENCES
- 1. Buckingham J. (2002) Dictionary of Natural Products (on-line web edition), Chapman & Hall/CRC Press [Google Scholar]
- 2. Christianson D. W. (2006) Structural biology and chemistry of the terpenoid cyclases. Chem. Rev. 106, 3412–3442 [DOI] [PubMed] [Google Scholar]
- 3. Croteau R., Kutchan T. M., Lewis N. G. (2000) Natural products (secondary metabolites) in Biochemistry & Molecular Biology of Plants (Buchanan B., Gruissem W., Jones R. eds.) pp. 1250–1318, American Society of Plant Biologists, Rockville, MD [Google Scholar]
- 4. Wendt K. U., Schulz G. E. (1998) Isoprenoid biosynthesis. Manifold chemistry catalyzed by similar enzymes. Structure 6, 127–133 [DOI] [PubMed] [Google Scholar]
- 5. Peters R. J. (2010) Two rings in them all. The labdane-related diterpenoids. Nat. Prod. Rep. 27, 1521–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Keeling C. I., Bohlmann J. (2006) Diterpene resin acids in conifers. Phytochemistry 67, 2415–2423 [DOI] [PubMed] [Google Scholar]
- 7. Trapp S. C., Croteau R. B. (2001) Genomic organization of plant terpene synthases and molecular evolutionary implications. Genetics 158, 811–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bohlmann J., Meyer-Gauen G., Croteau R. (1998) Plant terpenoid synthases. Molecular biology and phylogenetic analysis. Proc. Natl. Acad. Sci. U.S.A. 95, 4126–4133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Starks C. M., Back K., Chappell J., Noel J. P. (1997) Structural basis for cyclic terpene biosynthesis by tobacco 5-epi-aristolochene synthase. Science 277, 1815–1820 [DOI] [PubMed] [Google Scholar]
- 10. Lesburg C. A., Zhai G., Cane D. E., Christianson D. W. (1997) Crystal structure of pentalenene synthase. Mechanistic insights on terpenoid cyclization reactions in biology. Science 277, 1820–1824 [DOI] [PubMed] [Google Scholar]
- 11. Rynkiewicz M. J., Cane D. E., Christianson D. W. (2001) Structure of trichodiene synthase from Fusarium sporotrichioides provides mechanistic inferences on the terpene cyclization cascade. Proc. Natl. Acad. Sci. U.S.A. 98, 13543–13548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shishova E. Y., Di Costanzo L., Cane D. E., Christianson D. W. (2007) X-ray crystal structure of aristolochene synthase from Aspergillus terreus and evolution of templates for the cyclization of farnesyl diphosphate. Biochemistry 46, 1941–1951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Caruthers J. M., Kang I., Rynkiewicz M. J., Cane D. E., Christianson D. W. (2000) Crystal structure determination of aristolochene synthase from the blue cheese mold, Penicillium roqueforti. J. Biol. Chem. 275, 25533–25539 [DOI] [PubMed] [Google Scholar]
- 14. Whittington D. A., Wise M. L., Urbansky M., Coates R. M., Croteau R. B., Christianson D. W. (2002) Bornyl diphosphate synthase. Structure and strategy for carbocation manipulation by a terpenoid cyclase. Proc. Natl. Acad. Sci. U.S.A. 99, 15375–15380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hyatt D. C., Youn B., Zhao Y., Santhamma B., Coates R. M., Croteau R. B., Kang C. (2007) Structure of limonene synthase, a simple model for terpenoid cyclase catalysis. Proc. Natl. Acad. Sci. U.S.A. 104, 5360–5365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kampranis S. C., Ioannidis D., Purvis A., Mahrez W., Ninga E., Katerelos N. A., Anssour S., Dunwell J. M., Degenhardt J., Makris A. M., Goodenough P. W., Johnson C. B. (2007) Rational conversion of substrate and product specificity in a Salvia monoterpene synthase. Structural insights into the evolution of terpene synthase function. Plant Cell 19, 1994–2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gennadios H. A., Gonzalez V., Di Costanzo L., Li A., Yu F., Miller D. J., Allemann R. K., Christianson D. W. (2009) Crystal structure of (+)-δ-cadinene synthase from Gossypium arboreum and evolutionary divergence of metal binding motifs for catalysis. Biochemistry 48, 6175–6183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Aaron J. A., Lin X., Cane D. E., Christianson D. W. (2010) Structure of epi-isozizaene synthase from Streptomyces coelicolor A3(2), a platform for new terpenoid cyclization templates. Biochemistry 49, 1787–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cao R., Zhang Y., Mann F. M., Huang C., Mukkamala D., Hudock M. P., Mead M. E., Prisic S., Wang K., Lin F. Y., Chang T. K., Peters R. J., Oldfield E. (2010) Diterpene cyclases and the nature of the isoprene fold. Proteins 78, 2417–2432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhou K., Peters R. J. (2009) Investigating the conservation pattern of a putative second terpene synthase divalent metal binding motif in plants. Phytochemistry 70, 366–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Peters R. J., Ravn M. M., Coates R. M., Croteau R. B. (2001) Bifunctional abietadiene synthase. Free diffusive transfer of the (+)-copalyl diphosphate intermediate between two distinct active sites. J. Am. Chem. Soc. 123, 8974–8978 [DOI] [PubMed] [Google Scholar]
- 22. Kawaide H., Sassa T., Kamiya Y. (2000) Functional analysis of the two interacting cyclase domains in ent-kaurene synthase from the fungus Phaeosphaeria sp. L487 and a comparison with cyclases from higher plants. J. Biol. Chem. 275, 2276–2280 [DOI] [PubMed] [Google Scholar]
- 23. Peters R. J., Croteau R. B. (2002) Abietadiene synthase catalysis. Conserved residues involved in protonation-initiated cyclization of geranylgeranyl diphosphate to (+)-copalyl diphosphate. Biochemistry 41, 1836–1842 [DOI] [PubMed] [Google Scholar]
- 24. Prisic S., Xu J., Coates R. M., Peters R. J. (2007) Probing the role of the DXDD motif in Class II diterpene cyclases. ChemBioChem 8, 869–874 [DOI] [PubMed] [Google Scholar]
- 25. Xu M., Hillwig M. L., Prisic S., Coates R. M., Peters R. J. (2004) Functional identification of rice syn-copalyl diphosphate synthase and its role in initiating biosynthesis of diterpenoid phytoalexin/allelopathic natural products. Plant J. 39, 309–318 [DOI] [PubMed] [Google Scholar]
- 26. Wendt K. U., Poralla K., Schulz G. E. (1997) Structure and function of a squalene cyclase. Science 277, 1811–1815 [DOI] [PubMed] [Google Scholar]
- 27. Thoma R., Schulz-Gasch T., D'Arcy B., Benz J., Aebi J., Dehmlow H., Hennig M., Stihle M., Ruf A. (2004) Insight into steroid scaffold formation from the structure of human oxidosqualene cyclase. Nature 432, 118–122 [DOI] [PubMed] [Google Scholar]
- 28. Morrone D., Chambers J., Lowry L., Kim G., Anterola A., Bender K., Peters R. J. (2009) Gibberellin biosynthesis in bacteria. Separate ent-copalyl diphosphate and ent-kaurene synthases in Bradyrhizobium japonicum. FEBS Lett. 583, 475–480 [DOI] [PubMed] [Google Scholar]
- 29. Kawaide H., Imai R., Sassa T., Kamiya Y. (1997) ent-Kaurene synthase from the fungus Phaeosphaeria sp. L487. cDNA isolation, characterization, and bacterial expression of a bifunctional diterpene cyclase in fungal gibberellin biosynthesis. J. Biol. Chem. 272, 21706–21712 [DOI] [PubMed] [Google Scholar]
- 30. Oikawa H., Toyomasu T., Toshima H., Ohashi S., Kawaide H., Kamiya Y., Ohtsuka M., Shinoda S., Mitsuhashi W., Sassa T. (2001) Cloning and functional expression of cDNA encoding aphidicolan-16β-ol synthase. A key enzyme responsible for formation of an unusual diterpene skeleton in biosynthesis of aphidicolin. J. Am. Chem. Soc. 123, 5154–5155 [DOI] [PubMed] [Google Scholar]
- 31. Vogel B. S., Wildung M. R., Vogel G., Croteau R. (1996) Abietadiene synthase from grand fir (Abies grandis). cDNA isolation, characterization, and bacterial expression of a bifunctional diterpene cyclase involved in resin acid biosynthesis. J. Biol. Chem. 271, 23262–23268 [DOI] [PubMed] [Google Scholar]
- 32. Schepmann H. G., Pang J., Matsuda S. P. (2001) Cloning and characterization of ginkgo biloba levopimaradiene synthase which catalyzes the first committed step in ginkgolide biosynthesis. Arch. Biochem. Biophys. 392, 263–269 [DOI] [PubMed] [Google Scholar]
- 33. Martin D. M., Fäldt J., Bohlmann J. (2004) Functional characterization of nine Norway Spruce TPS genes and evolution of gymnosperm terpene synthases of the TPS-d subfamily. Plant Physiol. 135, 1908–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hayashi K., Kawaide H., Notomi M., Sakigi Y., Matsuo A., Nozaki H. (2006) Identification and functional analysis of bifunctional ent-kaurene synthase from the moss Physcomitrella patens. FEBS Lett. 580, 6175–6181 [DOI] [PubMed] [Google Scholar]
- 35. Keeling C. I., Dullat H. K., Yuen M., Ralph S. G., Jancsik S., Bohlmann J. (2010) Identification and functional characterization of monofunctional ent-copalyl diphosphate and ent-kaurene synthases in white spruce reveal different patterns for diterpene synthase evolution for primary and secondary metabolism in gymnosperms. Plant Physiol. 152, 1197–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ravn M. M., Coates R. M., Jetter R., Croteau R. (1998) Stereospecific intramolecular transfer in the cyclization of geranylgeranyl diphosphate to (−)-abietadiene catalyzed by recombinant cyclase from grand fir (Abies grandis). Chem. Commun. 1998, 21–22 [Google Scholar]
- 37. Ravn M. M., Coates R. M., Flory J. E., Peters R. J., Croteau R. (2000) Stereochemistry of the cyclization-rearrangement of (+)-copalyl diphosphate to (−)-abietadiene catalyzed by recombinant abietadiene synthase from Abies grandis. Org. Lett. 2, 573–576 [DOI] [PubMed] [Google Scholar]
- 38. Peters R. J., Flory J. E., Jetter R., Ravn M. M., Lee H. J., Coates R. M., Croteau R. B. (2000) Abietadiene synthase from grand fir (Abies grandis). Characterization and mechanism of action of the “pseudomature” recombinant enzyme. Biochemistry 39, 15592–15602 [DOI] [PubMed] [Google Scholar]
- 39. Ravn M. M., Peters R. J., Coates R. M., Croteau R. (2002) Mechanism of abietadiene synthase catalysis. Stereochemistry and stabilization of the cryptic pimarenyl carbocation intermediates. J. Am. Chem. Soc. 124, 6998–7006 [DOI] [PubMed] [Google Scholar]
- 40. Peters R. J., Carter O. A., Zhang Y., Matthews B. W., Croteau R. B. (2003) Bifunctional abietadiene synthase. Mutual structural dependence of the active sites for protonation-initiated and ionization-initiated cyclizations. Biochemistry 42, 2700–2707 [DOI] [PubMed] [Google Scholar]
- 41. Peters R. J., Croteau R. B. (2002) Abietadiene synthase catalysis. Mutational analysis of a prenyl diphosphate ionization-initiated cyclization and rearrangement. Proc. Natl. Acad. Sci. U.S.A. 99, 580–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wilderman P. R., Peters R. J. (2007) A single residue switch converts abietadiene synthase into a pimaradiene specific cyclase. J. Am. Chem. Soc. 129, 15736–15737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mann F. M., Prisic S., Davenport E. K., Determan M. K., Coates R. M., Peters R. J. (2010) A single residue switch for Mg2+-dependent inhibition characterizes plant class II diterpene cyclases from primary and secondary metabolism. J. Biol. Chem. 285, 20558–20563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Minor W., Cymborowski M., Otwinowski Z., Chruszcz M. (2006) HKL-3000. The integration of data reduction and structure solution–from diffraction images to an initial model in minutes. Acta Crystallogr. D Biol. Crystallogr. 62, 859–866 [DOI] [PubMed] [Google Scholar]
- 45. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX. A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. McRee D. E. (1999) XtalView/Xfit. A versatile program for manipulating atomic coordinates and electron density. J. Struct. Biol. 125, 156–165 [DOI] [PubMed] [Google Scholar]
- 47. Collaborative Computational Project, Number 4 (1994) The CCP4 suite. Programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 48. Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Crystallography & NMR system. A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 49. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 51. Cyr A., Wilderman P. R., Determan M., Peters R. J. (2007) A modular approach for facile biosynthesis of labdane-related diterpenes. J. Am. Chem. Soc. 129, 6684–6685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hamelberg D., Mongan J., McCammon J. (2004) Accelerated molecular dynamics. A promising and efficient simulation method for biomolecules. J. Chem. Phys. 120, 11919–11929 [DOI] [PubMed] [Google Scholar]
- 53. Wang Y., Harrison C., Schulten K., McCammon J. (2011) Comp. Sci. Discov. 4, 015002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Phillips J. C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., Chipot C., Skeel R. D., Kalé L., Schulten K. (2005) Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. MacKerell A. D., Jr., Banavali N., Foloppe N. (2000) Development and current status of the CHARMM force field for nucleic acids. Biopolymers 56, 257–265 [DOI] [PubMed] [Google Scholar]
- 56. Vanommeslaeghe K., Hatcher E., Acharya C., Kundu S., Zhong S., Shim J., Darian E., Guvench O., Lopes P., Vorobyov I., MacKerell A. D., Jr. (2010) CHARMM general force field: a force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J. Comput. Biol. 31, 671–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Jorgensen W. L., Chandrasekhar J., Madura J. D., Impey R. W., Klein M. L. (1983) Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926–935 [Google Scholar]
- 58. Essmann U., Perera L., Benkowitz M. L. (1995) A smooth particle mesh Ewald method. J. Chem. Phys. 103, 8577–8593 [Google Scholar]
- 59. Ryckaert J.-P., Ciccotti G., Berendsen J. C. (1977) Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 23, 327–341 [Google Scholar]
- 60. Humphrey W., Dalke A., Schulten K. (1996) VMD: visual molecular dynamics. J. Molec. Graphics 14, 33–38 [DOI] [PubMed] [Google Scholar]
- 61. Glykos N. M. (2006) Software news and updates. Carma. A molecular dynamics analysis program. J. Comput. Chem. 27, 1765–1768 [DOI] [PubMed] [Google Scholar]
- 62. Shortle D., Simons K. T., Baker D. (1998) Clustering of low-energy conformations near the native structures of small proteins. Proc. Natl. Acad. Sci. U.S.A. 95, 11158–11162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Köksal M., Jin Y., Coates R. M., Croteau R., Christianson D. W. (2011) Taxadiene synthase structure and evolution of modular architecture in terpene biosynthesis. Nature 469, 116–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Köksal M., Hu H., Coates R. M., Peters R. J., Christianson D. W. (2011) Structure and mechanism of the diterpene cyclase ent-copalyl diphosphate synthase. Nat. Chem. Biol. 7, 431–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Sato T., Hoshino T. (1999) Functional analysis of the DXDDTA motif in squalene-hopene cyclase by site-directed mutagenesis experiments. Initiation site of the polycyclization reaction and stabilization site of the carbocation intermediate of the initially cyclized A-ring. Biosci. Biotechnol. Biochem. 63, 2189–2198 [DOI] [PubMed] [Google Scholar]
- 66. Wendt K. U. (2005) Angew. Chem. Int. Ed. 44, 3966–3971 [DOI] [PubMed] [Google Scholar]
- 67. Prisic S., Peters R. J. (2007) Synergistic substrate inhibition of ent-copalyl diphosphate synthase. A potential feed-forward inhibition mechanism limiting gibberellin metabolism. Plant Physiol. 144, 445–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Xu M., Wilderman P. R., Peters R. J. (2007) Following evolution's lead to a single residue switch for diterpene synthase product outcome. Proc. Natl. Acad. Sci. U.S.A. 104, 7397–7401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zhou K., Peters R. J. (2011) Electrostatic effects on (di)terpene synthase product outcome. Chem. Commun. 47, 4074–4080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Chen F., Tholl D., Bohlmann J., Pichersky E. (2011) The family of terpene synthases in plants. A mid-size family of genes for specialized metabolism that is highly diversified throughout the kingdom. Plant J. 66, 212–229 [DOI] [PubMed] [Google Scholar]
- 71. Hillwig M. L., Xu M., Toyomasu T., Tiernan M. S., Wei G., Cui G., Huang L., Peters R. J. (2011) Domain loss has independently occurred multiple times in plant terpene synthase evolution. Plant J. 68, 1051–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]